Abstract
Aminobacterium colombiense Baena et al. 1999 is the type species of the genus Aminobacterium. This genus is of large interest because of its isolated phylogenetic location in the family Synergistaceae, its strictly anaerobic lifestyle, and its ability to grow by fermentation of a limited range of amino acids but not carbohydrates. Here we describe the features of this organism, together with the complete genome sequence and annotation. This is the second completed genome sequence of a member of the family Synergistaceae and the first genome sequence of a member of the genus Aminobacterium. The 1,980,592 bp long genome with its 1,914 protein-coding and 56 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: strictly anaerobic, fermentation of amino acids, gram-negative firmicute, syntrophic organism, Synergistaceae, GEBA
Introduction
Strain ALA-1T (= DSM 12261) is the type strain of the species Aminobacterium colombiense, which is the type species of the genus Aminobacterium [1,2]. The name of the genus relates to its ability to ferment amino acids and the species name refers to origin of the isolate, Columbia [1]. Currently, the genus Aminobacterium consists of only two species [1,3,4]. Strain ALA-1T has been isolated from an anaerobic dairy wastewater lagoon in 1998 or before [1]. At the moment, strain ALA-1T is the only known isolate of this species. Highly similar (98%) nearly complete (>1,400 bp) uncultured 16S gene clone sequences were frequently obtained from anaerobic habitats, e.g., from anaerobic municipal solid waste samples in France [5], from a biogas fermentation enrichment culture in China (GU476615), from a swine wastewater anaerobic digestion in a UASB reactor in China (FJ535518), and from a mesophilic anaerobic BSA digester in Japan [6], suggesting quite a substantial contribution of Aminobacterium to anaerobic prokaryotic communities. The type strain of the only other species in the genus, A. mobile [3] shares 95% 16S rRNA sequence identity with A. colombiense, whereas the type strains of the other species in the family Synergistaceae share between 84.3 and 88.3% 16S rRNA sequence identity [7]. Environmental samples and metagenomic surveys detected only one significantly similar phylotype (BABF01000111, 92% sequence similarity) in a human gut microbiome [7], with all other phylotypes sharing less than 84% 16S rRNA gene sequence identity, indicating a rather limited general ecological importance of the members of the genus Aminobacterium (status April 2010). Here we present a summary classification and a set of features for A. colombiense ALA-1T, together with the description of the complete genomic sequencing and annotation.
Classification and features
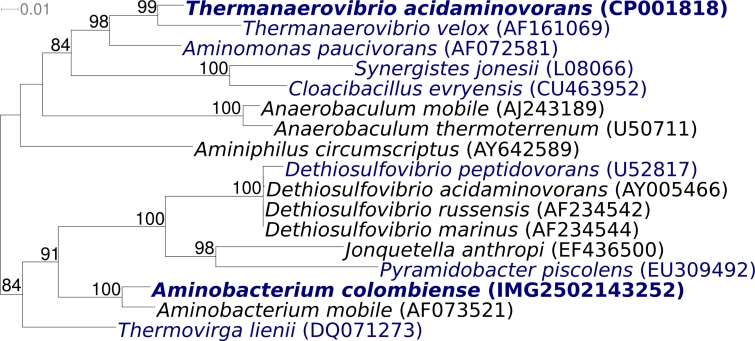
Figure 1 shows the phylogenetic neighborhood of A. colombiense ALA-1T in a 16S rRNA based tree. The sequences of the three identical copies of the 16S rRNA gene in the genome differ by 14 nucleotides (0.9%) from the previously published 16S rRNA sequence generated from DSM 12661 (AF069287). which contains 3 ambiguous base calls. These differences are most likely due to sequencing errors in AF069287.
Figure 1.
Phylogenetic tree highlighting the position of A. colombiense ALA-1T relative to the other type strains within the phylum Synergistetes. The tree was inferred from 1,282 aligned characters [8,9] of the 16S rRNA gene sequence under the maximum likelihood criterion [10] and rooted in accordance with the current taxonomy [11]. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 250 bootstrap replicates [12] if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [13] are shown in blue, published genomes in bold, e.g. the recently published GEBA genome of Thermanaerovibrio acidaminovorans [14].
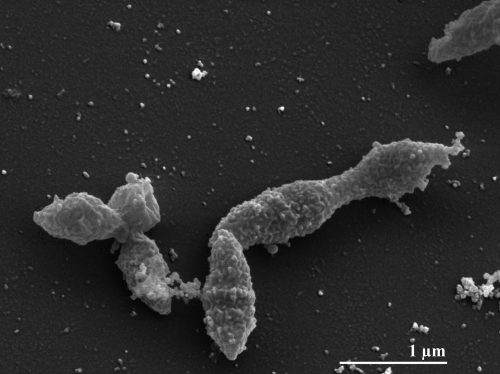
The cells are rod-like, occasionally slightly curved with 3-4 µm in length and 0.2-0.3 µm in width (Figure 2 and Table 1) [1]. The colonies are up to 1.0 mm in diameter and are round, smooth, lens-shaped, and white [1]. Strain ALA-1T requires yeast extract for growth and ferments serine, glycine, threonine, and pyruvate in its presence [1]. Poor growth is obtained on casamino acids, peptone, biotrypcase, cysteine and α-ketoglutarate [1]. The fermentation and end-products include acetate and H2, and also propionate in the case of α-ketoglutarate fermentation. Carbohydrates (such as glucose, saccharose, ribose, xylose, cellobiose, mellobiose, maltose, galactose, mannose, arabinose, rhamnose, lactose, sorbose and mannitol), gelatin, casein, glycerol, ethanol, acetate, propionate, butyrate, lactate, citrate, fumarate, malate, succinate and the other amino acids tested are not utilized [1].
Figure 2.
Scanning electron micrograph of A. colombiense ALA-1T
Table 1. Classification and general features of A. colombiense ALA-1T according to the MIGS recommendations [15].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [16] | |
| Phylum Synergistetes | TAS [17] | ||
| Class Synergistia | TAS [17] | ||
| Order Synergistales | TAS [17] | ||
| Family Synergistaceae | TAS [17] | ||
| Genus Aminobacterium | TAS [1,18] | ||
| Species Aminobacterium colombiense | TAS [1,18] | ||
| Type strain ALA-1 | TAS [1] | ||
| Gram stain | negative | TAS [1] | |
| Cell shape | slightly curved to rod shaped | TAS [1] | |
| Motility | nonmotile | TAS [1] | |
| Sporulation | non-sporulating | TAS [1] | |
| Temperature range | mesophile, 20°C – 42°C, no growth at 18°C and 45°C |
TAS [1] | |
| Optimum temperature | 37 °C | TAS [1] | |
| Salinity | no NaCl required, tolerates less than 1.5% NaCl |
TAS [1] | |
| MIGS-22 | Oxygen requirement | strictly anaerobic | TAS [1] |
| Carbon source | serine, threonine, glycine and pyruvate, not carbohydrates |
TAS [1] | |
| Energy source | serine, threonine, glycine and pyruvate, not carbohydrates |
TAS [1] | |
| MIGS-6 | Habitat | anaerobic sludge | TAS [1] |
| MIGS-15 | Biotic relationship | free-living | TAS [1] |
| MIGS-14 | Pathogenicity | pathogenicity is not reported | NAS |
| Biosafety level | 1 | TAS [19] | |
| Isolation | anaerobic dairy wastewater lagoon | TAS [1] | |
| MIGS-4 | Geographic location | Santa Fe de Bogota, Colombia | TAS [1] |
| MIGS-5 | Sample collection time | 1998 or before | TAS [1] |
| MIGS-4.1 MIGS-4.2 |
Latitude Longitude |
4.63 -74.08 |
NAS |
| MIGS-4.3 | Depth | unknown | |
| MIGS-4.4 | Altitude | about 2,640 m | NAS |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [20]. If the evidence code is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
As typical for anoxic habitats, strain ALA-1T is engaged in syntrophic interactions: alanine, glutamate, valine, isoleucine, leucine, methionine, aspartate and malate are oxidized only in the presence of the hydrogenotroph, Methanobacterium formicicum, strain DSM 1525 [1]. In addition, the utilization of cysteine, threonine and α-ketoglutarate are also improved in the presence of M. formicicum [1]. An 80% hydrogen atmosphere (supplied as H2-CO, (80:20) at 2 bar pressure) inhibits growth of strain ALA-1T on threonine and α-ketoglutarate, whereas glycine degradation is not affected [1]. Serine and pyruvate degradation are partially affected by the presence of hydrogen. Sulfate, thiosulfate, elemental sulfur, sulfite, nitrate, and fumarate are not utilized as electron acceptors [1]. Strain ALA-1T does not perform the Stickland reaction when alanine is provided as an electron donor and glycine, serine, arginine or proline are provided as electron acceptor.
As noted above, alanine is oxidized only in the presence of the hydrogenotroph M. formicicum, which utilizes the produced H2 [1]. In the absence of an H2-consuming organism, the H2 partial pressure would rapidly reach a level that thermodynamically inhibits further fermentation [21]. Adams and colleagues used a H2-purging culture vessel to replace the H2-consuming syntrophic partner, in order to study in detail the energetic characteristics of alanine consumption of strain ALA-1T in a pure culture [21].
Strain ALA-1T is non-motile [1], whereas interestingly the other species in the genus, A. mobile, is motile by means of lateral flagella [3]. A parallel situation is in the genus Anaerobaculum (Figure 1), where A. thermoterrenum is non-motile [22] but A. mobile is motile by means of lateral flagella [23]. In fact, the phenotype of non-motility versus motility by means of lateral flagella is heterogeneously distributed among the organisms depicted in Figure 1. This may suggest that the last common ancestor of the group shown in Figure 1 was motile by flagella and that the selection pressure for a functioning flagella might be currently more relaxed in this group, leading in individual strains to mutational inactivation of the flagella. Interestingly, the annotation of the genome does not give any indication of the presence of any genes related to flagellar assembly. The only genes related to cellular motility refer to type II secretory pathway and to pilus assembly. This is surprising, as it is hardly probable that strain ALA-1T lost all genes for flagellar assembly after the evolutionary separation of strain ALA-1T and its closely related sister species A. mobile from their last common ancestor. A similar situation has been observed in the non-motile strain Alicyclobacillus acidocaldarius 104-IAT in comparison to several motile sister species in the genus Alicyclobacillus [24]. Here, the genome of the non-motile strain A. acidocaldarius 104-IAT still contains most of the genes needed for flagellar assembly [24]. Thus, the genotypic status of flagellar motility in the genus Aminobacterium remains unclear.
Chemotaxonomy
Ultrathin sections of strain ALA-1T revealed a thick cell wall with an external S-layer similar to that of Gram-positive type cell walls [1]. Unfortunately, no chemotaxonomic data have been published for the genus Aminobacterium. Among the organisms depicted in Figure 1, chemotaxonomic data are available for Dethiosulfovibrio peptidovorans, Jonquetella anthropi, Pyramidobacter piscolens, Cloacibacillus evryensis, and Synergistes jonesii, though the data are not always present in the original species description publications [25-27]. In major phenotypes, such as being strictly anaerobic and Gram-negative in staining within the usually Gram-positive Firmicutes, mostly also in their ability to degrade amino acids, the organisms shown in Figure 1 are highly similar, which may justify also a comparison in their chemotaxonomic features. The major fatty acids in different strains of Jonquetella are iso-C15:0 (25-43%) and C16:0 (14-21%), other iso-branched and unbranched fatty acids are present in smaller amounts, and anteiso-C15:0 is below 5% [26]. In Dethiosulfovibrio, the major fatty acid is iso-C15:0 (59.7%), followed by C18:0 (9.0%) and C16:0 (8.5%) [26]. Dethiosulfovibrio differs qualitatively from Jonquetella by the absence of anteiso branched fatty acids and by the presence of C18:1ω9c (3.0%) [26]. The major fatty acids in two strains of P. piscolens are C14:0 (16-19%) and C13:0 (12-14%) [27]. The cellular fatty acids of C. evryensis are characterized by a mixture of saturated, unsaturated, hydroxy- and cyclopropane fatty acids [25]. The major fatty acids were iso-C15:0 (16.6%), iso-C15:0 3-OH (12.4%) and C17:1ω6c (9.5%) [25]; the major fatty acids in its closest relative, Synergistes jonesii, were C15:0 (16.0%), C20cyc (14:0) and C17:1ω6c (9.0%) [25]. The polar fatty acid profile of C. evryensis (data not shown in the original publication) revealed diphosphatidylglycerol, phosphatidylglycerol, phosphatidyl-ethanolamine and phosphatidylmonomethylamine [25].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [28], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [29]. The genome project is deposited in the Genome OnLine Database [13] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Three genomic libraries: one 454 pyrosequence standard library; one 454 12kb pyrosequence library; one Illumina 250bp library |
| MIGS-29 | Sequencing platforms | 454 GS FLX Titanium; Illumina GAii |
| MIGS-31.2 | Sequencing coverage | 85.5× 454 pyrosequence; 909 Mb Illumina data |
| MIGS-30 | Assemblers | Newbler version 2.0.0-PostRelease-10/28/2008, phrap |
| MIGS-32 | Gene calling method | Prodigal, GenePRIMP |
| INSDC ID | CP001997 | |
| Genbank Date of Release | April 5, 2010 | |
| GOLD ID | Gc01257 | |
| NCBI project ID | 32587 | |
| Database: IMG-GEBA | 2502082107 | |
| MIGS-13 | Source material identifier | DSM 12261 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
A. colombiense ALA-1T, DSM 12661, was grown anaerobically in DSMZ medium 846 (Anaerobic serine/arginine medium) [30] at 37°C. DNA was isolated from 1-1.5 g of cell paste using MasterPure Gram Positive DNA Purification Kit (Epicentre MGP04100) adding additional 1µl lysozyme and 5 µl mutanolysin to the standard lysis solution for 40 min incubation at 37°C.
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 technologies. An Illumina GAii shotgun library with reads of 909 Mb, a 454 Titanium draft library with average read length of 283 bases, and a paired end 454 library with average insert size of 12 kb were generated for this genome. All general aspects of library construction and sequencing can be found at http://www.jgi.doe.gov/. Draft assemblies were based on 169 Mb 454 draft data and 454 paired end data (543,550 reads). Newbler (version 2.0.0-PostRelease-10/28/2008 was used) parameters are -consed -a 50 -l 350 -g -m -ml 20. The initial Newbler assembly contained 18 contigs in 1 scaffold. The initial 454 assembly was converted into a phrap assembly by making fake reads from the consensus, collecting the read pairs in the 454 paired end library. Illumina sequencing data was assembled with VELVET [31], and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. The Phred/Phrap/Consed software package (www.phrap.com) was used for sequence assembly and quality assessment in the following finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolution (http://www.jgi.doe.gov/), Dupfinisher, or sequencing cloned bridging PCR fragments with subcloning or transposon bombing [32]. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR primer walks (J-F.Cheng, unpublished). A total of 113 additional Sanger reactions were necessary to close gaps and to raise the quality of the finished sequence. The error rate of the completed genome sequence is less than 1 in 100,000.
Genome annotation
Genes were identified using Prodigal [33] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [34]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [35].
Genome properties
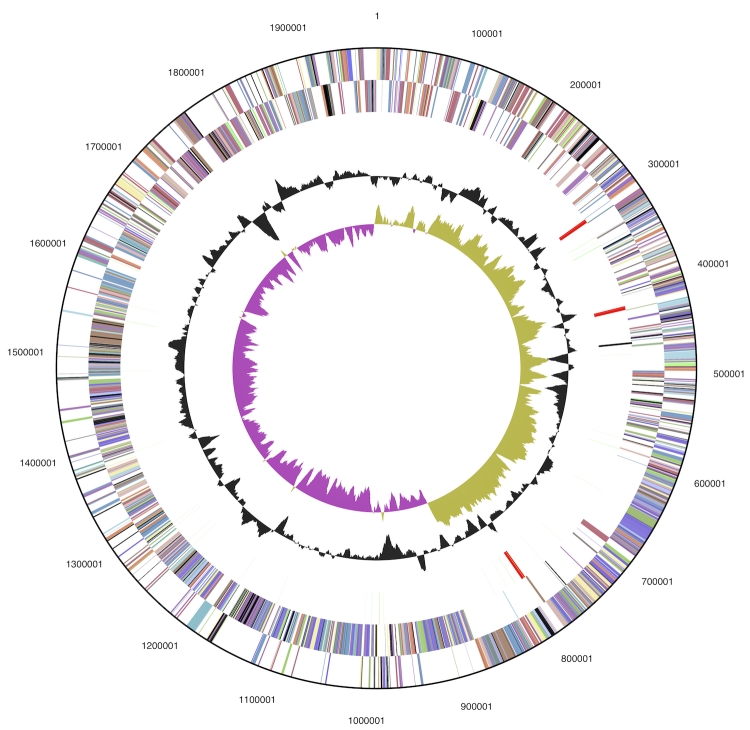
The genome consists of a 1,980,592 bp long chromosome with an overall GC content of 45.3% (Table 3 and Figure 3). Of the 1,970 genes predicted, 1,914 were protein-coding genes, and 56 RNAs; 38 pseudogenes were also identified. The majority of the protein-coding genes (77.2%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 1,980,592 | 100.00% |
| DNA coding region (bp) | 1,837,142 | 92.76% |
| DNA G+C content (bp) | 897,344 | 45.31% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 1,970 | 100.00% |
| RNA genes | 56 | 2.84% |
| rRNA operons | 3 | |
| Protein-coding genes | 1,914 | 97.16% |
| Pseudo genes | 38 | 1.93% |
| Genes with function prediction | 1,521 | 77.21% |
| Genes in paralog clusters | 225 | 12.94% |
| Genes assigned to COGs | 1,592 | 80.81% |
| Genes assigned Pfam domains | 1,617 | 82.08% |
| Genes with signal peptides | 337 | 17.11% |
| Genes with transmembrane helices | 540 | 27.41% |
| CRISPR repeats | 1 |
Figure 3.
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 150 | 8.8 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 105 | 6.1 | Transcription |
| L | 81 | 4.7 | Replication, recombination and repair |
| B | 1 | 0.1 | Chromatin structure and dynamics |
| D | 23 | 1.3 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 23 | 1.3 | Defense mechanisms |
| T | 51 | 3.0 | Signal transduction mechanisms |
| M | 106 | 6.2 | Cell wall/membrane biogenesis |
| N | 5 | 0.3 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 32 | 1.9 | Intracellular trafficking, secretion, and vesicular transport |
| O | 57 | 3.3 | Posttranslational modification, protein turnover, chaperones |
| C | 129 | 7.5 | Energy production and conversion |
| G | 118 | 6.9 | Carbohydrate transport and metabolism |
| E | 199 | 11.6 | Amino acid transport and metabolism |
| F | 66 | 3.9 | Nucleotide transport and metabolism |
| H | 67 | 3.9 | Coenzyme transport and metabolism |
| I | 42 | 2.5 | Lipid transport and metabolism |
| P | 98 | 5.7 | Inorganic ion transport and metabolism |
| Q | 26 | 1.5 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 207 | 12.1 | General function prediction only |
| S | 126 | 7.4 | Function unknown |
| - | 378 | 19.2 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Maren Schröder (DSMZ) for growing A. colombiense cells. This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, and UT-Battelle Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-2 and SI 1352/1-2.
References
- 1.Baena S, Fardeau ML, Labat M, Ollivier B, Thomas P, Garcia JL, Patel BKC. Aminobacterium colombiense gen. nov. sp. nov., an amino acid-degrading anaerobe isolated from anaerobic sludge. Anaerobe 1998; 4:241-250 10.1006/anae.1998.0170 [DOI] [PubMed] [Google Scholar]
- 2.Validation of publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1999; 49:1325-1326 10.1099/00207713-49-4-1325 [DOI] [Google Scholar]
- 3.Baena S, Fardeau ML, Labat M, Ollivier B, Garcia JL, Patel BKC. Aminobacterium mobile sp. nov., a new anaerobic amino-acid-degrading bacterium. Int J Syst Evol Microbiol 2000; 50:259-264 [DOI] [PubMed] [Google Scholar]
- 4.Euzéby JP. List of bacterial names with standing in nomenclature: A folder available on the Internet. Int J Syst Bacteriol 1997; 47:590-592 10.1099/00207713-47-2-590 [DOI] [PubMed] [Google Scholar]
- 5.Li T, Mazéas L, Sghir A, Leblon G, Bouchez T. Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ Microbiol 2009; 11:889-904 10.1111/j.1462-2920.2008.01810.x [DOI] [PubMed] [Google Scholar]
- 6.Tang Y, Shigematsu T, Morimura S, Kida K. Microbial community analysis of mesophilic anaerobic protein degradation process using bovine serum albumin (BSA)-fed continuous cultivation. J Biosci Bioeng 2005; 99:150-164 10.1263/jbb.99.150 [DOI] [PubMed] [Google Scholar]
- 7.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 2007; 57:2259-2261 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- 8.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 9.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 10.Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 11.Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 12.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [PubMed] [Google Scholar]
- 13.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346-D354 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chovatia M, Sikorski J, Schröder M, Lapidus A, Nolan M, Tice H, Glavina Del Rio T, Copeland A, Cheng JF, Chen F, et al. Complete genome sequence of Thermanaerovibrio acidaminovorans type strain (SU883T). Stand Genomic Sci 2009; 1:254-261 10.4056/sigs.40645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jumas-Bilak E, Roudière L, Marchandin H. Description of 'Synergistetes' phyl. nov. and emended description of the phylum 'Deferribacteres' and of the family Syntrophomonadaceae, phylum 'Firmicutes'. Int J Syst Evol Microbiol 2009; 59:1028-1035 10.1099/ijs.0.006718-0 [DOI] [PubMed] [Google Scholar]
- 18.Validation List No 71. Validation and publication of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 1999; 49:1325-1326 [Google Scholar]
- 19.Classification of Bacteria and Archaea in risk groups. www.baua.de TRBA 466.
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams CJ, Redmond MC, Valentine DL. Pure-culture growth of fermentative bacteria, facilitated by H2 removal: bioenergetics and H2 production. Appl Environ Microbiol 2006; 72:1079-1085 10.1128/AEM.72.2.1079-1085.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rees GN, Patel BKC, Grassia GS, Sheehy AJ. Anaerobaculum thermoterrenum gen. nov., sp. nov., a novel, thermophilic bacterium which ferments citrate. Int J Syst Bacteriol 1997; 47:150-154 10.1099/00207713-47-1-150 [DOI] [PubMed] [Google Scholar]
- 23.Menes RJ, Muxi L. Anaerobaculum mobile sp. nov., a novel anaerobic, moderately thermophilic, peptide-fermenting bacterium that uses crotonate as an electron acceptor, and emended description of the genus Anaerobaculum. Int J Syst Evol Microbiol 2002; 52:157-164 [DOI] [PubMed] [Google Scholar]
- 24.Mavromatis K, Sikorski J, Lapidus A, Rio TGD, Copeland A, Tice H, Cheng JF, Lucas S, Chen F, Nolan M, et al. Complete genome sequence of Alicyclobacillus acidocaldarius type strain (104-IAT). Stand Genomic Sci 2010; 2:9-18 10.4056/sigs.591104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganesan A, Chaussonnerie S, Tarrade A, Dauga C, Bouchez T, Pelletier E, Le Paslier D, Sghir A. Cloacibacillus evryensis gen. nov., sp. nov., a novel asaccharolytic, mesophilic, amino-acid-degrading bacterium within the phylum 'Synergistetes', isolated from an anaerobic sludge digester. Int J Syst Evol Microbiol 2008; 58:2003-2012 10.1099/ijs.0.65645-0 [DOI] [PubMed] [Google Scholar]
- 26.Jumas-Bilak E, Carlier JP, Jean-Pierre H, Citron D, Bernard K, Damay A, Gay B, Teyssier C, Campos J, Marchandin H. Jonquetella anthropi gen. nov., sp. nov., the first member of the candidate phylum 'Synergistetes' isolated from man. Int J Syst Evol Microbiol 2007; 57:2743-2748 10.1099/ijs.0.65213-0 [DOI] [PubMed] [Google Scholar]
- 27.Downes J, Vartoukian SR, Dewhirst FE, Izard J, Chen T, Yu WH, Sutcliffe IC, Wade WG. Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum 'Synergistetes' isolated from the human oral cavity. Int J Syst Evol Microbiol 2009; 59:972-980 10.1099/ijs.0.000364-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yarza P, Richter M, Peplies J, Euzeby JP, Amann R, Schleifer KH, Ludwig W, Glöckner FO, Rossello-Mora R. The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 2008; 31:241-250 10.1016/j.syapm.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 29.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/media_list.php
- 31.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sims D, Brettin T, Detter J, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 35.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]