Abstract
Streptosporangium roseum Crauch 1955 is the type strain of the species which is the type species of the genus Streptosporangium. The ‘pinkish coiled Streptomyces-like organism with a spore case’ was isolated from vegetable garden soil in 1955. Here we describe the features of this organism, together with the complete genome sequence and annotation. This is the first completed genome sequence of a member of the family Streptosporangiaceae, and the second largest microbial genome sequence ever deciphered. The 10,369,518 bp long genome with its 9421 protein-coding and 80 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: Sporangia, vegetative and aerial mycelia, aerobic, non-motile, non-motile spores, Gram-positive, Streptosporangiaceae, S. cloviforme
Introduction
Strain NI 9100T (= DSM 43021 = ATCC 12428 = JCM 3005) is the type strain of the species Streptosporangium roseum, which is the type species of the genus Streptosporangium, the type genus of the actinobacterial suborder Streptosporanineae [1-4]. S. roseum NI 9100T was isolated from vegetable garden soil and first described by Crouch in 1955 [2,4]. The name derives from ‘strepto’ from Greek meaning ‘coiled’ combined with ‘sporangium’, Latin for ‘spore case’, to mean ‘streptomyces-like’ but with sporangia [2,4]. The species epithet ‘roseum’ derives from the pinkish color on potato dextrose agar [2]. Here we present a summary classification and a set of features for S. roseum NI 9100T, together with the description of the complete genomic sequencing and annotation.
Classification and features
The 16S rRNA genes of the thirteen other validly named species currently ascribed to the genus Streptosporangium share 96-100% (S. vulgare [5]) sequence identity with NI 9100T, but S. claviforme (94%) [6,7] apparently does not belong to this genus (but to the genus Herbidospora) and thus has been excluded from phylogenetic analysis (see below). Two reference strains, DSM 43871 (X89949), and DSM 44111 (X89947), differ by just one nucleotide from strain NI 9100T, whereas the effectively published named species ‘S. koreanum’ DSM 44110 [99.9%, 5], ‘S. brasiliense’ DSM 44109 [99.4%, 5] and ‘S. rubrum’ DSM 44095 [99.4%, 5] appear to members of the genus. Members of the species and genus are rare in nature, at least based on the habitats screened thus far as 16S rRNA in environmental samples and metagenomic surveys do not exceed 88-91% sequence similarity to the 16S rRNA gene sequence of strain NI 9100T (U48996, X70425, X89947; status August 2009).
Figure 1a and Figure 1b show the phylogenetic neighborhood of S. roseum NI 9100T in a 16S rRNA based tree. The sequence of the six 16S rRNA gene copies in the genome do not differ from each other, and are identical to the previously published sequence generated from DSM 43021 (X89947), whereas the sequence generated in the same year from the JCM 3005 version of strain 9100T (U48996) differs by 24 nucleotides (1.7%).
Figure 1a.
Phylogenetic tree highlighting the position of S. roseum NI 9100T relative to the type strains of the other species within the genus (1a) except for S. claviforme (see text). The tree was inferred from 1,411 and aligned characters [8,9] of the 16S rRNA gene sequence under the maximum likelihood criterion [10] and either rooted with the results of Figure 1b (Figure 1a) or rooted in accordance with the current taxonomy. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 1,000 bootstrap replicates if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [11] are shown in blue, published genomes in bold.
Figure 1b.
Phylogenetic tree highlighting the position of S. roseum NI 9100T relative to the type strains of the other genera within the family Streptosporanginea . The tree was inferred from 1,369 aligned characters [8,9] of the 16S rRNA gene sequence under the maximum likelihood criterion [10] and either rooted with the results of Figure 1b (Figure 1a) or rooted in accordance with the current taxonomy. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 1,000 bootstrap replicates if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [11] are shown in blue, published genomes in bold.
A summary of the classification and features for S. roseum is listed in Table 1. We draw attention to the reader that we find quite an amount of contradictive results between old and more recent literature (see below). A potential but not ultimate source for this observation could be the usage of different experimental methods. A variety of media were used in the original description pertaining to cellular and mycelium morphology on (Figure 2) The color of the substrate mycelium is red-brown to yellow-brown [2,24]. Strain NI 9100T utilizes glucose, arabinose, sucrose, xylose, fructose, and raffinose, but not inositol, mannose, rhamnose, or cellulose [19,20]. The strain is positive for arginine dihydrolase and acetoin production (Voges Proskauer test), weakly positive for citrate utilization, lysine decarboxylase, and ornithine decarboxylase, and negative for Kohn's gelatin gelatinase, urease, o-nitro-phenyl-galactoside β-galactosidase, tryptophan desaminase, tryptophan indole production, H2S production from sodium thiosulfate [19,20]. Starch hydrolysis and nitrate reduction are positive, but growth at 42°C and iodinin production are negative [24]. Mertz and Yao [18] reported that strain NI 9100T can utilize glycerol, arabinose, rhamnose and inositol, which is in part contradictory to other results [20,21]. Gelatin is liquefied, milk is peptonized and red-brown to purple-brown soluble pigments are produced [18]. Zhang et al. [21] describe strain NI 9100T as utilizing sorbitol and sorbose but to be negative for L-arabinose, erythrose, D-fructose, D-galactose, inositol, D-mannose, maltose, raffinose, and rhamnose, which again is in part in conflict with other studies [18-20]. Strain NI 9100T produces a secondary metabolite, the antibiotic angucycline WS 79089B, which is an inhibitor of the endothelin-converting enzyme [20]. In contrast to S. carneum, strain NI 9100T does not produce an antibiotic against Staphylococcus aureus [18].
Table 1. Classification and general features of S. roseum NI 9100T according to the MIGS recommendations [12].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [13] | |
| Phylum Actinobacteria | TAS [14] | ||
| Class Actinobacteria | TAS [15] | ||
| Subclass Actinobacteridae | TAS [15] | ||
| Order Actinomycetales | TAS [15] | ||
| Suborder Streptosporangineae | TAS [15] | ||
| Family Streptosporangiaceae | TAS [16,17] | ||
| Genus Streptosporangium | TAS [1-4] | ||
| Species Streptosporangium roseum | TAS [1-4] | ||
| Type strain NI 9100 | |||
| Gram stain | not tested, probably positive | NAS [15,16] | |
| Cell shape | produces aerial mycelium | TAS [2] | |
| Motility | non-motile | TAS [2] | |
| Sporulation | non-motile spores | TAS [2] | |
| Temperature range | mesophile, temperature range not determined, does not grow at 42°C | TAS [1,18] | |
| Optimum temperature | 28°C | TAS [1,18] | |
| Salinity | 2.5% NaCl | TAS [19,20] | |
| MIGS-22 | Oxygen requirement | aerobic | TAS [2] |
| Carbon source | several (see text), but be aware of contradicting results | TAS [19-21] | |
| Energy source | carbohydrates | TAS [19-21] | |
| MIGS-6 | Habitat | soil | TAS [2] |
| MIGS-15 | Biotic relationship | free living | TAS [2] |
| MIGS-14 | Pathogenicity | non pathogenic | NAS |
| Biosafety level | 1 | TAS [22] | |
| Isolation | vegetable garden soil | TAS [2] | |
| MIGS-4 | Geographic location | most probably Chapel Hill, North Carolina, USA | TAS [2] |
| MIGS-5 | Sample collection time | 1955 or before | TAS [2] |
| MIGS-4.1 MIGS-4.2 | Latitude, Longitude | 35.913, -79.055 | |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [23]. If the evidence code is IDA, then the property was directly observed for a live isolate by one of the authors or an expert or mentioned in the acknowledgements.
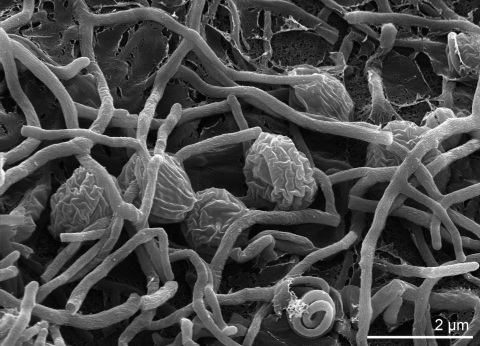
Figure 2.
Scanning electron micrograph of S. roseum NI 9100T
The characteristics of the ribosomal protein AT-L30 of strain S. roseum JCM2178T in comparison to other bacteria of the genus Streptosporangium is described elsewhere [25]. These data should be taken cautiously, as according to the Japanese Collection of Microorganisms (JCM) catalogue the strain number “JCM2178” is affiliated with Aspergillus oryzae (accessed to JCM in August 09), hence the true nature of strain S. roseum JCM2178T in the study of Ochi [25] is unclear.
Chemotaxonomy
The major fatty acids (relative ratio %) are iso-C16:0 (40.0), C17:0 10-methyl (23.0), C16:0 (1.95), C16:0 10-methyl (6.0), iso-C14:0 (14.0) (Reiner Kroppenstedt, personal communication). Partly different fatty acid patterns are reported elsewhere [18-20,26,27]. The proportions of diaminopimelic acid (A2pm) in the cell wall of strain S. roseum NI 9100T are 71% meso-A2pm and 29% LL-A2pm [26]. The phospholipids of strain S. roseum NI 9100T are phosphatidylethanolamine, hydroxyphosphatidylethanolamine, ninhydrin-positive and sugar-positive phospholipids, disphosphatidylglycerol, and posphatidylinositol [1]. The menaquinone compositions are MK-9 (III, VIII-H4) (56.5%), MK-9 (H2) (37.8%), MK-9 (H0) (5.0%), and MK-9 (H6) (0.7%) [1]. Galactose and madurose are present in whole cell sugars extracts, rhamnose is absent [1]. In general, the genus Streptosporangium is characterized by the whole-cell sugar type B or C, the phospholipid type IV and of the fatty acid type 3c [1].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position, and is part of the Genomic Encyclopedia of Bacteria and Archaea project. The genome project is deposited in the Genome OnLine Database [11] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Two Sanger libraries: 6kb pMCL200 and fosmid pcc1Fos One 454 Pyrosequence standard library |
| MIGS-29 | Sequencing platforms | ABI3730, 454 GS FLX |
| MIGS-31.2 | Sequencing coverage | 8.45× Sanger; 27.6× Pyrosequence |
| MIGS-30 | Assemblers | Newbler, phrap |
| MIGS-32 | Gene calling method | Prodigal, GenePrimp |
| INSDC ID | CP001814 (genome), CP001815 (plasmid) | |
| Genbank Date of Release | 12/10/2009 | |
| GOLD ID | Gc01156 | |
| NCBI project ID | 21083 | |
| Database: IMG-GEBA | 2501799901 | |
| MIGS-13 | Source material identifier | DSM 43021 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
S. roseum NI 9100T, DSM 43021, was grown in DSMZ medium 535, Trypticase Soy Broth [28], at 28°C. DNA was isolated from 0.5-1 g of cell paste using the JGI CTAP procedure with modification ALM as described in [29].
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger and 454 sequencing platforms. All general aspects of library construction and sequencing performed at the JGI can be found at http://www.jgi.doe.gov/. 454 Pyrosequencing reads were assembled using the Newbler assembler version 1.1.02.15 (Roche). Large Newbler contigs were broken into 11,709 overlapping fragments of 1,000 bp and entered into assembly as pseudo-reads. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and to adjust inflated q-scores. A hybrid 454/Sanger assembly was made using the parallel phrap assembler (High Performance Software, LLC). Possible mis-assemblies were corrected with Dupfinisher [30] or transposon bombing of bridging clones (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing in Consed, custom primer walk or PCR amplification. A total of 2,837 Sanger finishing reads were produced to close gaps, to resolve repetitive regions, and to raise the quality of the finished sequence. The error rate of the completed genome sequence is less than 1 in 100,000. Together all sequence types provided 36.05× coverage of the genome. The final assembly contains 128,042 Sanger and 1,033,578 Pyrosequence reads.
Genome annotation
Genes were identified using Prodigal [31] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline (http://geneprimp.jgi-psf.org) [32]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [33].
Genome properties
The genome consists of a 10,341,314 bp long chromosome and a small 28,204 bp plasmid with a 70.9% GC content (Table 3 and Figure 3). Of the 9,501 genes predicted, 9,421 were protein coding genes, and 80 RNAs. In addition, 446 pseudogenes were identified. The majority of protein-coding genes (62.5%) were assigned a putative function while those remaining were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 10,369,518 | 100.00% |
| DNA coding region (bp) | 9,121,910 | 87.97% |
| DNA G+C content (bp) | 7,348,162 | 70.86% |
| Number of replicons | 2 | |
| Extrachromosomal elements | 1 | |
| Total genes | 9,501 | 100.00% |
| RNA genes | 80 | 0.84% |
| rRNA operons | 6 | |
| Protein-coding genes | 9,421 | 99.16% |
| Pseudo genes | 446 | 4.49% |
| Genes with function prediction | 5,939 | 62.47% |
| Genes in paralog clusters | 2,792 | 29.37% |
| Genes assigned to COGs | 6,224 | 65.47% |
| Genes assigned Pfam domains | 6,596 | 69.38% |
| Genes with signal peptides | 2,248 | 23.65% |
| Genes with transmembrane helices | 2,235 | 23.51% |
| CRISPR repeats | 0 |
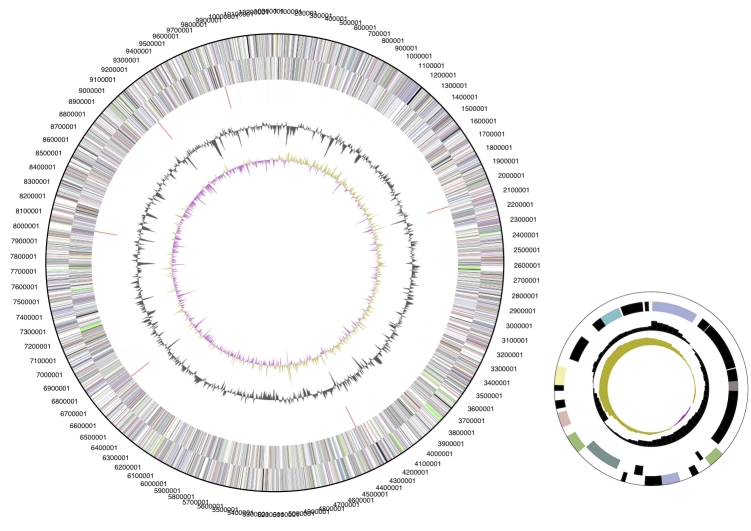
Figure 3.
Graphical circular map of the genome; plasmid not to scale. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 226 | 2.4 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.0 | RNA processing and modification |
| K | 966 | 10.3 | Transcription |
| L | 293 | 3.1 | Replication, recombination and repair |
| B | 1 | 0.0 | Chromatin structure and dynamics |
| D | 38 | 0.4 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 189 | 2.0 | Defense mechanisms |
| T | 511 | 5.4 | Signal transduction mechanisms |
| M | 298 | 3.2 | Cell wall/membrane biogenesis |
| N | 2 | 0.0 | Cell motility |
| Z | 1 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 43 | 0.5 | Intracellular trafficking and secretion |
| O | 167 | 1.8 | Posttranslational modification, protein turnover, chaperones |
| C | 424 | 4.5 | Energy production and conversion |
| G | 639 | 6.8 | Carbohydrate transport and metabolism |
| E | 600 | 6.4 | Amino acid transport and metabolism |
| F | 124 | 1.3 | Nucleotide transport and metabolism |
| H | 254 | 2.7 | Coenzyme transport and metabolism |
| I | 306 | 3.2 | Lipid transport and metabolism |
| P | 320 | 3.4 | Inorganic ion transport and metabolism |
| Q | 315 | 3.3 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 974 | 10.3 | General function prediction only |
| S | 473 | 5.0 | Function unknown |
| - | 3187 | 33.8 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Susanne Schneider (DSMZ) for DNA extraction and quality analysis. This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, as well as German Research Foundation (DFG) INST 599/1-1.
References
- 1.Stackebrandt E, Kroppenstedt RM, Jahnke KD, Kemmerling C, Gürtler H. Transfer of Streptosporangium viridogriseum (Okuda et al. 1966), Streptosporangium viridogriseum subsp. kofuense (Nonomura and Ohara 1969), and Streptosporangium albidum (Furumai et al. 1968) to Kutzneria gen. nov. as Kutzneria viridogrisea comb. nov., Kutzneria kofuensis comb. nov., and Kutzneria albida comb. nov., respectively, and emendation of the Genus Streptosporangium. Int J Syst Bacteriol 1994; 44:265-269 [Google Scholar]
- 2.Couch JN. A new genus and family of the Actinomycetales, with a revision of the genus Actinoplanes. J Elisha Mitchell Sci Soc 1955; 71:148-155 [Google Scholar]
- 3.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 [PubMed] [Google Scholar]
- 4.Couch JN, Bland CE. Genus III. Streptosporangium Couch 1955, 148. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 711-715. [Google Scholar]
- 5.Nonomura H, Ohara Y. Distribution of the actinomycetes in soil. IV. The isolation and classification of the genus Streptosporangium. J Ferment Technol 1960; 38:405-409 [Google Scholar]
- 6.Petrolini B, Quaroni S, Sardi P, Saracchi M, Andriolollo N. A sporangiate actinomycete with unusual morphological features: Streptosporangium claviforme sp. nov. Actinomycetes 1992; 3:45-50 [Google Scholar]
- 7.Ward-Rainey N, Rainey FA, Stackebrandt E. The phylogenetic structure of the genus Streptosporangium. Syst Appl Microbiol 1996; 19:50-55 [Google Scholar]
- 8.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 9.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 10.Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 11.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thompson N, Allen MJ, Anguiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 15.Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a New Hierarchic Classification System, Actinobacteria classis nov. Int J Syst Bacteriol 1997; 47:479-491 [Google Scholar]
- 16.Goodfellow M, Stanton LJ, Simpson KE, Minnikin DE. Numerical and chemical classification of Actinoplanes and some related actinomycetes. J Gen Microbiol 1990; 136:19-36 [Google Scholar]
- 17.List Editor Validation List no. 34. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1990; 40:320-321 [Google Scholar]
- 18.Mertz FP, Yao RC. Streptosporangium carneum sp. nov. Isolated from soil. Int J Syst Bacteriol 1990; 40:247-253 [DOI] [PubMed] [Google Scholar]
- 19.Wink JM.2009. http://www.gbif-prokarya.de/microorganisms/files/Methods.pdf
- 20.Wink JM. Compendium of Actinobacteria http://www.dsmz.de/microorganisms/ wink_pdf/DSM43021.pdf 2009
- 21.Zhang LP, Zhang LM, Zhang XM. Streptosporangium canum sp. nov., isolated from soil. Int J Syst Evol Microbiol 2009; 59:1715-1719 10.1099/ijs.0.007401-0 [DOI] [PubMed] [Google Scholar]
- 22.Biological Agents. Technical rules for biological agents www.baua.de TRBA 466
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shearer MC, Colman PM, Nash CH., III Streptosporangium fragile sp. nov. Int J Syst Bacteriol 1983; 33:364-368 [Google Scholar]
- 25.Ochi K, Miyadoh S. Polyacrylamide Gel Electrophoresis Analysis of Ribosomal Protein AT-L30 from an actinomycete Genus, Streptosporangium. Int J Syst Bacteriol 1992; 42:151-155 [DOI] [PubMed] [Google Scholar]
- 26.Gyobu Y, Miyadoh S. Proposal to transfer Actinomadura carminata to a new subspecies of the genus Nonomuraea as Nonomuraea roseoviolacea subsp. carminata comb. nov. Int J Syst Evol Microbiol 2001; 51:881-889 [DOI] [PubMed] [Google Scholar]
- 27.Kudo T, Seino A. Transfer of Streptosporangium indianense Gupta 1965 to the Genus Streptomyces as Streptomyces indiaensis (Gupta 1965) comb. nov. Int J Syst Bacteriol 1987; 37:241-244 [Google Scholar]
- 28.List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/ media_list.php
- 29.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sims D, Brettin T, Detter JC, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anonymous <http://compbio.ornl.gov/prodigal/>
- 32.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A gene prediction improvement pipeline for microbial genomes. (Submitted). [DOI] [PubMed] [Google Scholar]
- 33.Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. Expert review of functional annotations for microbial genomes. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]