Abstract
Ilyobacter polytropus Stieb and Schink 1984 is the type species of the genus Ilyobacter, which belongs to the fusobacterial family Fusobacteriaceae. The species is of interest because its members are able to ferment quite a number of sugars and organic acids. I. polytropus has a broad versatility in using various fermentation pathways. Also, its members do not degrade poly-β-hydroxybutyrate but only the monomeric 3-hydroxybutyrate. This is the first completed genome sequence of a member of the genus Ilyobacter and the second sequence from the family Fusobacteriaceae. The 3,132,314 bp long genome with its 2,934 protein-coding and 108 RNA genes consists of two chromosomes (2 and 1 Mbp long) and one plasmid, and is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: strictly anaerobic, none-motile, Gram-negative, 3-hydroxybutyrate, mesophilic, chemoorganotrophic, Fusobacteriaceae, GEBA
Introduction
Strain CuHbu1T (= DSM 2926 = ATCC 51220 = LMG 16218) is the type strain of I. polytropus, which is the type species of the genus Ilyobacter [1,2]. Currently, there are four species placed in the genus Ilyobacter [1]. The generic name derives from the Greek word ‘ilus’ meaning ‘mud’ and the Neo-Latin word ‘bacter’ meaning ‘a rod’, referring to a mud-inhabiting rod [2]. The species epithet is derived from the Neo-Latin word ‘polytropus’ meaning ‘versatile’, referring to metabolic versatility of the species [2]. I. polytropus strain CuHbu1T was isolated from marine anoxic mud in Cuxhaven, Germany, and described by Stieb and Schink in 1984 [2]. No further isolates have been obtained for I. polytropus. Members of the genus Ilyobacter were isolated from anoxic marine sediments in Germany [2], Italy [3,4] and of estuarine origin [5]. Here we present a summary classification and a set of features for I. polytropus CuHbu1T, together with the description of the complete genomic sequencing and annotation.
Classification and features
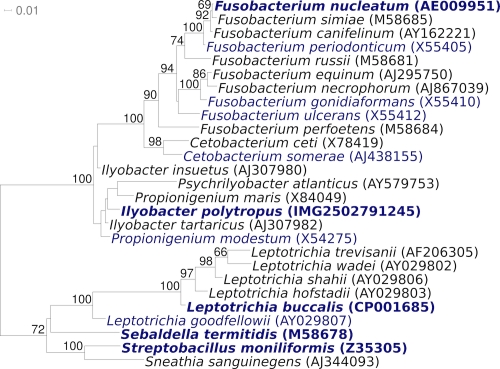
The 16S rRNA gene sequence of I. polytropus shares the highest degree of sequence similarity with the type strains of the other two members of the genus, I. insuentus (97.3%) and I. tartaricus (98.3%), the latter was isolated from anoxic marine sediment of Canal Grande and Rio Martin in Venice, Italy. The degree of sequence identity with the type strains of the other members of the family Fusobacteriaceae varies between 89.5% and 97.8%, with Propionigenium modestum as most similar species [6] (Figure 1). The genome survey sequence database (gss) contains the 16S rRNA gene sequence of human gut metagenome clone 5192b-5192b-A-con-04 (FI579563) as the best hit, which is 91% identical to the 16S rRNA gene sequence of strain CuHbu1T. No phylotypes from environmental samples database (env_nt) could be linked to the species I. polytropus or even the genus Ilyobacter, indicating a rather rare occurrence of these in the habitats screened so far (as of October 2010). A representative genomic 16S rRNA sequence of I. polytropus was compared using NCBI BLAST under default values (e.g., considering only the best 250 hits) with the most recent release of the Greengenes database [17] and the relative frequencies of taxa and keywords, weighted by BLAST scores, of taxa and keywords were determined. The four most frequent genera were Fusobacterium (70.2%), Ilyobacter (13.8%), Propionigenium (12.4%) and Clostridium (3.6%). Regarding hits to sequences from other members of the genus, the average identity within HSPs (high-scoring segment pairs) was 96.4%, whereas the average coverage by HSPs was 98.4%. The species yielding the highest score was I. tartaricus. The five most frequent keywords within the labels of environmental samples which yielded hits were 'microbiome' (6.5%), 'fecal' (6.1%), 'feces' (5.7%), 'calves/microorganisms/neonatal/shedding' (5.5%) and 'evolution/gut/mammals/microbes' (2.5%). These keywords suggest further, animal associated habitats for I. polytropus, beyond the anaerobic muds of marine origin as stated in the original description [2]. Environmental samples which yielded hits of a higher score than the highest scoring species were not found.
Figure 1.
Phylogenetic tree highlighting the position of I. polytropus CuHbu1T relative to the other type strains within the family Fusobacteriaceae. The tree was inferred from 1,399 aligned characters [7,8] of the 16S rRNA gene sequence under the maximum likelihood criterion [9] and rooted in accordance with the current taxonomy. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 900 bootstrap replicates [10] if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [11] are shown in blue, published genomes in bold [12-15]. Note that Ilyobacter appears as polyphyletic in the tree [16], but none of the relevant branches obtains any bootstrap support. Thus, the current classification is not in significant conflict with our phylogenetic analysis.
Figure 1 shows the phylogenetic neighborhood of I. polytropus CuHbu1T in a 16S rRNA based tree. The sequences of the eight 16S rRNA gene copies in the genome of I. polytropus differ from each other by up to three nucleotides, and differ by up to three nucleotides from the previously published 16S rRNA sequence (AJ307981), which contains two ambiguous base calls.
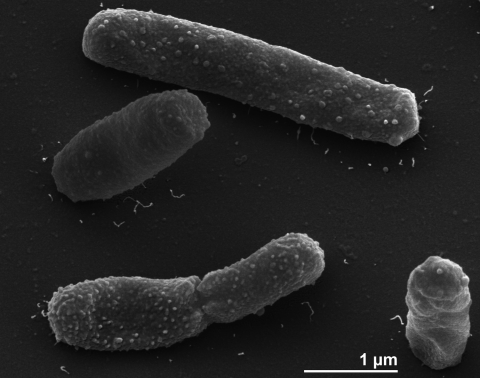
The cells of I. polytropus are generally rod-shaped (0.7×1.5-3.0 µm) with rounded ends (Figure 2). Cells of I. polytropus show irregularly elongated rods, when grown on glucose and fructosecontaining media [2]. The cells are usually arranged in pairs or chains [2]. I. polytropus is a Gram-negative and non spore-forming bacterium (Table 1). The organism is nonmotile and no flagellar genes have been found in the genome If at all, active movement by twitching motility could be possible, as some genes related to this phenotype were identified (this paper, see below). Interestingly, the original description states that “the originally motile rods lost motility after several transfers” [2]. The organism is a strictly anaerobic chemoorganotroph [2]. I. polytropus requires 1% NaCl in media for good growth [25]. The selective medium for I. polytropus is a NaCl-containing mineral media, which contains 3-hydroxybutyrate as a sole carbon and energy source [2]. The organism also grows in salt water medium or brackish water medium containing 1% NaCl and 0.15% MgCl2.6H2O [2]. Vitamins are not required in the enrichment media for at least five subsequent transfers [2]. Phosphate (up to 50 mM) does not inhibit growth of I. polytropus, when grown on 3-hydroxybutyrate [2]. The temperature range for growth is between 10°C and 35°C, with an optimum at 30°C [2]. The organism does not grow at 4°C or at 40°C [2]. The pH range for growth is 6.5-8.5, with an optimum at pH 7.0-7.5 [2]. No cytochromes are detected from I. polytropus [2]. I. polytropus is able to utilize 3-hydroxybutyrate, crotonate, glycerol, pyruvate, citrate, oxaloacetate, glucose, fructose, malate and fumarate and to ferment a variety of sugars and organic acids [2]. The organism does not utilize lactose, sucrose, mannitol, sorbitol, xylitol, 1,2-butanediol, 1,3-butanediol, 2,3-butanediol, maltose, arabinose, cellobiose, mannose, melezitose, raffinose, sorbose, rhamnose, trehalose, xylose, acetone, diacetyl acetoin, acetoacetyl ethylester, acetoacetyl amide, peptone, casamino acids, yeast extract, glyoxylate, glycolate, lactate, succinate, L-tartrate, poly-β-hydroxybutyrate, starch, methanol plus acetate and formate plus acetate [2]. I. polytropus is able to ferment 3-hydroxybutyrate and crotonate to acetate and butyrate [2]. Glycerol is fermented to 1,3-propanediol and 3-hydroxypropionate [2]. Malate and fumarate are fermented to acetate, formate and propionate [2]. I. polytropus is able to ferment glucose and fructose to acetate, formate and ethanol [2]. The organism does not reduce sulfate, sulfur, thiosulfate and nitrate [2]. I. polytropus grows in mineral media with a reductant [2]. It does not hydrolyze gelatin or urea and does not produce indole [2]. I. polytropus shows acetate kinase, phosphate acetyl transferase and hydrogenase activities, which are sufficient for involvement in dissimilatory metabolism [2]. Also, pyruvate formate lyase activity was shown in crude cell extracts, however, activity was extremely low and ascribed to a potential instability of this enzyme if traces of oxygen are present during the enzyme activity measurement [2]. I. polytropus maintains its energy metabolism exclusively by substrate-linked phosphorylation reactions [2]. I. polytropus differs from other anaerobes because the organism exhibits broad versatility in its use of various fermentation pathways. However, pathway regulation was reported as enigmatic because neither propionate nor butyrate were formed during glucose or fructose fermentation, although the necessary enzymes are present [2]. I. polytropus is of ecological interest because the organism does not degrade poly-β-hydroxybutyrate but only the monomeric of 3- hydroxybutyrate [2]. Metabolism of the polymer appears to be confined to aerobic microbial communities [26].
Figure 2.
Scanning electron micrograph of I. polytropus CuHbu1T
Table 1. Classification and general features of I. polytropus CuHbu1T according to the MIGS recommendations [18].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [19] | |
| Phylum “Fusobacteria” | TAS [20,21] | ||
| Class “Fusobacteria” | TAS [20] | ||
| Order “Fusobacteriales” | TAS [20] | ||
| Family “Fusobacteriaceae” | TAS [20] | ||
| Genus Ilyobacter | TAS [2,22] | ||
| Species Ilyobacter polytropus | TAS [2,22] | ||
| Type strain CuHbu1 | TAS [2] | ||
| Gram stain | negative | TAS [2] | |
| Cell shape | rod-shaped with rounded ends, single or in pairs | TAS [2] | |
| Motility | non-motile | TAS [2] | |
| Sporulation | none | TAS [2] | |
| Temperature range | 10°C–35°C | TAS [2] | |
| Optimum temperature | 30°C | TAS [2] | |
| Salinity | 1% NaCl | TAS [2] | |
| MIGS-22 | Oxygen requirement | strictly anaerobic | TAS [2] |
| Carbon source | carbohydrates | TAS [2] | |
| Energy source | chemoorganotroph | TAS [2] | |
| MIGS-6 | Habitat | marine anoxic mud | TAS [2] |
| MIGS-15 | Biotic relationship | free-living | NAS |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [23] | |
| Isolation | marine anoxic mud | TAS [2] | |
| MIGS-4 | Geographic location | Cuxhaven, Germany | TAS [2] |
| MIGS-5 | Sample collection time | 1983 or before | TAS [2] |
| MIGS-4.1 | Latitude | 53.87 | NAS |
| MIGS-4.2 | Longitude | 8.69 | NAS |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | sea level | NAS |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [24]. If the evidence code is IDA, then the property was directly observed by one of the authors or an expert mentioned in the acknowledgements.
Chemotaxonomy
No chemotaxonomic data are currently available for I. polytropus or for the genus Ilyobacter.
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [27], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [28]. The genome project is deposited in the Genome OnLine Database [11,29] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Tree genomic libraries: one 454 pyrosequence standard library, one 454 PE library (14 kb insert size), one Illumina library |
| MIGS-29 | Sequencing platforms | Illumina GAii, 454 GS FLX Titanium |
| MIGS-31.2 | Sequencing coverage | 124.8 × Illumina; 91.0 × pyrosequence |
| MIGS-30 | Assemblers | Newbler version 2.0.00.20- PostRelease-10-28-2008-g-3.4.6, phrap |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| INSDC ID | CP002281 chromosome I CP002282 chromosome II CP002283 plasmid |
|
| Genbank Date of Release | November 1, 2010 | |
| GOLD ID | Gc01413 | |
| NCBI project ID | 32577 | |
| Database: IMG-GEBA | 2503538000 | |
| MIGS-13 | Source material identifier | DSM 2926 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
I. polytropus CuHbu1T, DSM 2926, was grown anaerobically in medium 314 (Ilyobacter polytropus medium) [30] at 30°C. DNA was isolated from 0.5-1 g of cell paste using MasterPure Gram-positive DNA purification kit (Epicentre MGP04100) following the standard protocol as recommended by the manufacturer, with modification st/LALM for cell lysis as described in Wu et al. [28].
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website [31]. Pyrosequencing reads were assembled using the Newbler assembler version 2.0.00.20-PostRelease-10-28-2008-g++-3.4.6 (Roche). The initial Newbler assembly consisting of 85 contigs in 1 scaffold was converted into a phrap assembly by [32] making fake reads from the consensus, to collect the read pairs in the 454 paired end library. Illumina GAii sequencing data (387 Mb) was assembled with Velvet [33] and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. The 454 draft assembly was based on 284.7 Mb 454 draft data and all of the 454 paired end data. Newbler parameters are -consed -a 50 -l 350 -g -m -ml 20.
The Phred/Phrap/Consed software package [32] was used for sequence assembly and quality assessment in the subsequent finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolution [31], Dupfinisher, or sequencing cloned bridging PCR fragments with subcloning or transposon bombing (Epicentre Biotechnologies, Madison, WI) [34]. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR primer walks (J.-F.Chang, unpublished). A total of 719 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. Illumina reads were also used to correct potential base errors and increase consensus quality using a software Polisher developed at JGI [35]. The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Illumina and 454 sequencing platforms provided 215.8 × coverage of the genome. The final assembly contained 656,481 pyrosequence and 10,750,000 Illumina reads
Genome annotation
Genes were identified using Prodigal [36] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [37]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [38].
Genome properties
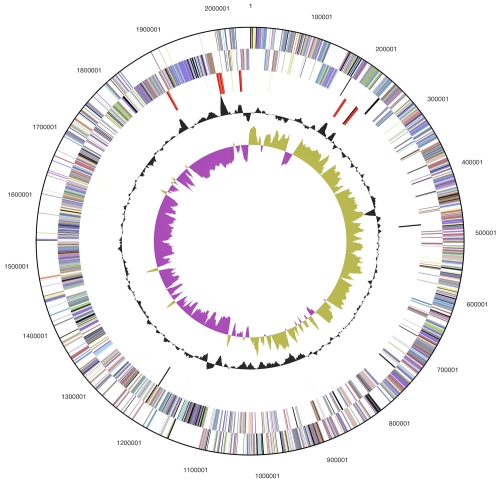
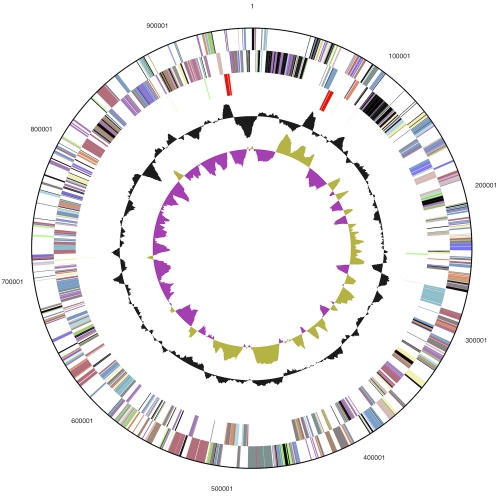
The genome consists of a 2,046,464 bp long chromosome I with a GC content of 35%, a 961,624 bp long chromosome II with 34% GC content, and a 124,226 bp long plasmid with 32% GC content (Table 3 and Figure 3a, Figure 3b, and Figure 3c). Of the 3,042 genes predicted, 2,934 were protein-coding genes, and 108 RNAs; 108 pseudogenes were also identified. The majority of the protein-coding genes (73.3%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,132,314 | 100.00% |
| DNA Coding region (bp) | 2,690,412 | 85.89% |
| DNA G+C content (bp) | 1,076,435 | 34.37% |
| Number of replicons | 3 | |
| Extrachromosomal elements | 1 | |
| Total genes | 3,042 | 100.00% |
| RNA genes | 108 | 3.55% |
| rRNA operons | 8 | |
| Protein-coding genes | 2,934 | 96.45% |
| Pseudo genes | 54 | 1.78% |
| Genes with function prediction | 2,230 | 73.31% |
| Genes in paralog clusters | 676 | 22.22% |
| Genes assigned to COGs | 2,283 | 75.05% |
| Genes assigned Pfam domains | 2,359 | 77.55% |
| Genes with signal peptides | 719 | 23.64% |
| Genes with transmembrane helices | 638 | 20.97% |
| CRISPR repeats | 1 |
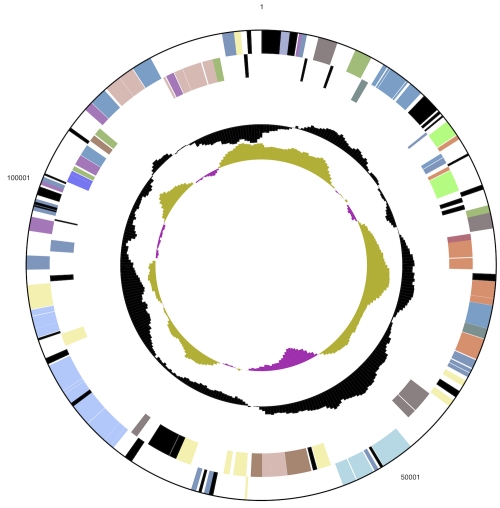
Figure 3a.
Graphical circular map of chromosome I. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Figure 3b.
Graphical circular map of chromosome II. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew. Chromosome II was identified as a chromosome due to its two 16S rRNA gene copies.
Figure 3c.
Graphical circular map of the plasmid. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew. Chromosome II was identified as a chromosome due to its two 16S rRNA gene copies.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | % age | Description |
|---|---|---|---|
| J | 146 | 5.8 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 152 | 6.1 | Transcription |
| L | 124 | 5.0 | Replication, recombination and repair |
| B | 1 | 0.0 | Chromatin structure and dynamics |
| D | 27 | 1.1 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 43 | 1.7 | Defense mechanisms |
| T | 140 | 5.6 | Signal transduction mechanisms |
| M | 161 | 6.4 | Cell wall/membrane/envelope biogenesis |
| N | 21 | 0.8 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 61 | 2.4 | Intracellular trafficking, secretion, and vesicular transport |
| O | 69 | 2.8 | Posttranslational modification, protein turnover, chaperones |
| C | 244 | 9.7 | Energy production and conversion |
| G | 151 | 6.0 | Carbohydrate transport and metabolism |
| E | 245 | 9.8 | Amino acid transport and metabolism |
| F | 73 | 2.9 | Nucleotide transport and metabolism |
| H | 134 | 5.4 | Coenzyme transport and metabolism |
| I | 72 | 2.9 | Lipid transport and metabolism |
| P | 104 | 4.2 | Inorganic ion transport and metabolism |
| Q | 38 | 1.5 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 311 | 12.4 | General function prediction only |
| S | 190 | 7.6 | Function unknown |
| - | 759 | 25.0 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Katja Steenblock (DSMZ) for growing I. polytropus cultures. This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-2 and Thailand Research Fund Royal Golden Jubilee Ph.D. Program No. PHD/0019/2548 for MY.
References
- 1.Garrity G. NamesforLife. BrowserTool takes expertise out of the database and puts it right in the browser. Microbiol Today 2010; 7:1 [Google Scholar]
- 2.Stieb M, Schink B. A new 3-hydroxybutyrate fermenting anaerobe, Ilyobacter polytropus, gov. nov. sp. nov., possessing various fermentation pathways. Arch Microbiol 1984; 140:139-146 10.1007/BF00454916 [DOI] [Google Scholar]
- 3.Brune A, Evers S, Kaim G, Ludwig W, Schink B. Ilyobacter insuetus sp. nov., a fermentative bacterium specialized in the degradation of hydroaromatic compounds. Int J Syst Evol Microbiol 2002; 52:429-432 [DOI] [PubMed] [Google Scholar]
- 4.Schink B. Fermentation of tartrate enantiomers by anaerobic bacteria, and description of two new species of strict anaerobes, Ruminococcus pasteurii and Ilyobacter tartaricus. Arch Microbiol 1984; 139:409-414 10.1007/BF00408388 [DOI] [Google Scholar]
- 5.Janssen PH, Harfoot CG. Ilyobacter delafieldii sp. nov., a metabolically restricted anaerobic bacterium fermenting PHB. Arch Microbiol 1990; 154:253-259 10.1007/BF00248964 [DOI] [Google Scholar]
- 6.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 2007; 57:2259-2261 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- 7.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 8.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 9.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 10.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [PubMed] [Google Scholar]
- 11.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2009; 38:D346-D354 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, Bhattacharyya A, Bartman A, Gardner W, Grechkin G, et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 2002; 184:2005-2018 10.1128/JB.184.7.2005-2018.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanova N, Gronow S, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Lucas S, Chen F, Tice H, Cheng JF, et al. Complete genome sequence of Leptotrichia buccalis type strain (C-1013-bT). Stand Genomic Sci 2009; 1:126-132 10.4056/sigs.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolan M, Gronow S, Lapidus A, Ivanova N, Copeland A, Lucas S, Glavina Del Rio T, Chen F, Tice H, Pitluck S, et al. Complete genome sequence of Streptobacillus moniliformis type strain (9901T). Stand Genomic Sci 2009; 1:300-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmon-Smith M, Celia L, Chertkov O, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Lucas S, Tice H, Cheng JF, et al. Complete genome sequence of Sebaldella termitidis type strain (NCTC 11300T). Stand Genomic Sci 2010; 2:220-227 10.4056/sigs.811799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farris JS. Formal definitions of paraphyly and polyphyly. Syst Zool 1974; 23:548-554 10.2307/2412474 [DOI] [Google Scholar]
- 17.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrity GM, Holt JG. 2001. Taxonomic outline of the Archaea and Bacteria, p. 155-166. In G. M. Garrity, D. R. Boone, and R. W. Castenholz (ed.), Bergey's Manual of Systematic Bacteriology, 2nd ed, vol. 1. Springer, New York. [Google Scholar]
- 21.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 22.Validation List no. 18. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1985; 35:375-376 10.1099/00207713-35-3-375 [DOI] [Google Scholar]
- 23.Classification of Bacteria and Archaea in risk groups. http://www.baua.de
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST. 1994. Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore. [Google Scholar]
- 26.Herron JS, King JD, White DC. Recovery of poly-β-hydroxybutyrate from estuarine microflora. Appl Environ Microbiol 1978; 35:251-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klenk HP, Goeker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 28.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.List of growth media used at DSMZ: http//www.dsmz.de/microorganisms/media_list.php
- 31.DOE Joint Genome Institute. http://www.jgi.doe.gov
- 32.Phrap and Phred for Windows, MacOS, Linux, and Unix. http://www.phrap.com
- 33.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sims D, Brettin T, Detter JC, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008. [Google Scholar]
- 36.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 38.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]