Abstract
Veillonella parvula (Veillon and Zuber 1898) Prévot 1933 is the type species of the genus Veillonella in the family Veillonellaceae within the order Clostridiales. The species V. parvula is of interest because it is frequently isolated from dental plaque in the human oral cavity and can cause opportunistic infections. The species is strictly anaerobic and grows as small cocci which usually occur in pairs. Veillonellae are characterized by their unusual metabolism which is centered on the activity of the enzyme methylmalonyl-CoA decarboxylase. Strain Te3T, the type strain of the species, was isolated from the human intestinal tract. Here we describe the features of this organism, together with the complete genome sequence, and annotation. This is the first complete genome sequence of a member of the large clostridial family Veillonellaceae, and the 2,132,142 bp long single replicon genome with its 1,859 protein-coding and 61 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: opportunistic infections, human oral microflora, dental plaque, intergeneric coaggregation, methylmalonyl-CoA decarboxylase, Veillonellaceae
Introduction
Strain Prévot Te3T (= DSM 2008 = ATCC 10790 = JCM 12972) is the type strain of the species Veillonella parvula and was first described in 1898 by Veillon and Zuber [1] as “Staphylococcus parvulus” before it was renamed as Veillonella parvula by Prévot in 1933 [2]. Although it is a Gram-negative organism harboring lipopolysaccharide [3] it is more closely related to Gram-positive species like Sporomusa, Megasphaera or Selenomonas. Together, they share the unusual presence of cadaverine and putrescine in their cell walls [4]. The genus Veillonella comprises 11 species (status July 2009) which are all known to inhabit the oral cavity and the gastrointestinal tract of homeothermic vertebrates. Six of the species, among them V. parvula, have been isolated from man, the others are typical for rodents [5]. In general, veillonellae are harmless inhabitants of most body cavities, however, occasionally they can participate in multispecies infections at diverse body sites and in rare cases cause severe infections also as pure cultures [6].
Here we present a summary classification and a set of features for V. parvula Te3T, together with the description of the complete genomic sequencing and annotation.
Classification and features
The natural habitat is human dental plaque and V. parvula can amount to up to 98% of the cultivable veillonellae in healthy subgingival sites [7]. Additionally, veillonellae are common inhabitants of the gastrointestinal tract. Although the other species of the genus Veillonella are found in large numbers throughout the oral cavity, V. parvula is the only species of the genus involved in oral diseases such as gingivitis. It has also been isolated in rare cases of endocarditis, meningitis, discitis [8] or bacteremia as pure culture but more often V. parvula is involved in multispecies infections (reviewed in [6]). Medline indexes few cultivated strains with a high degree of 16S rRNA gene sequence similarity to Te3T, e.g. DJF_B315 from porcine intestine (EU728725, Hojberg and Jensen, unpublished, 99.9% identity). The other type strains of the genus Veillonella vary from 94.1% (V. ratti) to 99.2% (V. dispar). A vast number of phylotypes with significant 16S rRNA sequence similarity to V. parvula were observed from intubated patients [9], carious dentine from advanced caries (AY995757; 99.7% identity), and the human skin microbiome [10]. Curiously, only one sample from a human gut metagenome analysis [11] scored above 96% sequence similarity in screenings of environmental samples (status September 2009).
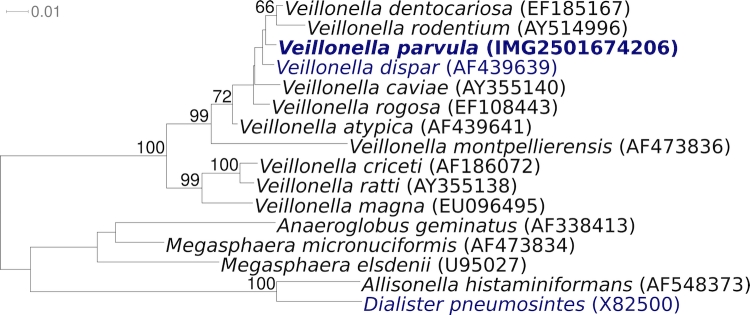
Figure 1 shows the phylogenetic neighborhood of V. parvula strain Te3T in a 16S rRNA based tree. The sequences of the four copies of the 16S rRNA gene in the genome differ by up to seven nucleotides, and differ by up to four nucleotides from the previously published sequence generated from ATCC 10790 (AY995767).
Figure 1.
Phylogenetic tree highlighting the position of V. parvula strain Te3T relative to all other type strains within the genus Veillonella. The tree was inferred from 1,378 aligned characters [12,13] of the 16S rRNA gene sequence under the maximum likelihood criterion [14]. The tree was rooted with the type strains of other genera within the family Veillonellaceae. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 1,000 bootstrap replicates if greater than 60%. Lineages with type strain genome sequencing projects registered in GOLD [15] are shown in blue, published genomes in bold.
V. parvula is a Gram-negative, non-motile, non-sporeforming, anaerobic coccus (approximately 0.3 to 0.5 µm in diameter) that grows in pairs or short chains (Table 1 and Figure 2). Veillonellae are characterized by an unusual metabolism using methylmalonyl-CoA decarboxylase to convert the free energy derived from decarboxylation reactions into an electrochemical gradient of sodium ions [29]. They utilize the metabolic end products of co-existing carbohydrate-fermenting bacteria, i.e. lactic acid bacteria in the gastrointestinal tract, and thereby play an important role in a natural microbial food chain [6]. Another characteristic trait of veillonellae is their ability to form intergeneric coaggregates with other bacteria which occur in the same ecological niche [30]. Although Veillonella cannot adhere to surfaces itself, the bacterium is able to attach to specific surface structures present on other cells, often mediated by lectin-carbohydrate interactions [31]. The coaggregation creates a functional community providing nutrients and protection for all participants.
Table 1. Classification and general features of V. parvula Te3T according to the MIGS recommendations [16].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [17] | |
| Phylum Firmicutes | TAS [18,19] | ||
| Class Clostridia | TAS [18] | ||
| Order Clostridiales | TAS [20,21] | ||
| Family Veillonellaceae | TAS [22,23] | ||
| Genus Veillonella | TAS [2] | ||
| Species Veillonella parvula | TAS [2] | ||
| Type strain Te3 | TAS [2] | ||
| Gram stain | negative | TAS [1] | |
| Cell shape | small cocci | TAS [1] | |
| Motility | nonmotile | TAS [1] | |
| Sporulation | nonsporulating | TAS [1] | |
| Temperature range | mesophile | TAS [1] | |
| Optimum temperature | 37°C | TAS [1] | |
| Salinity | normal | TAS [1] | |
| MIGS-22 | Oxygen requirement | anaerobic | TAS [1] |
| Carbon source | acid production from lactate and other organic acids | TAS [24] | |
| Energy source | lactate; succinate | TAS [25,26] | |
| MIGS-6 | Habitat | human gastrointestinal tract | TAS [1] |
| MIGS-15 | Biotic relationship | human pathogen | TAS [1] |
| MIGS-14 | Pathogenicity | subgingival plaque formation; opportunistic pathogen | TAS [1] |
| Biosafety level | 2 | TAS [27] | |
| Isolation | human intestinal tract | TAS [1] | |
| MIGS-4 | Geographic location | unknown, probably France | TAS [1] |
| MIGS-5 | Sample collection time | before 1898 | TAS [1] |
| MIGS-4.1 MIGS-4.2 |
Latitude, Longitude | unknown | |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [28]. If the evidence code is IDA, then the property was observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
Figure 2.
Scanning electron micrograph of V. parvula Te3T
Strain Te3T produces propionic and acetic acid, carbon dioxide and hydrogen from lactate and other organic acids like pyruvate, malate or fumarate. V. parvula cannot grow on succinate as a sole carbon source but can decarboxylate succinate during fermentation of lactate or malate [25]. Veillonellae are unable to use glucose or other carbohydrates for fermentation [26] and they do not possess a functional hexokinase [24]. Nitrate is reduced and arginine dihydrolase is produced.
Veillonellae show resistance to tetracycline (>25 µg/ml), erythromycin (>25 µg/ml) gentamicin (>25 µg/ml) and kanamycin (>25 µg/ml) and they are susceptible to penicillin G (0.4 µg/ml), cephalotin (1.6 µg/ml) and clindamycin (0.1 µg/ml). Their resistance is intermediate for chloramphenicol (3.1 µg/ml) and lincomycin (6.2 µg/ml) [32].
Chemotaxonomy
The cell wall of V. parvula comprises an outer membrane, clearly demonstrating the presence of lipopolysaccharide [33]. The peptidoglycan of veillonellae is of the A1γ-type with glutamic acid in D configuration, diaminopimelic acid in meso configuration and covalently bound cadaverine or putrescine attached in α-linkage to glutamic acid [34]. As major fatty acids straight-chain saturated C13:0 (24%), C15:0 (12%) and C16:0 (7%) and unsaturated C16:1 (5%), C17:1 (22%) and C18:1 (6%) are synthesized [35]. Another characteristic feature of V. parvula is the presence of plasmalogens such as plasmenylethanolamine and plasmenylserine as major constituents of the cytoplasmic membrane. These ether lipids replace phospholipids and play an important role in the regulation of membrane fluidity [36].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position, and is part of the Genomic Encyclopedia of Bacteria and Archaea project. The genome project is deposited in the Genome OnLine Database [15] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Two Sanger libraries: 8kb pMCL200 and fosmid pcc1Fos One 454 pyrosequence standard library |
| MIGS-29 | Sequencing platforms | ABI3730, 454 GS FLX |
| MIGS-31.2 | Sequencing coverage | 7.67× Sanger; 50.6× pyrosequence |
| MIGS-30 | Assemblers | Newbler, Phrap |
| MIGS-32 | Gene calling method | Prodigal, GenePRIMP |
| INSDC ID | CP001820 | |
| Genbank Date of Release | 23-November 2009 | |
| GOLD ID | Gc01152 | |
| NCBI project ID | 21091 | |
| Database: IMG-GEBA | 2501651195 | |
| MIGS-13 | Source material identifier | DSM 2008 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
V. parvula strain Te3T, DSM 2008, was grown anaerobically in DSMZ medium 104 (modified PYG-Medium, with the addition of lactate and putrescine) at 37°C [37]. DNA was isolated from 1.5-2 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol, with cell lysis protocol L as described in Wu et al. [38].
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger and 454 sequencing platforms. All general aspects of library construction and sequencing performed at the JGI can be found at http://www.jgi.doe.gov/. 454 Pyrosequencing reads were assembled using the Newbler assembler version 1.1.02.15 (Roche). Large Newbler contigs were broken into 1,716 overlapping fragments of 1,000 bp and entered into assembly as pseudo-reads. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and to adjust inflated q-scores. A hybrid 454/Sanger assembly was made using the parallel phrap assembler (High Performance Software, LLC). Possible mis-assemblies were corrected with Dupfinisher [39] or transposon bombing of bridging clones (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing in Consed, custom primer walk or PCR amplification. A total of 1,082 Sanger finishing reads were produced to close gaps, to resolve repetitive regions, and to raise the quality of the finished sequence. The error rate of the completed genome sequence is less than 1 in 100,000. Together all sequence types provided 51.2 × coverage of the genome. The final assembly contains 16,169 Sanger and 445,271 pyrosequence reads.
Genome annotation
Genes were identified using Prodigal [40] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline (http://geneprimp.jgi-psf.org) [41]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation were performed within the Integrated Microbial Genomes Expert Review (IMG-ER) platform (http://img.jgi.doe.gov/er) [42].
Genome properties
The genome is 2,132,142 bp long and comprises one main circular chromosome with a 38.6% GC content (Table 3 and Figure 3). Of the 1,920 genes predicted, 1,859 were protein coding genes, and 61 RNAs; 15 pseudogenes were also identified. The majority (73.6%) of the genes were assigned a putative function while those remaining were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 2,132,142 | 100.00% |
| DNA coding region (bp) | 1,886,054 | 88.46% |
| DNA G+C content (bp) | 823,631 | 38.63% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 1,920 | 100.00% |
| RNA genes | 61 | 3.18% |
| rRNA operons | 4 | |
| Protein-coding genes | 1,859 | 96.82% |
| Pseudo genes | 15 | 0.78% |
| Genes with function prediction | 1,413 | 73.59% |
| Genes in paralog clusters | 174 | 9.06% |
| Genes assigned to COGs | 1,474 | 76.77% |
| Genes assigned Pfam domains | 1,490 | 77.06% |
| Genes with signal peptides | 306 | 15.94% |
| Genes with transmembrane helices | 492 | 25.62% |
| CRISPR repeats | 5 |
Figure 3.
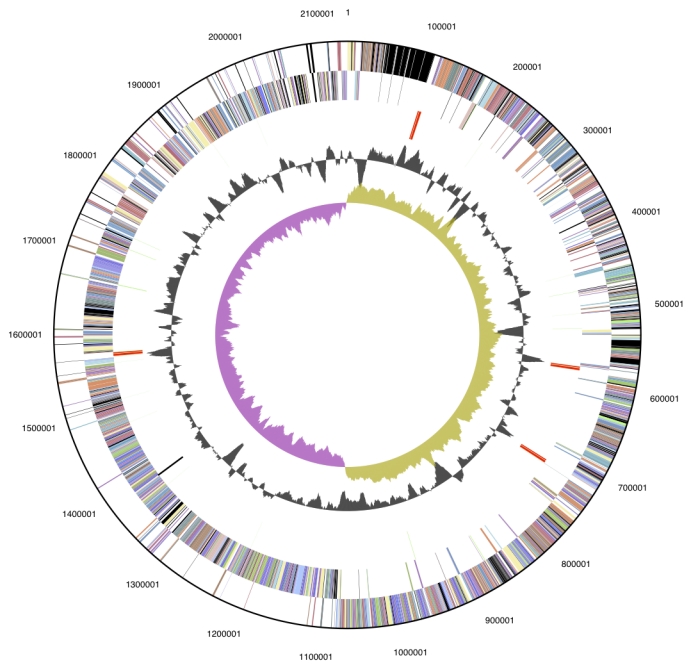
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 142 | 7.6 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 71 | 3.8 | Transcription |
| L | 92 | 4.9 | Replication, recombination and repair |
| B | 2 | 0.1 | Chromatin structure and dynamics |
| D | 20 | 1.1 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 26 | 1.4 | Defense mechanisms |
| T | 29 | 1.6 | Signal transduction mechanisms |
| M | 106 | 5.7 | Cell wall/membrane biogenesis |
| N | 6 | 0.3 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 35 | 1.9 | Intracellular trafficking and secretion |
| O | 48 | 2.6 | Posttranslational modification, protein turnover, chaperones |
| C | 100 | 5.4 | Energy production and conversion |
| G | 68 | 3.6 | Carbohydrate transport and metabolism |
| E | 160 | 8.6 | Amino acid transport and metabolism |
| F | 63 | 3.4 | Nucleotide transport and metabolism |
| H | 117 | 6.3 | Coenzyme transport and metabolism |
| I | 33 | 1.8 | Lipid transport and metabolism |
| P | 124 | 6.7 | Inorganic ion transport and metabolism |
| Q | 19 | 1.8 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 177 | 9.5 | General function prediction only |
| S | 143 | 7.7 | Function unknown |
| - | 385 | 20.7 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Susanne Schneider (DSMZ) for DNA extraction and quality analysis. This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, as well as German Research Foundation (DFG) INST 599/1-1.
References
- 1.Veillon A, Zuber MM. Recherches sur quelques microbes strictement anaérobies et leur rôle en pathologie. Arch Med Exp 1898; 10:517-545 [Google Scholar]
- 2.Prévot AR. Études de systématique bactérienne. I: Lois générale. II: Cocci anaérobies. Ann Sci Nat Bot 1933; 15:23-260 [Google Scholar]
- 3.Mergenhagen SE. Polysaccharide-lipid complexes from Veillonella parvula. J Bacteriol 1965; 90:1730-1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stackebrandt E, Pohla H, Kroppenstedt R, Hippe H, Woese CR. 16S rRNA analysis of Sporomusa, Selenomonas and Megasphaera on the phylogenetic origin of Gram-positive Eubacteria. Arch Microbiol 1985; 143:270-276 10.1007/BF00411249 [DOI] [Google Scholar]
- 5.Kraatz M, Taras D. Veillonella magna sp. nov., isolated from the jejunal mucosa of a healthy pig, and emended description of Veillonella ratti. Int J Syst Evol Microbiol 2008; 58:2755-2761 10.1099/ijs.0.2008/001032-0 [DOI] [PubMed] [Google Scholar]
- 6.Kolenbrander P. The Genus Veillonella Prokaryotes . Springer, New York; 2006. [Google Scholar]
- 7.Moore WEC, Holdeman LV, Cato EP, Smibert RM, Burmeister JA, Palcanis KG, Ranney RR. Comparative bacteriology of juvenile periodontitis. Infect Immun 1985; 48:507-519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marriott D, Stark D, Harkness J. Veillonella parvula discitis and secondary bacteremia: a rare infection complicating endoscopy and colonoscopy? J Clin Microbiol 2007; 45:672-674 10.1128/JCM.01633-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan JL, Brodie EL, Weng L, Lynch SV, Garcia O, Bron R, Hugenholtz P, Desantis TZ, Andersen GL, Wiener-Kronish JP, Bristow J. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol 2007; 45:1954-1962 10.1128/JCM.02187-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grice EA, Kong HH, Conlan S, Deming S, Davis J, Young AC, Bouffard GG, Blakesley RW, Muray PR, Green ED, et al. Topographical and temporal diversity of the human skin microbiome. Science 2009; 324:1190-1192 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivasta TP, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 2007; 14:169-181 10.1093/dnares/dsm018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 13.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 14.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 15.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. [2112744] Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garrity GM, Holt JG. In: Garrity GM, Boone DR, Castenholz RW (2001) Taxonomic Outline of the Archaea and Bacteria Bergey's Manual of Systematic Bacteriology 155-166. [Google Scholar]
- 19.Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol 1978; 28: 1-6 [Google Scholar]
- 20.Prévot AR. in: Hauduroy P, Ehringer G, Guillot G, Magrou J, Prevot, AR, Rosset A, Urbain, A. Dictionnaire des bactéries pathogènes. 2nd ed. Masson, Paris. 1953, pp 692. [Google Scholar]
- 21.Murray RGE. The Higher Taxa, or, a Place for Everything...? In: Holt JG (ed), Bergey's Manual of Systematic Bacteriology, First Edition, Volume 1, The Williams and Wilkins Co., Baltimore, 1984, p. 31-34. [Google Scholar]
- 22.Rogosa M. Transfer of Veillonella Prévot and Acidaminococcus Rogosa from Neisseriaceae to Veillonellaceae fam. nov., and the inclusion of Megasphaera Rogosa in Veillonellaceae. Int J Syst Evol Microbiol 1971; 21:231-233 10.1099/00207713-21-3-231 [DOI] [Google Scholar]
- 23.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30: 225-420 [PubMed] [Google Scholar]
- 24.Rogosa M, Krichevsky MI, Bishop FS. Truncated glycolytic system in Veillonella. J Bacteriol 1965; 90:164-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denger K, Schink B. Energy conservation by succinate decarboxylation in Veillonella parvula. J Gen Microbiol 1992; 138:967-971 [DOI] [PubMed] [Google Scholar]
- 26.Dimroth P. Bacterial sodium ion-coupled energetics. Antonie Van Leeuwenhoek 1994; 65:381-395 10.1007/BF00872221 [DOI] [PubMed] [Google Scholar]
- 27.Anonymous. Biological Agents: Technical rules for biological agents www.baua.de TRBA 466.
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimroth P. Biotin-dependent decarboxylases as energy transducing systems. Ann N Y Acad Sci 1985; 447:72-85 10.1111/j.1749-6632.1985.tb18426.x [DOI] [PubMed] [Google Scholar]
- 30.Gibbons RJ, Nygaard M. Interbacterial aggregation of plaque bacteria. Arch Oral Biol 1970; 15:1397-1400 10.1016/0003-9969(70)90031-2 [DOI] [PubMed] [Google Scholar]
- 31.Hughes CV, Kolenbrander PE, Andersen RN, Moore LV. Coaggregation properties of human oral Veillonella spp.: relationship to colonization site and oral ecology. Appl Environ Microbiol 1988; 54:1957-1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin WJ, Gardner M, Washington JA., II In vitro antimicrobial susceptibility of anaerobic bacteria isolated from clinical specimens. Antimicrob Agents Chemother 1972; 1:148-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bladen HA, Mergenhagen SE. Ultrastructure of Veillonella and morphological correlation of an outer membrane with particles associated with endotoxic activity. J Bacteriol 1964; 88:1482-1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamio Y. Structural specificity of diamines covalently linked to peptidoglycan for cell growth of Veillonella alcalescens and Selenomonas ruminantium. J Bacteriol 1987; 169:4837-4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jumas-Bilak E, Carlier JP, Jean-Pierre H, Teyssier C, Gay B, Campos J, Marchandin H. Veillonella montpellierensis sp. nov., a novel, anaerobic, Gram-negative coccus isolated from human clinical samples. Int J Syst Evol Microbiol 2004; 54:1311-1316 10.1099/ijs.0.02952-0 [DOI] [PubMed] [Google Scholar]
- 36.Olsen I. Salient structural features in the chemical composition of oral anaerobes, with particular emphasis on plasmalogens and sphingolipids. Rev Med Microbiol 1997; 8(Suppl 1):S3-S6 [Google Scholar]
- 37.List of media used at DSMZ for cell growth http://www.dsmz.de/microorganisms/ media_list.php
- 38.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova N, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sims D, Brettin T, Detter JC, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome of Kytococcus sedentarius type strain (541T). Standards in Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anonymous. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. Oak Ridge National Laboratory and University of Tennessee 2009. [Google Scholar]
- 41.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. (Submitted) [DOI] [PubMed] [Google Scholar]
- 42.Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. Expert IMG ER: A system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]