Abstract
Segniliparus rotundus Butler 2005 is the type species of the genus Segniliparus, which is currently the only genus in the corynebacterial family Segniliparaceae. This family is of large interest because of a novel late-emerging genus-specific mycolate pattern. The type strain has been isolated from human sputum and is probably an opportunistic pathogen. Here we describe the features of this organism, together with the complete genome sequence and annotation. This is the first completed genome sequence of the family Segniliparaceae. The 3,157,527 bp long genome with its 3,081 protein-coding and 52 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: aerobic, non-sporeforming, novel mycolic acid, opportunistic pathogen, Corynebacterineae, GEBA
Introduction
Strain CDC 1076T (= DSM 44985 = ATCC BAA-972 = JCM 13578) is the type strain of the species Segniliparus rotundus [1], which is the type species of the genus Segniliparus. Besides S. rotundus, the genus Segniliparus contains currently only one additional species: S. rugosus at present[1]. Segniliparus is currently the only genus in the family Segniliparaceae. The generic name of the genus derives from the Latin word ‘segnis’, meaning ‘slow’, and the Greek word ‘liparos’, fat/fatty, meaning ‘one with slow fats’, to indicate the possession of slow reacting fatty acids, i.e., late eluting mycolic acids detected with HPLC [1]. The species name is derived from the Latin word ‘rotundus’, rounded, referring to the smooth, round-domed colony forms [1]. Strain CDC 1076T was isolated from human sputum in Tennessee, USA [1]. Currently, only one additional strain of the species, CDC 413 (with identical 16S rRNA gene sequence), is known, which has been isolated from the human nasal region in Missouri, USA [1]. The 16S rRNA gene sequence of the type strain for the second species in the genus, S. rugosus [1], differs by only 1.1% from that of strain CDC 1076T. S. rugosus strains have been isolated from patients with cystic fibrosis in Australia and most probably USA [2,3], suggesting that S. rotundus could also be an opportunistic pathogen. The next closest relatives of S. rotundus outside the genus are the members of the genus Rhodococcus, which share 93.3 to 94.8% 16S rRNA genes sequence similarity with strain CDC 1076T [4]. Environmental screens and metagenomic surveys did not detected a single phylotype with more than 90-92% 16S rRNA gene sequence similarity, indicating a rather limited ecological distribution of the members of the genus Segniliparus (status February 2010). Here we present a summary classification and a set of features for S. rotundus CDC 1076T, together with the description of the complete genomic sequencing and annotation.
Classification and features
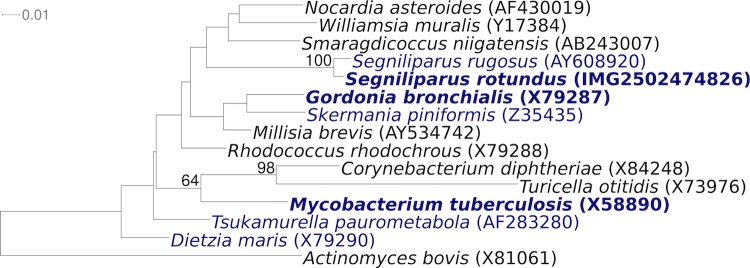
Figure 1 shows the phylogenetic neighborhood of for S. rotundus CDC 1076T in a 16S rRNA based tree. The sequence of the sole 16S rRNA gene in the genome is identical with the previously published 16S rRNA sequence generated from DSM 44985 (AY608918).
Figure 1.
Phylogenetic tree highlighting the position of S. rotundus CDC 1076T relative to the other type strains within the suborder Corynebacterineae. The tree was inferred from 1,436 aligned characters [5,6] of the 16S rRNA gene sequence under the maximum likelihood criterion [7] and rooted with the type strains of the order Actinomycetales. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 350 bootstrap replicates [8] if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [9] are shown in blue, published genomes in bold [10,11].
CDC 1076T cells are short rods with 0.4µm width by 1.0-1.3 µm length (Table 1 and Figure 2), forming round, smooth, dense and domed colonies [1]. Occasionally, v-forms are produced, but no true branching, mycelium, or spores have been reported. The colonies are non-pigmented, non-photochromogenic and do not produce a diagnostic odor [1]. It is negative for arylsulfatase after three days but positive after 14 days. Strain CDC 1076T does not grow on MacConkey agar, is weakly positive for iron uptake, Tween opacity and Tween hydrolysis, but negative for nitrate and tellurite reduction and for growth in lysozyme (21 days) [1]. Strain CDC 1076T does not produce niacin and develops bubbles in the semi-quantitative catalase test [1]. Using the API CORYNE test kit, strain CDC 1076T is positive for β-glucosidase and pyrazinamidase activities and negative for alkaline phosphatase, β-galactosidase, β-glucuronidase, α-glucosidase, N-acetyl-β-glucosaminidase and pyrrolidonyl arylamidase activity at 33°C [1]. Strain CDC 1076T is susceptible to amikacin, cefoxitan, clarithromycin, ciprofloxacin, doxycycline, imipenem and sulfamethoxazole at or below the respective MIC breakpoints but intermediate to tobramycin [1]. Glucose, maltose, D-fructose and trehalose are used as carbon source for growth with acid production, but not adonitol, L-arabinose, cellobiose, dulcitol, i-erythritol, galactose, i-myo-inositol, lactose, mannose, melibiose, raffinose, L-rhamnose, salicin, D-mannitol, D-sorbitol and sodium citrate [1]. Strain CDC 1076T hydrolyzes urea but not acetamide, adenine, casein, citrate, aesculin, hypoxanthine, tyrosine and xanthine [1].
Table 1. Classification and general features of S. rotundus CDC 1076T according to the MIGS recommendations [12].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [13] | |
| Phylum Actinobacteria | TAS [14] | ||
| Class Actinobacteria | TAS [15] | ||
| Subclass Actinobacteridae | TAS [15] | ||
| Order Actinomycetales | TAS [15] | ||
| Suborder Corynebacterineae | TAS [15] | ||
| Family Segniliparaceae | TAS [1] | ||
| Genus Segniliparus | TAS [1] | ||
| Species Segniliparus rotundus | TAS [1] | ||
| Type strain CDC 1076 | TAS [1] | ||
| Gram stain | Gram-negative | NAS | |
| Cell shape | short rods | TAS [1] | |
| Motility | nonmotile | TAS [1] | |
| Sporulation | non-sporulating | TAS [1] | |
| Temperature range | mesophile, 28°C - 37°C | TAS [1] | |
| Optimum temperature | 33°C | TAS [1] | |
| Salinity | not determined | ||
| MIGS-22 | Oxygen requirement | aerobic | TAS [1] |
| Carbon source | glucose, maltose, D-fructose, trehalose | TAS [1] | |
| Energy source | chemoorganotroph | TAS [1] | |
| MIGS-6 | Habitat | unknown, but probably host associated | TAS [1] |
| MIGS-15 | Biotic relationship | unknown | |
| MIGS-14 | Pathogenicity | most probably opportunistic pathogen | TAS [1-3] |
| Biosafety level | 2 | TAS [16] | |
| Isolation | human sputum | TAS [1] | |
| MIGS-4 | Geographic location | Tennessee, USA | TAS [1] |
| MIGS-5 | Sample collection time | 2005 or before | TAS [1] |
| MIGS-4.1 MIGS-4.2 |
Latitude Longitude |
unknown | |
| MIGS-4.3 | Depth | unknown | |
| MIGS-4.4 | Altitude | unknown |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [17]. If the evidence code is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
Figure 2.
Scanning electron micrograph of S. rotundus CDC 1076T
Chemotaxonomy
The cell wall of strain CDC 1076T contains mycolic acids and meso-diaminopimelic acid [1]. The mycolic acid HPLC pattern is a triple cluster of contiguous eluting peaks starting at approx. 6.0 min and ending with the last peak co-eluting with the internal standard. The TLC mycolic acid pattern reveals α+- and α-mycolates [1]. The fatty acids composition of the strain is dominated by straight-chain saturated acids such as the taxon-specific C10:0 (21.0%), C16:0 (18.5%), C14:0 (15.3%), 10-methyl-C18:0 (7.4%, tuberculostearic acid), C20:0 (4.9%), C12:0 (2.4%), C18:0 (1.9%), with some by straight-chain desaturated acids, C18:1 cis (15.1%) and C16:1ω9t (9.7%); (personal communication with R.M. Kroppenstedt). Quinones are mainly MK 8(H4) and MK 8(H2) with some MK 8(H6) and traces of MK 9(H2) (R.M. Kroppenstedt, personal communication).
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position, and is part of the Genomic Encyclopedia of Bacteria and Archaea project [18]. The genome project is deposited in the Genome OnLine Database [9] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Two genomic 454 libraries: one standard and one 4kb PE; one Illumina shotgun library |
| MIGS-29 | Sequencing platforms | 454 GS FLX Titanium, Illumina GAii |
| MIGS-31.2 | Sequencing coverage | 58.1× 454 pyrosequence, 73.3× Illumina |
| MIGS-30 | Assemblers | Newbler version 12.0.1 PreRelease 3/30/2009.1.02.15, Velvet, phrap |
| MIGS-32 | Gene calling method | Prodigal |
| INSDC ID | CP001958 | |
| GenBank Date of Release | not yet | |
| GOLD ID | Gc01232 | |
| NCBI project ID | 37711 | |
| Database: IMG-GEBA | 2502422312 | |
| MIGS-13 | Source material identifier | DSM 44985 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
S. rotundus CDC 1076T, DSM 44985, was grown in DSMZ medium 645 (Middlebrook Medium) [19] at 28°C. DNA was isolated from 1-1.5 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany) with lysis modification LALMP according to Wu et al. [18].
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 technologies [20]. An Illumina GAii shotgun library with reads of 443 Mb, a 454 Titanium draft library with average read length of 304 bases, and a paired-end 454 library with average insert size of 4 Kb were generated for this genome. All general aspects of library construction and sequencing can be found at http://www.jgi.doe.gov/. Illumina sequencing data was assembled with VELVET [21] and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. Draft assemblies were based on 183 Mb 454 data, and 454 paired-end data. Newbler parameters are -consed -a 50 -l 350 -g -m -ml 20. The initial assembly contained 26 contigs in one scaffold. We converted the initial 454 assembly into a phrap assembly by making fake reads from the consensus, collecting the read pairs in the 454 paired-end library. The Phred/Phrap/Consed software package (www.phrap.com) was used for sequence assembly and quality assessment [18] in the following finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolution (unpublished, http://www.jgi.doe.gov/), Dupfinisher [22], or sequencing cloned bridging PCR fragments with subcloning or transposon bombing (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR (J-F Cheng, unpublished) primer walks. A total of 108 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. The completed genome sequences had an error rate less than one in 100,000 bp.
Genome annotation
Genes were identified using Prodigal [23] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [24]. The predicted CDSs were translated and used to search the National Center for Biotechnology In-formation (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and manual functional annotation was performed within the Integrated Microbial Genomes Expert Review (IMG-ER) platform [25].
Genome properties
The genome consists of a 3,157,527 bp long chromosome (Table 3 and Figure 3). Of the 3,133 genes predicted, 3,081 were protein-coding genes, and 52 RNAs; 75 pseudogenes were also identified. The majority of the protein-coding genes (63.0%) were assigned with a putative function while those remaining were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,157,527 | 100.00% |
| DNA coding region (bp) | 2,914,227 | 92.29% |
| DNA G+C content (bp) | 2,108,953 | 66.79% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 3,133 | 100.00% |
| RNA genes | 52 | 1.66% |
| rRNA operons | 1 | |
| Protein-coding genes | 3,081 | 98.34% |
| Pseudo genes | 75 | 2.39% |
| Genes with function prediction | 1,974 | 63.01% |
| Genes in paralog clusters | 442 | 14.11% |
| Genes assigned to COGs | 1,861 | 59.40% |
| Genes assigned Pfam domains | 2,097 | 66.93% |
| Genes with signal peptides | 848 | 27.07% |
| Genes with transmembrane helices | 671 | 21.42% |
| CRISPR repeats | 0 |
Figure 3.
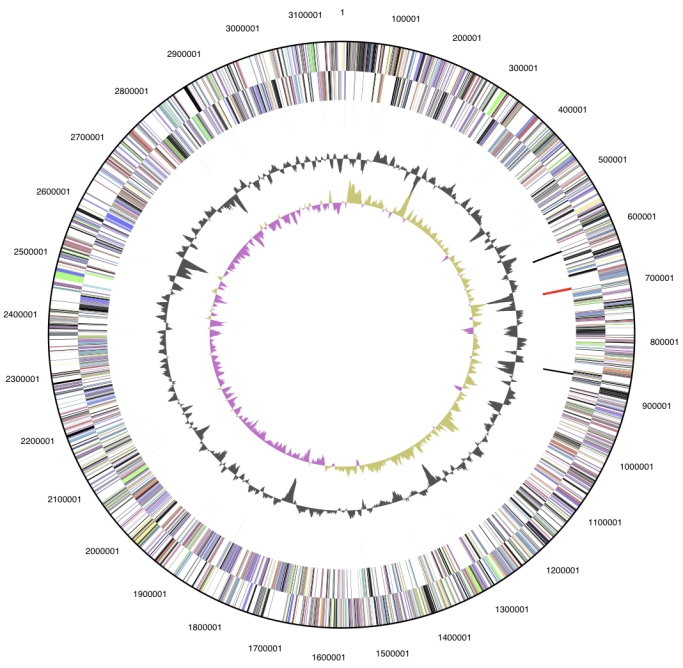
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 134 | 4.3 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.0 | RNA processing and modification |
| K | 126 | 4.1 | Transcription |
| L | 114 | 3.7 | Replication, recombination and repair |
| B | 0 | 0.0 | Chromatin structure and dynamics |
| D | 22 | 0.7 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 20 | 0.7 | Defense mechanisms |
| T | 58 | 1.9 | Signal transduction mechanisms |
| M | 97 | 3.1 | Cell wall/membrane biogenesis |
| N | 4 | 0.1 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 23 | 0.7 | Intracellular trafficking, secretion, and vesicular transport |
| O | 82 | 2.7 | Posttranslational modification, protein turnover, chaperones |
| C | 141 | 4.6 | Energy production and conversion |
| G | 125 | 4.1 | Carbohydrate transport and metabolism |
| E | 209 | 6.8 | Amino acid transport and metabolism |
| F | 77 | 2.5 | Nucleotide transport and metabolism |
| H | 116 | 3.8 | Coenzyme transport and metabolism |
| I | 117 | 3.8 | Lipid transport and metabolism |
| P | 103 | 3.3 | Inorganic ion transport and metabolism |
| Q | 85 | 2.8 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 247 | 8.0 | General function prediction only |
| S | 149 | 4.8 | Function unknown |
| - | 1,272 | 41.3 | Not in COGs |
Acknowledgements
This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-1 and SI 1352/1-2.
References
- 1.Butler WR, Floyd MM, Brown JM, Toney SR, Daneshvar MI, Cooksey RC, Carr J, Steigerwalt AG, Charles N. Novel mycolic acid-containing bacteria in the family Segniliparaceae fam. nov., including the genus Segniliparus gen. nov., with descriptions of Segniliparus rotundus sp. nov. and Segniliparus rugosus sp. nov. Int J Syst Evol Microbiol 2005; 55:1615-1624 10.1099/ijs.0.63465-0 [DOI] [PubMed] [Google Scholar]
- 2.Butler WR, Sheils CA, Brown-Elliott BA, Charles N, Colin AA, Gant MJ, Goodill J, Hindman D, Toney SR, Wallace RJ, Jr, et al. First Isolations of Segniliparus rugosus from patients with cystic fibrosis. J Clin Microbiol 2007; 45:3449-3452 10.1128/JCM.00765-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen T, Van Kerckhof J, Jelfs P, Wainwright C, Ryan P, Coulter C. Segniliparus rugosus infection, Australia. Emerg Infect Dis 2009; 15:611-613 10.3201/eid1504.081479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 2007; 57:2259-2261 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- 5.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 6.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 7.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 8.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [PubMed] [Google Scholar]
- 9.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346-D354 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanova N, Sikorski J, Jando M, Lapidus A, Nolan M, Lucas S, Glavina Del Rio T, Tice H, Copeland A, Cheng JF, et al. Complete genome sequence of Gordonia bronchialis type strain (3410T). Stand Genomic Sci 2010; 2:19-28 10.4056/sigs.611106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998; 393:537-544 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 12.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 15.Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 1997; 47:479-491; . 10.1099/00207713-47-2-479 [DOI] [Google Scholar]
- 16.Classification of bacteria and archaea in risk groups. www.baua.de TRBA 466.
- 17.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova N, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/media_list.php
- 20.Bennett S. Solexa Ltd. Pharmacogenomics 2004; 5:433-438 10.1517/14622416.5.4.433 [DOI] [PubMed] [Google Scholar]
- 21.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sims D, Brettin T, Detter J, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Genomics 2010; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. Nat Methods (In press). [DOI] [PubMed] [Google Scholar]
- 25.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]