Abstract
Methanothermus fervidus Stetter 1982 is the type strain of the genus Methanothermus. This hyperthermophilic genus is of a thought to be endemic in Icelandic hot springs. M. fervidus was not only the first characterized organism with a maximal growth temperature (97°C) close to the boiling point of water, but also the first archaeon in which a detailed functional analysis of its histone protein was reported and the first one in which the function of 2,3-cyclodiphosphoglycerate in thermoadaptation was characterized. Strain V24ST is of interest because of its very low substrate ranges, it grows only on H2 + CO2. This is the first completed genome sequence of the family Methanothermaceae. Here we describe the features of this organism, together with the complete genome sequence and annotation. The 1,243,342 bp long genome with its 1,311 protein-coding and 50 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: hyperthermophile, strictly anaerobic, motile, Gram-positive, chemolithoautotroph, Methanothermaceae, Euryarchaeota, GEBA
Introduction
Strain V24ST (= DSM 2088 = ATCC 43054 = JCM 10308) is the type strain of Methanothermus fervidus [1]. Together with M. sociabilis, there are currently two species placed in the genus Methanothermus. The strain V24ST was isolated from an anaerobic Icelandic spring [1,2] and M. sociabilis from a continental solfatara field in Iceland [2]. Since any attempt to isolate Methanothermus from similar places (Italy, the Azores, Yellowstone National Park) was without success, Lauerer et al. (1986) have speculated that strains of Methanothermus may exist endemically within Iceland [2]. The genus name derives from the Latin word “methanum”, methane, and from the Greek adjective “therme”, meaning heat, which refers to a methane producing organism living in a hot niche [1]. The species epithet fervidus comes from the Latin adjective “fervidus”, glowing hot, burning, fervent, because of its growth in almost-boiling water [1]. No further cultivated strains belonging to the species M. fervidus have been described so far. Here we present a summary classification and a set of features for M. fervidus strain V24ST, together with the description of the complete genomic sequencing and annotation.
Classification and features
The original 16S rRNA gene sequence of strain V24ST (M59145) shows 92% sequence identity with the 16S rRNA gene of M. sociabilis (AF095273) [2] (Figure 1) and 88% identity with an uncultured clone, NRA12 (HM041913). The highest sequence similarities of the strain V24ST 16S rRNA to metagenomic libraries (env_nt) were 87% or less (status August 2010), indicating that members of the species, genus and even family are poorly represented in the habitats screened so far. The 16S rRNA gene sequence of strain V24ST was compared with the most recent release of the Greengenes database using BLAST [13] and the relative frequencies weighted by BLAST scores, of taxa and keywords within the 250 best hits were determined. The five most frequent genera were Methanobacterium (55.3%), Methanothermobacter (23.5%), Methanobrevibacter (12.8%), Methanothermus (5.7%) and Thermococcus (1.7%). The five most frequent keywords within the labels of environmental samples which yielded hits were 'anaerobic' (7.1%), 'sludge' (4.7%), 'microbial' (3.7%), 'archaeal' (3.5%) and 'temperature' (3.4%). Besides 'sludge', these keywords fit well to what is known from the taxonomy, ecology, and physiology of strain V24ST. Environmental samples which yielded hits of a higher score than the highest scoring species were not found. The genome of M. fervidus contains two rRNA operons. One of these operons has a closely linked 7S RNA gene, encoding the RNA component of signal recognition particle [14].
Figure 1.
Phylogenetic tree highlighting the position of M. fervidus V24ST relative to the other type strains within the family Methanothermaceae. The tree was inferred from 1,249 aligned characters [3,4] of the 16S rRNA gene sequence under the maximum likelihood criterion [5] and rooted in accordance with the current taxonomy [6]. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are *support values from 100 bootstrap replicates [7] if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [8] are shown in blue, published genomes in bold [9-12].
Figure 1 shows the phylogenetic neighborhood of M. fervidus V24ST in a 16S rRNA based tree. The sequences of the two 16S rRNA gene copies in the genome of Methanothermus fervidus DSM 2088 differ from each other by up to four nucleotides, and differ by up to 17 nucleotides from the previously published 16S rRNA sequence (M59145), which contains 87 ambiguous base calls.
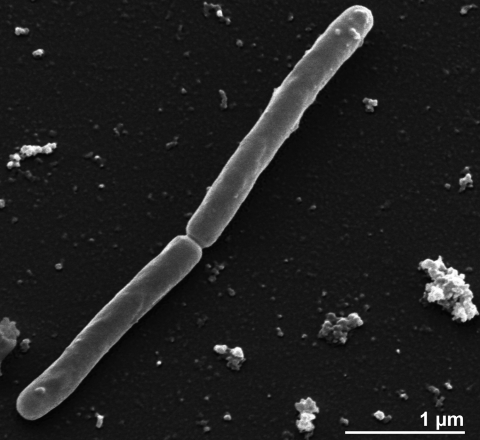
Although the cells of the strain V24ST do not contain a typical bacterial peptidoglycan, they stain Gram-positive. Cells are curved rods, 1-3 µm long and 0.3 - 0.4 µm in width (Figure 2 and Table 1), occurring singly and in pairs, with a doubling time of 170 minutes [1]. Round, smooth, opaque, and slightly grayish colonies of 1 to 3 mm in diameter were observed on modified MM-medium plates containing trace amounts of solid sodium dithionite, sodium silicate solution and resazurin [1]. Strain V24ST is strictly anaerobic and strictly autotrophic [26]. Due to the low melting point of agar, strain V24ST could do not be grown on agar. Cells did not grow at temperatures below 61°C or above 97°C; the optimal temperature was 83°C [1]. Growth occurs at a slightly acidic pH and equal to 6.5, while no growth could be observed at pH above 7.0 [1]. In comparison, M. sociabilis grows at the temperatures ranged between 65°C and 97°C, with the optimal temperature at 88°C, its pH for growth being acidic to neutral (pH 5.5 to 7.5) [27]. Strain V24ST produces methane from H2 + CO2, whereas acetate and formate are not used [1,27]. The addition of 2-mercaptoethanesulfonic acid (coenzyme M) enhances growth, especially when small inoculates are used [1]. In artificial medium, yeast extract is required as an organic factor for growth [1]. Strain V24ST gains energy by oxidizing H2 to reduce CO2 as the terminal electron acceptor [26,28]. At the time of isolation, strain V24ST was described to be nonmotile [1]. Later, strain V24ST as well as M. sociabilis were described to be motile via bipolar peritrichous ‘flagella’, which was taken to indicate motility [28]. These cell surface appendages, however, were recently determined to have a diameter of 5-6 nm, and therefore, very probably, represent not organelles used for motility, but for adhesion [R Wirth et al., unpublished]. The genome does not contain any flagellar genes. M. fervidus produces large intracellular potassium concentrations and amounts of 2,3-cyclic diphosphoglycerate, which are both thought to be involved in the thermoadaptation of M. fervidus [29,30]. Moreover, the DNA-binding protein HMf (histone M. fervidus), which binds to double stranded DNA molecules and increases their resistance to thermal denaturation, has been of interest in M. fervidus [31]. A partial amino acid sequence analysis of the D-glyceraldehyde-3-phosphate dehydrogenase of M. fervidus shows high sequence similarity to the enzymes from eubacteria and from the cytoplasm of eukaryotes [32]. This enzyme reacts with both NAD+ and NADP+ [32,33] and is not inhibited by pentalenolactone [32]. However, the enzyme activity is low at temperatures below 40°C, but it is intrinsically stable only up to 75°C [32], which is interesting as growth of M. fervidus may occur up to 97°C [1]. Also, the biochemistry of triose-phosphate isomerase, which catalyzes the interconversion of dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (GAP) in the reversible Embden-Meyerhof-Parnas (EMP) pathway, has been studied to some detail in M. fervidus [34].
Figure 2.
Scanning electron micrograph of M. fervidus V24ST
Table 1. Classification and general features of M. fervidus V24ST according to the MIGS recommendations [15].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Archaea | TAS [16] | |
| Phylum Euryarchaeota | TAS [17,18] | ||
| Class Methanobacteria | TAS [18,19] | ||
| Order Methanobacteriales | TAS [20-22] | ||
| Family Methanothermaceae | TAS [1,23] | ||
| Genus Methanothermus | TAS [1,23] | ||
| Species Methanothermus fervidus | TAS [1,23] | ||
| Type strain V24S | TAS [1] | ||
| Gram stain | positive | TAS [1] | |
| Cell shape | straight to curved, single and in pair rods | TAS [1] | |
| Motility | non-motile | TAS [1] | |
| Sporulation | not reported | NAS | |
| Temperature range | 61°C–97°C | TAS [1] | |
| Optimum temperature | 83°C | TAS [1] | |
| Salinity | not reported | NAS | |
| MIGS-22 | Oxygen requirement | strict anaerobic | TAS [1] |
| Carbon source | CO2 | TAS [1] | |
| Energy source | H2 + CO2 | TAS [1] | |
| MIGS-6 | Habitat | solfataric fields | TAS [1] |
| MIGS-15 | Biotic relationship | not reported | NAS |
| MIGS-14 | Pathogenicity | no | NAS |
| Biosafety level | 1 | TAS [24] | |
| Isolation | Icelandic hot spring | TAS [1] | |
| MIGS-4 | Geographic location | Kerlingarfjöll mountains, Iceland | TAS [1] |
| MIGS-5 | Sample collection time | 1979 | NAS |
| MIGS-4.1 MIGS-4.2 |
Latitude Longitude |
64.65 19.25 |
NAS |
| MIGS-4.3 | Depth | surface | TAS [1] |
| MIGS-4.4 | Altitude | 1.477 m | NAS |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [25]. If the evidence code is IDA, then the property was directly observed by one of the authors or an expert mentioned in the acknowledgements
Chemotaxonomy
The cell envelope of the strain V24ST consists of a double-layer of pseudomurein and protein, while the cell wall contains pseudomurein consisting of N-acetyl-glucosamine, N-acetyl-galactosamine, N-talosaminuronic acid, glutamic acid, alanine, and lysine [1,2].
M. fervidus contains approximately 50% diethers, 25% diglycerol tetraethers and 25% of an unknown component moving slower than the tetraethers when examined by thin layer chromatography [2]. Here, M. fervidus differs from M. sociabilis, which lacks the unknown component while its diether and tetraethers were found at about equal proportions [2]. The diethers of M. fervidus contain only C20 phytanyl chains while the tetraethers include about 98-99% C40 biphytane and only some trace of C40 monocyclic biphytane [2]. Besides an unknown core lipid (FU, 31% of total core lipids, migrates slower than caldarchaeol by thin layer chromatography), other core lipids found in M. fervidus were caldarchaeol (60%), archaeol (4%) and others (5%) [35,36]. Interestingly, M. fervidus also differs from M. sociabilis regarding the glycolipid composition with four glycolipids and about equal proportions of five phospholipids and only three phospholipids for M. sociabilis [2].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [37], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [38]. The genome project is deposited in the Genome OnLine Database [8] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Three genomic libraries: one 454 pyrosequence standard library, one 454 PE library (17.8 kb insert size), one Illumina library |
| MIGS-29 | Sequencing platforms | Illumina GAii, 454 GS FLX |
| MIGS-31.2 | Sequencing coverage | 530 × Illumina; 75.0 × pyrosequence |
| MIGS-30 | Assemblers | Newbler version 2.0.00.20-PostRelease- 11-05-2008-gcc-3.4.6, phrap |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| INSDC ID | CP002278 | |
| Genbank Date of Release | November 5, 2010 | |
| GOLD ID | Gc01509 | |
| NCBI project ID | 33689 | |
| Database: IMG-GEBA | 2502422313 | |
| MIGS-13 | Source material identifier | DSM 2088 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
M. fervidus V24ST, DSM 2088, was grown anaerobically in culture vessels made of type III glass (alkali-rich soda lime glass) in DSMZ medium 203 (M. fervidus medium) [39] at 83°C. DNA was isolated from 0.5-1 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany) following the standard protocol as recommended by the manufacturer, with a modified cell lysis step. The modified lysis mixture contained only 100 µl lysozyme, but additional 58 µl achromopeptidase, lysostaphine, mutanolysin, each, for over night incubation at 35°C on a shaker. Proteinase K digestion was reduced to 200 µl for 1h 37°C.
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website [40]. Pyrosequencing reads were assembled using the Newbler assembler version 2.0.00.20-PostRelease-11-05-2008-gcc-3.4.6 (Roche). The initial Newbler assembly consisting of 24 contigs in one scaffold was converted into a phrap assembly [41] by making fake reads from the consensus, collecting the read pairs in the 454 paired end library. Illumina GAii sequencing data (636 Mb) was assembled with Velvet [42] and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. 454 Draft assembly was based on 96.5.0 Mb 454 draft data and all of the 454 paired end data. Newbler parameters are -consed -a 50 -l 350 -g -m -ml 20. The Phred/Phrap/Consed software package [41] was used for sequence assembly and quality assessment in the subsequent finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolution [40], Dupfinisher, or sequencing cloned bridging PCR fragments with subcloning or transposon bombing (Epicentre Biotechnologies, Madison, WI) [43]. Gaps between contigs were closed by editing in CONSED and additional sequencing reactions were necessary to close gaps and/or to raise the quality of the finished sequence. Illumina reads were also used to correct potential base errors and increase consensus quality using a software Polisher developed at JGI [44]. The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Illumina and 454 sequencing platforms provided 605 × coverage of the genome. The final assembly contained 267,328 pyrosequence and 17,666,667 Illumina reads.
Genome annotation
Genes were identified using Prodigal [45] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [46]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [47].
Genome properties
The genome consists of a 1,243,342 bp long chromosome with a 31.6% GC content (Table 3 and Figure 3). Of the 1,361 genes predicted, 1,311 were protein-coding genes, and 50 RNAs; twenty eight pseudogenes were also identified. The majority of the protein-coding genes (74.8%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 1,243,342 | 100.00% |
| DNA coding region (bp) | 1,163,294 | 93.56% |
| DNA G+C content (bp) | 393,356 | 31.64% |
| Number of replicons | 1 | 100.00% |
| Extrachromosomal elements | 0 | |
| Total genes | 1,361 | 100.00% |
| RNA genes | 50 | 3.67% |
| rRNA operons | 2 | 0.15% |
| Protein-coding genes | 1,311 | 96.33% |
| Pseudo genes | 28 | 2.06% |
| Genes with function prediction | 1,018 | 74.80% |
| Genes in paralog clusters | 100 | 7.35% |
| Genes assigned to COGs | 1,126 | 82.73% |
| Genes assigned Pfam domains | 1,144 | 84.06% |
| Genes with signal peptides | 109 | 8.01% |
| Genes with transmembrane helices | 248 | 18.22% |
| CRISPR repeats | 0 |
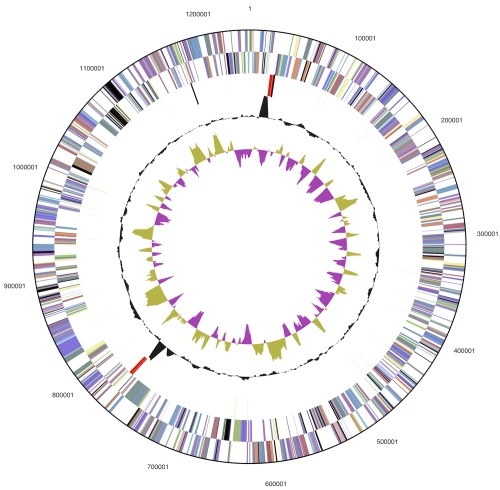
Figure 3.
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | % age | Description |
|---|---|---|---|
| J | 144 | 12.2 | Translation, ribosomal structure and biogenesis |
| A | 2 | 0.2 | RNA processing and modification |
| K | 47 | 4.0 | Transcription |
| L | 49 | 4.2 | Replication, recombination and repair |
| B | 4 | 0.3 | Chromatin structure and dynamics |
| D | 12 | 1.0 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 6 | 0.5 | Defense mechanisms |
| T | 11 | 0.9 | Signal transduction mechanisms |
| M | 52 | 4.4 | Cell wall/membrane/envelope biogenesis |
| N | 1 | 0.1 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 14 | 1.2 | Intracellular trafficking and secretion, and vesicular transport |
| O | 46 | 3.9 | Posttranslational modification, protein turnover, chaperones |
| C | 111 | 9.4 | Energy production and conversion |
| G | 39 | 3.3 | Carbohydrate transport and metabolism |
| E | 88 | 7.5 | Amino acid transport and metabolism |
| F | 47 | 4.0 | Nucleotide transport and metabolism |
| H | 115 | 9.8 | Coenzyme transport and metabolism |
| I | 17 | 1.4 | Lipid transport and metabolism |
| P | 50 | 4.2 | Inorganic ion transport and metabolism |
| Q | 5 | 0.4 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 172 | 14.6 | General function prediction only |
| S | 147 | 12.5 | Function unknown |
| - | 235 | 17.3 | Not in COGs |
Acknowledgements
This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-1.
References
- 1.Stetter KO, Thomm M, Winter J, Wildgruber G, Huber H, Zillig W, Jané-Covic D, König H, Palm P, Wunderl S. Methanothermus fervidus, sp. nov., a novel extremely thermophilic methanogen isolated from an icelandic hot spring. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig C2 1981; 2:166-178 [Google Scholar]
- 2.Lauerer G, Kristjansson JK, Langworthy TA, König H, Stetter KO. Methanothermus sociabilis sp. nov., a second species within the Methanothermaceae growing at 97°C. Syst Appl Microbiol 1986; 8:100-105 [Google Scholar]
- 3.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 4.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 5.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 6.Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Glöckner FO, Rosselló-Móra R. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 2008; 31:241-250 10.1016/j.syapm.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 7.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [Google Scholar]
- 8.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bult CJ, White O, Olsen G, Zhou L, Fleischmann R, Sutton G, Blake J, FitzGerald F, Clayton R, Gocayne J, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 1996; 273:1058-1073 10.1126/science.273.5278.1058 [DOI] [PubMed] [Google Scholar]
- 10.Fricke WF, Seedorf H, Henne A, Krüer M, Liesegang H, Hedderich R, Gottschalk G, Thauer RK. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol 2006; 188:642-658 10.1128/JB.188.2.642-658.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 1997; 390:364-370 10.1038/37052 [DOI] [PubMed] [Google Scholar]
- 12.Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM, Li D, Kong Z, McTavish S, Sang C, et al. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS ONE 2010; 5:e8926 10.1371/journal.pone.0008926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie E, Keller K, Huber T, Dalevi D, Hu P, Andersen G. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas ES, Brown JW, Daniels CJ, Reeve JN. Genes encoding the 7S RNA and tRNASer are linked to one of the two rRNA operons in the genome of the extremely thermophilic archaebacterium Methanothermus fervidus. Gene 1990; 90:51-59 10.1016/0378-1119(90)90438-W [DOI] [PubMed] [Google Scholar]
- 15.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1. Springer, New York 2001:119-169. [Google Scholar]
- 18.Validation list 85: Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int J Syst Evol Microbiol 2002; 52:685-690 10.1099/ijs.0.02358-0 [DOI] [PubMed] [Google Scholar]
- 19.Boone DR. Class I. Methanobacteria class. nov. In: Bergey's Manual of Systematic Bacteriology, 2nd ed., vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria) (D.R. Boone and R.W. Castenholz, eds.), Springer-Verlag, New York 2001:p. 213. [Google Scholar]
- 20.List 6. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1981; 31:215-218 10.1099/00207713-31-2-215 [DOI] [Google Scholar]
- 21.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: Reevaluation of a unique biological group. Microbiol Rev 1979; 43:260-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Judicial Commission of the International Committee on Systematics of Prokaryotes The nomenclatural types of the orders Acholeplasmatales, Halanaerobiales, Halobacteriales, Methanobacteriales, Methanococcales, Methanomicrobiales, Planctomycetales, Prochlorales, Sulfolobales, Thermococcales, Thermoproteales and Verrucomicrobiales are the genera Acholeplasma, Halanaerobium, Halobacterium, Methanobacterium, Methanococcus, Methanomicrobium, Planctomyces, Prochloron, Sulfolobus, Thermococcus, Thermoproteus and Verrucomicrobium, respectively. Opinion 79. Int J Syst Evol Microbiol 2005; 55:517-518 10.1099/ijs.0.63548-0 [DOI] [PubMed] [Google Scholar]
- 23.List No 8. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1982; 32:266-268 10.1099/00207713-32-2-266 [DOI] [Google Scholar]
- 24.Classification of bacteria and archaea in risk groups. http://www.baua.de TRBA 466.
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stetter KO, Fiala G, Huber G, Huber R, Segerer A. Hyperthermophilic microorganisms. FEMS Microbiol Rev 1990; 75:117-124 10.1111/j.1574-6968.1990.tb04089.x [DOI] [Google Scholar]
- 27.Stetter KO. Hyperthermophilic procaryotes. FEMS Microbiol Rev 1996; 18:149-158 10.1111/j.1574-6976.1996.tb00233.x [DOI] [Google Scholar]
- 28.Bonin AS, Boone DR. 2006. The order Methanobacteriales In: M Dworkin, S Falkow, E Rosenberg, KH Schleifer E Stackebrandt (eds), The Prokaryotes, 3. ed, vol. 7. Springer, New York, p. 231-243. [Google Scholar]
- 29.Hensel R, König H. Thermoadaptation of methanogenic bacteria by intracellular ion concentration. FEMS Microbiol Lett 1988; 49:75-79 10.1111/j.1574-6968.1988.tb02685.x [DOI] [Google Scholar]
- 30.Lehmacher A, Vogt AB, Hensel R. Biosynthesis of cyclic 2,3-diphosphoglycerate: Isolation and characterization of 2-phosphoglycerate kinase and cyclic 2,3-diphosphoglycerate synthetase from Methanothermus fervidus. FEBS Lett 1990; 272:94-98 10.1016/0014-5793(90)80456-S [DOI] [PubMed] [Google Scholar]
- 31.Sandman K, Krzycki JA, Dobrinski B, Lurz R, Reeve JN. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc Natl Acad Sci USA 1990; 87:5788-5791 10.1073/pnas.87.15.5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabry S, Hensel R. Purification and characterization of D-glyceraldehyde-3-phosphate dehydrogenase from the thermophilic archaebacterium Methanothermus fervidus. Eur J Biochem 1987; 165:147-155 10.1111/j.1432-1033.1987.tb11205.x [DOI] [PubMed] [Google Scholar]
- 33.Charron C, Talfournier F, Isupov MN, Littlechild JA, Branlant G, Vitoux B, Aubry A. The crystal structure of D-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Methanothermus fervidus in the presence of NADP+ at 2.1 Å resolution. J Mol Biol 2000; 297:481-500 10.1006/jmbi.2000.3565 [DOI] [PubMed] [Google Scholar]
- 34.Schramm A, Kohlhoff M, Hensel R. Triose-phosphate isomerase from Pyrococcus woesei and Methanothermus fervidus. Methods Enzymol 2001; 331:62-77 10.1016/S0076-6879(01)31047-9 [DOI] [PubMed] [Google Scholar]
- 35.Koga Y, Nishihara M, Morii H, Akagawa-Matsushita M. Ether polar lipids of methanogenic bacteria: structures, comparative aspects, and biosyntheses. Microbiol Rev 1993; 57:164-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morii H, Eguchi T, Nishihara M, Kakinuma K, König H, Koga Y. A novel ether core lipid with H-shaped C-isoprenoid hydrocarbon C80 chain from the hyperthermophilic methanogen Methanothermus fervidus. Biochim Biophys Acta 1998; 1390:339-345 [DOI] [PubMed] [Google Scholar]
- 37.Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 38.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/media_list.php
- 40.DOE Joint Genome Institute http://www.jgi.doe.gov/
- 41.The Phred/Phrap/Consed software package http://www.phrap.com
- 42.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sims D, Brettin T, Detter J, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008. [Google Scholar]
- 45.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Podigal ProkaryoticDynamic Programming Genefinding Algorithm. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A gene prediction improvement pipeline for microbial genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 47.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]