Abstract
Alicyclobacillus acidocaldarius (Darland and Brock 1971) is the type species of the larger of the two genera in the bacillal family ‘Alicyclobacillaceae’. A. acidocaldarius is a free-living and non-pathogenic organism, but may also be associated with food and fruit spoilage. Due to its acidophilic nature, several enzymes from this species have since long been subjected to detailed molecular and biochemical studies. Here we describe the features of this organism, together with the complete genome sequence and annotation. This is the first completed genome sequence of the family ‘Alicyclobacillaceae’. The 3,205,686 bp long genome (chromosome and three plasmids) with its 3,153 protein-coding and 82 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: thermophile, acidophilic, aerobic, non-pathogenic, food spoilage, non-motile but encodes flagellar genes, GEBA
Introduction
Strain 104-IAT (= DSM 446 = ATCC 27009 = JCM 5260 = NCIMB 11725) is the type strain of the species Alicyclobacillus acidocaldarius, which is the type species of the genus Alicyclobacillus [1]. The genus currently consists of 20 species and two subspecies. Strain 104-IAT was originally isolated as ‘Bacillus acidocaldarius’ in 1971 (or earlier) from a hot and acidic spring in Yellowstone National Park, USA. In 1992, it was reclassified on the basis of comparative 16S rRNA gene sequence analysis into the new genus Alicyclobacillus [1]. With the description of A. acidocaldarius subsp. rittmannii in 2002 [2] the subspecies name A. acidocaldarius subsp. acidocaldarius was automatically created following rule 46 of the bacteriological code [3], with 104-IAT as its type strain. (hereinafter nevertheless referred to as A. acidocaldarius, without subspecies epithet). The species name derives from ‘acidus’ from Latin meaning acidic combined with ‘caldarius’, Latin for ‘belonging to the hot’. Due to its thermoacidic nature, this species serves as a model organism for molecular and biochemical studies of its enzymes [4-19]. Strain 104-IAT has also been used to produce the restriction enzyme BacI [20]. Here we present a summary classification and a set of features for A. acidocaldarius 104-IAT, together with the description of the complete genomic sequencing and annotation.
Classification and features
The type strain 104-IAT and several other strains were isolated from acidic hot springs in the Yellowstone National Park, USA, from soil from an acid fumarole in the Hawaiian Volcano National Park [21], and also from acidic environments in Japan [22]. Other strains, as identified by 16S rRNA gene sequences and by metabolic traits, were isolated from orchard soil, mango juice, vinegar flies or pre-pasteurized pear puree in South Africa [23-25]. These findings are supported by the experimentally determined heat resistance of A. acidocaldarius strains in water, acidic buffer and orange juice [26]. Thus, A. acidocaldarius might be involved in food and fruit spoilage, which is a characteristic of several other species of the genus Alicyclobacillus [23-25,27]. Clones with high sequence similarity (99%, AB042056) with the 16S rRNA gene sequence of strain 104-IAT are reported by the NCBI BLAST server from a ‘simulated low level waste site’ in USA (GQ263212), but not with any metagenomic environmental samples (October 2009).
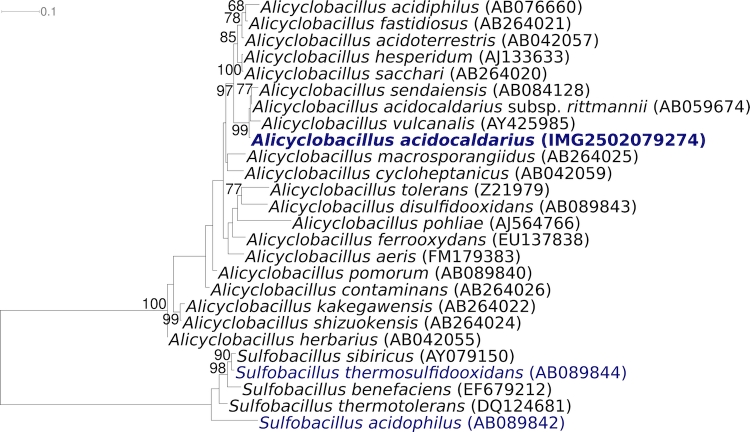
Figure 1 shows the phylogenetic neighborhood of for A. acidocaldarius 104-IAT in a 16S rRNA based tree. The sequences of the six 16S rRNA gene copies in the genome of A. acidocaldarius 104-IAT, differ from each other by up to six nucleotides, and differ by up to five nucleotides from the previously published 16S rRNA sequence derived from DSM 446 (AJ496806).
Figure 1.
Phylogenetic tree highlighting the position of A. acidocaldarius 104-IAT relative to the other type strains within the family. The tree was inferred from 1,419 aligned characters [28,29] of the 16S rRNA gene sequence under the maximum likelihood criterion [30] and rooted with the genus Sulfobacillus. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 1,000 bootstrap replicates if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [31] are shown in blue, published genomes in bold.
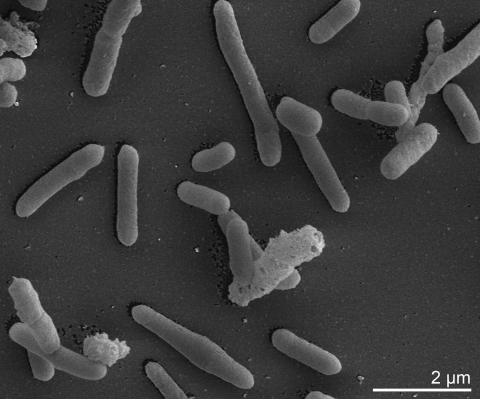
On B. acidocaldarius medium (BAM medium) [32] strain 104-IAT forms round, slightly mucous, creamy-white colonies after 72 hours of growth with a diameter of 1-4 mm and rod shaped cells that were 2.0-4.5 μm long and 0.5-1.0 μm wide (Table 1 and Figure 2) [22]. The endospores are terminal or subterminal and the sporangia are not swollen [22]. The upper and lower pH growth limits are pH 2 and pH 6 [21]. Strain 104-IAT grows on basal medium supplemented with glucose, galactose, casamino acids, starch, glycerol, sucrose, gluconate, inositol, ribose, rhamnose, and lactose, but not with ethanol, sorbitol, sodium acetate, succinic acid, and sodium citrate [21]. Strain 104-IAT hydrolyses gelatin and starch but is oxidase negative and does not reduce nitrate to nitrite [43]. Strain 104-IAT produces acid from glycerol, L-arabinose, D-xylose, D-galactose, rhamnose, mannitol, methyl-α-D-glucoside, arbutin, aesculin, salicin, cellobiose, maltose, lactose, melibiose, sucrose, trehalose, D-raffinose, starch, and glycogen, but it does not produce acid from erythritol, D-arabinose, L-xylose, L-sorbose, inositol, sorbitol, methyl-α-D-mannisode, amygdalin, melezitose, xylitol, β-gentibiose, D-turanose, D-lyxose, D-tagatose, D-fucose, and 5-ketogluconate [43]. These acid production characteristics are largely congruent with the results from [22], however, L-sorbose, salicin, D-raffinose, starch, and D-turanose deviate across the studies [22,43].
Table 1. Classification and general features of A. acidocaldarius 104-IAT in accordance with the MIGS recommendations [33].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [34] | |
| Phylum ‘Firmicutes’ | TAS [35-37] | ||
| Class Bacilli | TAS [36] | ||
| Order Bacillales | TAS [38,39] | ||
| Family ‘Alicyclobacillaceae’ | TAS [40] | ||
| Genus Alicyclobacillus | TAS [1] | ||
| Species Alicyclobacillus acidocaldarius | TAS [21] | ||
| Type strain 104-IA | TAS [21] | ||
| Gram stain | positive | TAS [21] | |
| Cell shape | small rods | TAS [1] | |
| Motility | not reported (relevant genes missing) | NAS | |
| Sporulation | refractile endospores | TAS [21] | |
| Temperature range | 45°C-70°C | TAS [21] | |
| Optimum temperature | 60°C-65°C | TAS [21] | |
| Salinity | does not grow with 5% (w/v) NaCl | TAS [22] | |
| MIGS-22 | Oxygen requirement | strictly aerobic | TAS [21] |
| Carbon source | saccharolytic | TAS [21] | |
| Energy source | carbohydrates | TAS [21] | |
| MIGS-6 | Habitat | hot acidic springs and soil | TAS [21] |
| MIGS-15 | Biotic relationship | free living | NAS |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [41] | |
| Isolation | acid hot spring | TAS [21] | |
| MIGS-4 | Geographic location | Nymph Creek, Yellowstone National Park, USA | TAS [21] |
| MIGS-5 | Sample collection time | about 1970 | TAS [21] |
| MIGS-4.1 | Latitude | 44.376 | NAS |
| MIGS-4.2 | Longitude | 110.690 | NAS |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [42]. If the evidence code is IDA, then the property was directly observed for a living isolate by one of the authors or an expert mentioned in the acknowledgements.
Figure 2.
Scanning electron micrograph of A. acidocaldarius 104-IAT
Motility has not been reported for strain 104-IAT, although closely related species from the genus Alicyclobacillus are motile [22,27,43-45], which suggests a recent loss of motility in A. acidocaldarius. Indeed, strain 104-IAT appears to have all genes necessary for a flagellum. However, essential genes for type 3 secretion system chaperones (flgN, fliJ, fliT) and for flagellar gene expression (flhC, flhD) are missing in the genome, which finally explains the non-motile phenotype.
Chemotaxonomy
Characteristic for several Alicyclobacillus species is the presence of a large amount of ω-alicyclic fatty acids [1,46]. As such, strain 104-IAT has approximately 51 ω-cyclohexane C17:0 and 33% ω-cyclohexane C19:0. Other fatty acids such as C16:0, C18:0, iso-C15:0, iso-C16:0, iso-C18:0, anteiso-C15:0, and anteiso-C17:0 amount at individual levels of approximately 1% to 5% [22,43]. Fatty acid composition is rather stable though not static across different temperature and pH values [47]. Moreover, strain 104-IAT produces hopanoids, a group of pentacyclic triterpenoids, which together with the fatty acids constitute the lipophilic core of the cytoplasmic membrane. The amount of hopanoids depends on the temperature more so than the pH value [48]. The main isoprenoid quinone is menaquinone with seven isoprene units (MK-7) [1].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position, and is part of the Genomic Encyclopedia of Bacteria and Archaea project. The genome project is deposited in the Genome OnLine Database [31] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | One Sanger 8 kb pMCL200 library and one 454 pyrosequencing standard library |
| MIGS-29 | Sequencing platforms | ABI3730, 454 GS FLX |
| MIGS-31.2 | Sequencing coverage | 7.0 ×Sanger; 27.3× pyrosequencing |
| MIGS-30 | Assemblers | Newbler, Phrap |
| MIGS-32 | Gene calling method | Prodigal, GenePRIMP |
| INSDC ID | CP001727 (chromosome) CP001728-30 (plasmids) |
|
| GenBank Date of Release | September 10-14, 2009 | |
| GOLD ID | Gc01110 | |
| NCBI project ID | 29405 | |
| Database: IMG-GEBA | 2501939636 | |
| MIGS-13 | Source material identifier | DSM 446 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
A. acidocaldarius 104-IAT, DSM 446, was grown in DSM Medium 402 [49] at 60°C. DNA was isolated from 0.5-1 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany) with cell lysis modification st/L [50] and one hour incubation at 37°C.
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger and 454 sequencing platforms. All general aspects of library construction and sequencing performed at the JGI can be found at http://www.jgi.doe.gov/. 454 Pyrosequencing reads were assembled using the Newbler assembler version 1.1.02.15 (Roche). Large Newbler contigs were broken into 3,478 overlapping fragments of 1,000 bp and entered into assembly as pseudo-reads. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and to adjust inflated q-scores. A hybrid 454/Sanger assembly was made using the parallel phrap assembler (High Performance Software, LLC). Possible mis-assemblies were corrected with Dupfinisher [51] or transposon bombing of bridging clones (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing in Consed, custom primer walk or PCR amplification. A total of 767 Sanger finishing reads were produced to close gaps, to resolve repetitive regions, and to raise the quality of the finished sequence. The error rate of the completed genome sequence is less than 1 in 100,000. The final assembly contains 24,980 Sanger and 363,136 Pyrosequencing reads. Together all sequence types provided 34.3 × coverage of the genome
Genome annotation
Genes were identified using Prodigal [52] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [53]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and manual functional annotation were performed within the Integrated Microbial Genomes Expert Review (IMG-ER) platform [54].
Genome properties
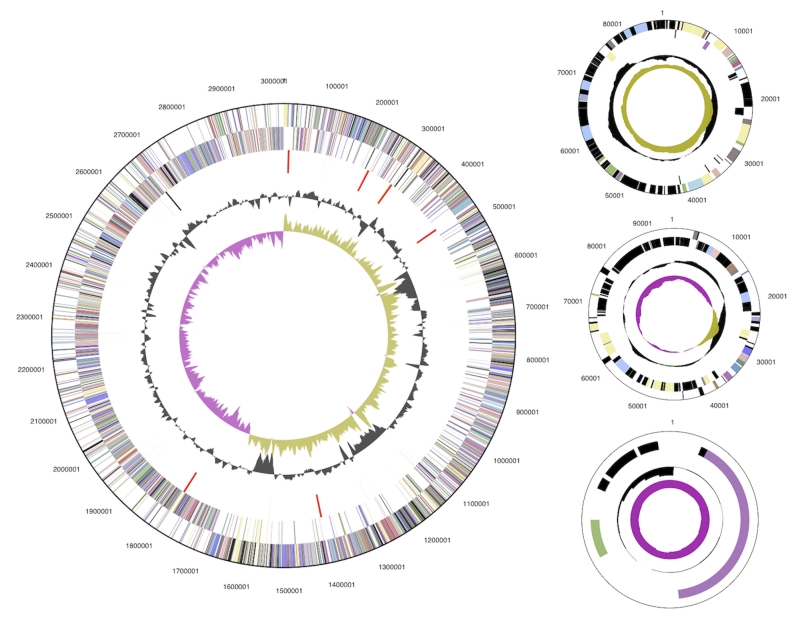
The genome consists of a 3,018,755 bp long chromosome and three plasmids of 91,726 bp, 87,298 bp, and 7,907 bp (Table 3 and Figure 3). Of the 3,235 genes predicted, 3,153 were protein-coding genes, and 82 RNAs; 69 pseudogenes were also identified. The majority of the protein-coding genes (68.4%) were assigned with a putative function while those remaining were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,205,686 | 100.00% |
| DNA coding region (bp) | 2,907,874 | 90.71% |
| DNA G+C content (bp) | 1,984,066 | 61.89% |
| Number of replicons | 4 | |
| Extrachromosomal elements | 3 | |
| Total genes | 3,235 | 100.00% |
| RNA genes | 82 | 2.53% |
| rRNA operons | 6 | |
| Protein-coding genes | 3,153 | 97.47% |
| Pseudo genes | 82 | 2.13% |
| Genes with function prediction | 2,214 | 68,44% |
| Genes in paralog clusters | 661 | 20.43% |
| Genes assigned to COGs | 2,221 | 68.66% |
| Genes assigned Pfam domains | 2,297 | 71.00% |
| Genes with signal peptides | 686 | 21.21% |
| Genes with transmembrane helices | 858 | 26.52% |
| CRISPR repeats | 4 |
Figure 3.
Graphical circular map of the chromosome and plasmids. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 147 | 4.7 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 191 | 6.1 | Transcription |
| L | 177 | 5.6 | Replication, recombination and repair |
| B | 0 | 0.0 | Chromatin structure and dynamics |
| D | 33 | 1.0 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 29 | 0.9 | Defense mechanisms |
| T | 110 | 3.5 | Signal transduction mechanisms |
| M | 124 | 3.9 | Cell wall/membrane biogenesis |
| N | 58 | 1.8 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 59 | 1.9 | Intracellular trafficking and secretion |
| O | 78 | 2.5 | Posttranslational modification, protein turnover, chaperones |
| C | 130 | 4.1 | Energy production and conversion |
| G | 203 | 6.4 | Carbohydrate transport and metabolism |
| E | 201 | 6.4 | Amino acid transport and metabolism |
| F | 61 | 1.9 | Nucleotide transport and metabolism |
| H | 117 | 3.7 | Coenzyme transport and metabolism |
| I | 120 | 3.8 | Lipid transport and metabolism |
| P | 104 | 3.3 | Inorganic ion transport and metabolism |
| Q | 58 | 1.8 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 266 | 8.4 | General function prediction only |
| S | 185 | 5.9 | Function unknown |
| - | 1014 | 32.2 | Not in COGs |
Acknowledgement
We would like to gratefully acknowledge the help of Susanne Schneider (DSMZ) for DNA extraction and quality analysis. This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, as well as German Research Foundation (DFG) INST 599/1-1 and SI 1352/1-2.
References
- 1.Wisotzkey JD, Jurtshuk P, Jr, Fox GE, Deinhard G, Poralla K. Comparative sequence analyses on the 16S rRNA (rDNA) of Bacillus acidocaldarius, Bacillus acidoterrestris, and Bacillus cycloheptanicus and proposal for creation of a new genus, Alicyclobacillus gen. nov. Int J Syst Bacteriol 1992; 42:263-269 [DOI] [PubMed] [Google Scholar]
- 2.Nicolaus B, Improta R, Manca MC, Lama L, Esposito E, Gambacorta A. Alicyclobacilli from an unexplored geothermal soil in Antarctica: Mount Rittmann. Polar Biol 1998; 19:133-141 10.1007/s003000050224 [DOI] [Google Scholar]
- 3.Anonymous. The Bacteriological Code (1990 Revision) http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=icnb
- 4.Agafonov DE, Rabe KS, Grote M, Huang Y, Sprinzl M. The esterase from Alicyclobacillus acidocaldarius as a reporter enzyme and affinity tag for protein biosynthesis. FEBS Lett 2005; 579:2082-2086 10.1016/j.febslet.2005.02.059 [DOI] [PubMed] [Google Scholar]
- 5.Bakker EP, Borchard A, Michels M, Altendorf K, Siebers A. High-affinity potassium uptake system in Bacillus acidocaldarius showing immunological cross-reactivity with the Kdp system from Escherichia coli. J Bacteriol 1987; 169:4342-4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer EW, Ingle MB, Mercer GD. Isolation and characterization of unusual bacterial amylases. Starch 1979; 31:166-171 10.1002/star.19790310508 [DOI] [Google Scholar]
- 7.Hafer J, Siebers A, Bakker EP. The high-affinity K+-translocating ATPase complex from Bacillus acidocaldarius consists of three subunits. Mol Microbiol 1989; 3:487-495 10.1111/j.1365-2958.1989.tb00195.x [DOI] [PubMed] [Google Scholar]
- 8.Hülsmann A, Lurz R, Scheffel F, Schneider E. Maltose and maltodextrin transport in the thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius is mediated by a high-affinity transport system that includes a maltose binding protein tolerant to low pH. J Bacteriol 2000; 182:6292-6301 10.1128/JB.182.22.6292-6301.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Lauro B, Rossi M, Moracci M. Characterization of a b-glycosidase from the thermoacidophilic bacterium Alicyclobacillus acidocaldarius. Extremophiles 2006; 10:301-310 10.1007/s00792-005-0500-1 [DOI] [PubMed] [Google Scholar]
- 10.Lee SJ, Lee DW, Choe EA, Hong YH, Kim SB, Kim BC, Pyun YR. Characterization of a thermoacidophilic L-arabinose isomerase from Alicyclobacillus acidocaldarius: Role of Lys-269 in pH optimum. Appl Environ Microbiol 2005; 71:7888-7896 10.1128/AEM.71.12.7888-7896.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manco G, Mandrich L, Rossi M. Residues at the active site of the esterase 2 from Alicyclobacillus acidocaldarius involved in substrate specificity and catalytic activity at high temperature. J Biol Chem 2001; 276:37482-37490 10.1074/jbc.M103017200 [DOI] [PubMed] [Google Scholar]
- 12.Matzke J, Schwermann B, Bakker EP. Acidostable and acidophilic proteins: the example of the alpha-amylase from Alicyclobacillus acidocaldarius. Comp Biochem Physiol Physiol 1997; 118:475-479 10.1016/S0300-9629(97)00008-X [DOI] [PubMed] [Google Scholar]
- 13.Matzke J, Herrmann A, Schneider E, Bakker EP. Gene cloning, nucleotide sequence and biochemical properties of a cytoplasmic cyclomaltodextrinase (neopullulanase) from Alicyclobacillus acidocaldarius, reclassification of a group of enzymes. FEMS Microbiol Lett 2000; 183:55-61 10.1111/j.1574-6968.2000.tb08933.x [DOI] [PubMed] [Google Scholar]
- 14.Michels M, Bakker EP. Generation of a large, protonophore-sensitive proton motive force and pH difference in the acidophilic bacteria Thermoplasma acidophilum and Bacillus acidocaldarius. J Bacteriol 1985; 161:231-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matzke J, Herrmann A, Schneider E, Bakker EP. Erratum to: "Gene cloning, nucleotide sequence and biochemical properties of a cytoplasmic cyclomaltodextrinase (neopullulanase) from Alicyclobacillus acidocaldarius, reclassification of a group of enzymes. FEMS Microbiol Lett 2000; 188:107. [DOI] [PubMed] [Google Scholar]
- 16.Michels M, Bakker EP. Low-affinity potassium uptake system in Bacillus acidocaldarius. J Bacteriol 1987; 169:4335-4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morana A, Esposito A, Maurelli L, Ruggiero G, Ionata E, Rossi M, La Cara F. A novel thermoacidophilic cellulase from Alicyclobacillus acidocaldarius. Protein Pept Lett 2008; 15:1017-1021 10.2174/092986608785849209 [DOI] [PubMed] [Google Scholar]
- 18.Schleussinger E, Schmid R, Bakker EP. New type of kdp region with a split sensor-kinase kdpD gene located within two divergent kdp operons from the thermoacidophilic bacterium Alicyclobacillus acidocaldarius. Biochimica et Biophysica Acta 2006; 1759:437-441 [DOI] [PubMed] [Google Scholar]
- 19.Schwermann B, Pfau K, Liliensiek B, Schleyer M, Fischer T, Bakker EP. Purification, properties and structural aspects of a thermoacidophilic a-amylase from Alicyclobacillus acidocaldarius ATCC 27009. Eur J Biochem 1994; 226:981-991 10.1111/j.1432-1033.1994.00981.x [DOI] [PubMed] [Google Scholar]
- 20.Roberts RJ. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res 1984; 12:r167-r204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darland G, Brock TD. Bacillus acidocaldarius sp.nov., an acidophilic thermophilic spore-forming bacterium. J Gen Microbiol 1971; 67:9-15 [Google Scholar]
- 22.Goto K, Tanimoto Y, Tamura T, Mochida K, Arai D, Asahara M, Suzuki M, Tanaka H, Inagaki K. Identification of thermoacidophilic bacteria and a new Alicyclobacillus genomic species isolated from acidic environments in Japan. Extremophiles 2002; 6:333-340 10.1007/s00792-001-0262-3 [DOI] [PubMed] [Google Scholar]
- 23.Gouws PA, Gie L, Pretorius A, Dhansay N. Isolation and identification of Alicyclobacillus acidocaldarius by 16s rDNA from mango juice and concentrate. Int J Food Sci Technol 2005; 40:789-792 10.1111/j.1365-2621.2005.01006.x [DOI] [Google Scholar]
- 24.Groenewald WH, Gouws P, Witthuhn R. Isolation and identification of species of Alicyclobacillus from orchard soil in the Western Cape, South Africa. Extremophiles 2008; 12:159-163 10.1007/s00792-007-0112-z [DOI] [PubMed] [Google Scholar]
- 25.Groenewald WH, Gouws PA, Witthuhn RC. Isolation, identification and typification of Alicyclobacillus acidoterrestris and Alicyclobacillus acidocaldarius strains from orchard soil and the fruit processing environment in South Africa. Food Microbiol 2009; 26:71-76 10.1016/j.fm.2008.07.008 [DOI] [PubMed] [Google Scholar]
- 26.Palop A, Alvarez I, Raso J, Condón S. Heat resistance of Alicyclobacillus acidocaldarius in water, various buffers, and orange juice. J Food Prot 2000; 63:1377-1380 [DOI] [PubMed] [Google Scholar]
- 27.Silva FVM, Gibbs P. Alicyclobacillus acidoterrestris spores in fruit products and design of pasteurization processes. Trends Food Sci Technol 2001; 12:68-74 10.1016/S0924-2244(01)00070-X [DOI] [Google Scholar]
- 28.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 29.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 30.Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 31.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deinhard G, Blanz P, Poralla K, Altan E. Bacillus acidoterrestris sp. nov., a new thermotolerant acidophile isolated from different soils. Syst Appl Microbiol 1987; 10:47-53 [Google Scholar]
- 33.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrity GM, Lilburn TG, Cole JR, Harrison SH, Euzéby J, Tindall BJ. Taxonomic outline of the Bacteria and Archaea, Release 7.7 March 6, 2007. Part 9 - The Bacteria: Phylum "Firmicutes": Class "Bacilli". http://www.taxonomicoutline.org/index.php/toba/article/viewFile/186/218 2007.
- 36.Garrity GM, Holt JG. In: Garrity GM, Boone DR, Castenholz RW (2001) Taxonomic Outline of the Archaea and Bacteria Bergey's Manual of Systematic Bacteriology 155-166. [Google Scholar]
- 37.Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol 1978; 28:1-6 [Google Scholar]
- 38.Prévot AR. In: Magrou J, Prévot AR, Rosset G (1953) Dictionnaire des Bactéries Pathogènes. 1-692. [Google Scholar]
- 39.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 [PubMed] [Google Scholar]
- 40.Ludwig W, Schleifer KH, Whitman WB. Revised road map to the phylum Firmicutes In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 1-12. [Google Scholar]
- 41.Biological Agents. Technical rules for biological agents www.baua.de TRBA 466.
- 42.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goto K, Mochida K, Asahara M, Suzuki M, Kasai H, Yokota A. Alicyclobacillus pomorum sp. nov., a novel thermo-acidophilic, endospore-forming bacterium that does not possess w-alicyclic fatty acids, and emended description of the genus Alicyclobacillus. Int J Syst Evol Microbiol 2003; 53:1537-1544 10.1099/ijs.0.02546-0 [DOI] [PubMed] [Google Scholar]
- 44.Goto K, Matsubara H, Mochida K, Matsumura T, Hara Y, Niwa M, Yamasato K. Alicyclobacillus herbarius sp. nov., a novel bacterium containing ω-cycloheptane fatty acids, isolated from herbal tea. Int J Syst Evol Microbiol 2002; 52:109-113 [DOI] [PubMed] [Google Scholar]
- 45.Matsubara H, Goto K, Matsumura T, Mochida K, Iwaki M, Niwa M, Yamasato K. Alicyclobacillus acidiphilus sp. nov., a novel thermo-acidophilic, ω-alicyclic fatty acid-containing bacterium isolated from acidic beverages. Int J Syst Evol Microbiol 2002; 52:1681-1685 10.1099/ijs.0.02169-0 [DOI] [PubMed] [Google Scholar]
- 46.De Rosa M, Gambacorta A, Minale L, Bu'lock JD. The formation of ω-cyclohexyl-fatty acids from shikimate in an acidophilic thermophilic Bacillus. A new biosynthetic pathway. Biochem J 1972; 128:751-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Rosa M, Gambacorta A, Bu'lock JD. Effects of pH and temperature on the fatty acid composition of Bacillus acidocaldarius. J Bacteriol 1974; 117:212-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poralla K, Härtner T, Kannenberg E. Effect of temperature and pH on the hopanoid content of Bacillus acidocaldarius. FEMS Microbiol Lett 1984; 23:253-256 10.1111/j.1574-6968.1984.tb01073.x [DOI] [Google Scholar]
- 49.List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/ media_list.php
- 50.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sims D, Brettin T, Detter J, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anonymous. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. Oak Ridge National Laboratory and University of Tennessee 2009 http://compbio.ornl.gov/prodigal
- 53.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. (Submitted). [DOI] [PubMed] [Google Scholar]
- 54.Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. Expert Review of Functional Annotations for Microbial Genomes. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]