Abstract
The tumor suppressor gene TP53 is frequently mutated in human cancers. Abnormality of the TP53 gene is one of the most significant events in lung cancers and plays an important role in the tumorigenesis of lung epithelial cells. Human lung cancers are classified into two major types, small cell lung cancer (SCLC) and nonsmall cell lung cancer (NSCLC). The latter accounts for approximately 80% of all primary lung cancers, and the incidence of NSCLC is increasing yearly. Most clinical studies suggest that NSCLC with TP53 alterations carries a worse prognosis and may be relatively more resistant to chemotherapy and radiation. A deep understanding of the role of TP53 in lung carcinogenesis may lead to a more reasonably targeted clinical approach, which should be exploited to enhance the survival rates of patients with lung cancer. This paper will focus on the role of TP53 in the molecular pathogenesis, epidemiology, and therapeutic strategies of TP53 mutation in NSCLC.
1. Introduction
The TP53 gene, first described in 1979, was the first tumor suppressor gene to be identified [1–4]. It was originally believed to be an oncogene, but genetic and functional data obtained 10 years after its discovery proved it to be a tumor suppressor. Inactivation of TP53 function or its attendant pathway is a common feature of human tumors that often correlates with increased malignancy, poor patient survival, and resistance to treatment [5–7]. The TP53 tumor suppressor gene is located at the short arm of chromosome 17 (17p13). It contains 11 exons spanning 20 kilobases and encodes a nuclear phosphoprotein of 53 kDa. The TP53 protein contains distinct functional domains: the N-terminus transactivation domain, the sequence-specific DNA-binding domain, the oligomerization domain, and the C-terminus negative regulatory domain [8]. The TP53 gene has been implicated in a growing number of biological processes, including DNA repair, cell-cycle arrest, apoptosis, autophagy, senescence, metabolism, and aging [6, 9–11]. The transcription factor TP53 can activate the transcription of numerous downstream genes, such as p21 and MDM2, by binding to specific sequences, which often mediates their biological functions [9, 12, 13]. Indeed, the activities of TP53, both transcription dependent and independent, are regulated via its mRNA and protein levels, cellular localization, and ability to bind over 100 cellular proteins and control the expression of thousands of potential target genes. Under normal conditions, TP53 is rapidly degraded and, thus, not present at detectable levels within the cell. Various types of cellular stress, such as DNA damage induced by UV and oncogene activation, result in the stabilization and activation of TP53, causing protein accumulation within the nucleus. The TP53 pathway is activated by such cellular stresses that alter the normal cell-cycle progression or can induce mutations of the genome, leading to the transformation of a normal cell into a cancerous cell.
Lung cancer, primarily caused by tobacco exposure, is one of the commonest neoplasms all over the world, with approximately 1.35 million new cases worldwide in 2002 [14]. It is the leading cause of cancer death in the world, with more than 1.18 million deaths in 2002 [14]. Despite recent advances in surgical and chemo/radiation therapies, the prognosis is very poor, with an overall survival rate of only 15%. NSCLC represents a heterogeneous group of cancers, consisting mainly of squamous cell carcinoma, adenocarcinoma, and large-cell carcinoma. Approximately 80% of human lung cancers are NSCLC, and their development involves multiple genetic abnormalities that lead to malignant transformation of the bronchial epithelial cells, followed by invasion and lymph node and distant metastases. Among such genetic abnormalities, the TP53 tumor suppressor gene appears to be the most frequent target, and abnormality of TP53 plays an important role in the tumorigenesis of lung epithelial cells. Indeed, mutations of the TP53 gene occur in about 50% of NSCLC [15, 16]. As TP53 mutants are present in almost half of NSCLC whose incidence rate increasing every year, the possibility of abolishing their oncogenic effects is undoubtedly important for a successful treatment of NSCLC.
2. TP53 Gene Mutations in Human Cancers
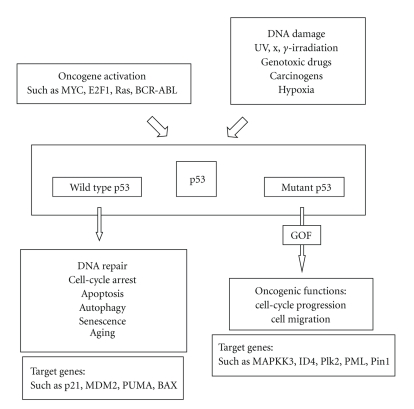
Mutations in the tumor suppressor TP53 gene are one of the most common genetic alterations present at high frequency in human cancers [6, 17–20]. Up to 50% of all human cancers contain mutations in both alleles of the TP53 gene [21–23]. Unlike most other tumor suppressor genes, such as RB, APC, and BRCA1 genes, they are inactivated by frameshift or nonsense mutations leading to disappearance or aberrant synthesis of the gene product, almost 80% of TP53 gene mutations are missense mutations [24]. Other alterations include frameshift insertions and deletions (9%), nonsense mutations (7%), silent mutations (5%), and other infrequent alterations [25]. In 1991, a database of published TP53 mutations was established by Dr. Curtis Harris and his collaborators to facilitate the retrieval and analysis of TP53 mutations [26]. Since 1994, this database has been maintained at the International Agency for Research on Cancer (IARC) [27] and is made freely available as a service to the scientific community (http://www.iarc.fr/p53). The current release is version R14 (November, 2009). It contains 26,597 somatic mutations and 535 germline TP53 mutations. Germline TP53 mutation causes predisposition to early-onset diverse mesenchymal and epithelial neoplasms, including breast carcinomas, sarcomas, brain tumors, and adrenal cortical carcinomas, defining the Li-Fraumenni syndrome, which is a rare autosomal dominant syndrome [28–30]. As reported, somatic TP53 missense mutations are found in approximately 50% of human cancers, and inactivating mutations in the TP53 gene are the most common genetic events in human cancers affecting a specific gene, with the vast majority arising from a single-point mutation in the segment encoding the DNA-binding domain of TP53 [21, 23]. The inactivating mutations render the mutant TP53 protein unable to carry out its normal functions, that is, transcriptional transactivation of downstream target genes that regulate cell cycle and apoptosis [31]. Several recent lines of evidence indicate that in addition to abrogating the tumor suppressor functions of wild-type TP53, the common types of cancer-associated TP53 mutations also endow the mutant protein with new activities, so-called “gain-of-function” (GOF) activities, which can contribute actively to various stages of tumor progression, including distant metastases, and to increased resistance to anticancer treatments. GOF activities of mutant TP53 are exerted by aberrant protein interaction or gene regulation, such as MAPKK3, inhibitor of DNA-binding 4 (ID4), polo-like kinase 2 (Plk2), promyelocytic leukemia protein (PML), and prolyl isomerase Pin1 (see Figure 1) [32–38]. Although the occurrence of TP53 mutations is not limited to a few particular sequences or codons along this gene, most mutations cluster in the TP53 DNA-binding domain, which encompasses exons five through eight and spans approximately 180 codons or 540 nucleotides [39]. Most TP53 missense mutations lead to the synthesis of a stable protein, which lacks its specific DNA-binding and transactivation function and accumulates in the nucleus of cells. These mutant accumulated proteins are retained in distant metastasis. In addition, the most frequent mutants have been shown to be capable of cooperating with oncogenes for cellular transformation [40]. About 30% of TP53 missense mutations found in cancer correspond to nucleotide substitutions at highly mutable CpG dinucleotides and at codons encoding residues that play essential structural and chemical roles in the contact between the TP53 protein and specific DNA sequences that constitute the TP53 response elements [41]. These mutations result in a significant loss of DNA-binding activity and transactivation capacity [31]. It is noteworthy that five of the six most prominent mutation hotspots in the TP53 gene are represented by G to T mutations at codons containing methylated CpG sequences, including codons 157, 158, 245, 248, and 273 [42]. The understanding of the tumor-specific mutational spectra of the TP53 gene is quite important for the realization of TP53-associated carcinogenesis [39]. Analysis of the spectrum of TP53 mutations in human cancer demonstrates a link between exposure to various types of carcinogens and the development of specific cancers [43]. For example, tobacco smoke carcinogens are linked to G to T mutations in lung cancer arising in smokers, as will hereinafter be described in detail [44].
Figure 1.
A synthetic view of the p53 pathway. Mutant p53 exerts new oncogenic activities, so-called “gain-of-function” (GOF) activities, which can contribute actively to various stages of tumor progression, including distant metastases, and to increased resistance to anticancer treatments.
3. TP53 Mutations in NSCLC
3.1. TP53 Alterations in Lung Cancer
The frequent detection of loss of heterozygosity (LOH) in lung cancer cell lines and tumor samples at the location of the TP53 gene on chromosome 17p13 suggested that this gene was likely to be involved in the pathogenesis of lung cancer, and genetic abnormality of the TP53 in lung cancers has been shown to be associated with a poorer survival prognosis and increased cellular resistance to therapy [45–47]. The highest frequency of TP53 alterations is found in SCLC specimens [48–51]. On the other hand, the frequency of TP53 mutations is the highest in squamous cell carcinomas and lower in adenocarcinomas among NSCLC tumor samples [52, 53]. It has been reported that somatic mutations and increased expression of TP53 were frequently found in ∼23% and ∼65% of NSCLC, respectively, [54, 55]. TP53 mutations are found in tumors both with and without allele loss at 17p13 and are mostly located within the DNA-binding domain of TP53 [56]. Because coding mutations of TP53 occur relatively early in the development of lung cancer and are potentially required for maintaining the malignant phenotype, the acquired TP53 mutations are preserved during tumor progression and metastatic spread [57, 58]. Chang et al. reported that the incidence of TP53 mutations in primary tumors and metastatic lymph nodes was 23.2% and 21.4%, respectively, and the TP53 gene status in primary tumors and metastatic lymph nodes showed 92.9% concordance among 56 patients with NSCLC who had undergone surgical resection, which explained the fact that TP53 mutations usually precede lymph node metastasis [59]. Most TP53 mutations occur before the tumor metastasizes. They are then preserved through subsequent stages of tumor development; as a result, no selection against TP53 mutations occurs during metastasis.
3.2. Prognosis of NSCLC Patients in Wild-Type TP53 and Mutant TP53
TP53 is the most extensively investigated prognostic marker in NSCLC, and the following meta-analyses have suggested a weak to modest negative prognostic role for TP53. Steels et al. identified aberrant TP53 as a prognostic factor in NCSCL at any stage (localized or advanced disease) and any histology (adenocarcinoma and squamous cell carcinoma). Moreover, no significant differences were found when comparing the different antibodies used in the Immunohistochemical (IHC) assays [47]. Mitsudomi et al. analyzed protein overexpression and DNA mutations separately. In lung cancer, the concordance between the two assays in 60%–70% aberrant TP53 was globally more prevalent in squamous cell carcinomas than in adenocarcinomas; however, its role as a prognostic marker was significant only in patients with this latter lung cancer type. In conclusion, the presence of an aberrant TP53 status could identify a disease with more aggressive features and, thus, a potentially shorter survival [60]. Cancer and Leukemia Group B (CALGB) 9633 was a phase III trial that randomized patients with stage IB NSCLC to either observation or four cycles of carboplatin and paclitaxel. A statistically significant effect in favor of adjuvant chemotherapy was seen for disease-free survival (DFS) and overall survival (OS) in the subgroup of patients with tumors ≥4 cm. Recently, a laboratory companion study was conducted to determine whether molecular and clinical factors could provide additional prognostic information. Formalin-fixed, paraffin-embedded blocks were obtained for 250 of the 344 patients enrolled. IHC staining for TP53, bcl-2, blood group antigen A, and mucin was correlated with DFS and OS. The results showed that the prevalence of TP53 and mucin expression was 47% and 45%, respectively. Univariate analysis for DFS showed a statistically significant effect for the presence of mucin (P = .0005) and TP53 (P = .05). In the multivariate Cox model, there was a statistically significant association between shorter DFS and the presence of mucin (P = .002; HR 2.05) and TP53 (P = .003; HR 1.95) and between shorter OS and the presence of mucin (P = .004; HR 2.03) and TP53 (P = .0005; HR 2.30) [61]. In another published study, TP53 protein expression was evaluated by IHC in tissue samples of 253 completely resected NSCLC patients. The incidence of TP53 protein overexpression was 52% (132 patients). Untreated TP53-positive patients had significantly shorter overall survival than patients with TP53-negative tumors (P = .03; HR 1.89; 95% CI, 1.07–3.34). However, these TP53-positive patients also had a significantly greater survival benefit from adjuvant chemotherapy (P = .02; HR 0.54) than patients with TP53-negative tumors (P = .26; interaction P = .02; HR 1.40), indicating that TP53 protein overexpression is a significant marker of poor prognosis as well as a significant predictive marker for a differentially greater benefit from adjuvant chemotherapy in completely resected NSCLC patients [62, 63]. In addition, 35 patients with advanced NSCLC who had received neoadjuvant chemotherapy in the context of a prospective phase II trial were analyzed for the TP53 genotype of their tumors. Response to induction therapy was then correlated to the TP53 genotype as assessed by complete direct DNA sequencing. Patients had received 3 cycles of cisplatin and etoposide and 1 cycle of simultaneous radiochemotherapy. The presence of a mutant TP53 genotype was highly indicative of resistance to induction chemotherapy (P < .002). The sensitivity of the mutant TP53 genotype to identify nonresponders was 94% (71.3–99.9 confidence interval). A normal TP53 gene was significantly associated with radical resection (P < .004) and survival advantage (P = .02) [64]. Thus, most of the evidence indicates that alterations in TP53 are associated with a poor prognosis. Nevertheless, there is no definitive evidence that the knowledge of TP53 status could play a role in the management of individual patients with NSCLC.
3.3. TP53 Mutations in Tobacco-Associated Lung Cancer
As already reported, human NSCLC is classified into several subtypes. The incidence of the different subtypes in NSCLC has changed over the last two decades. Since the introduction of filtered tobacco cigarettes and the reduction in the number of smokers, the percentage of individuals with adenocarcinoma has increased whereas the incidence of squamous cell carcinoma has decreased. Recent estimates indicate that 15% of men and 53% of women with lung cancer worldwide are never-smokers [14]. Otherwise, the leading risk factor for lung cancer is undoubtedly tobacco smoking, and the risk for lung cancer increases with the number of cigarettes smoked and the duration of smoking [65, 66]. Tobacco smoking accounts for more than 90% of lung cancers in men and 74% to 80% of lung cancers in women in the United States and the European Union [14, 67–70]. According to the prevailing research, it is the direct mutagenic action on DNA the means by which smoking causes lung cancer [71]. Because TP53 mutations are found in more than half of lung tumors, the TP53 gene should be considered to be one of the most frequent targets of tobacco smoking-related DNA mutations. In fact, TP53 mutations are the most extensively studied mutations in lung cancer, and a database of all published mutations is maintained mainly by the IARC, as noted above. Another database is curated by Dr. Therry Soussi, Curie Institute, Paris, France (http://p53.curie.fr/). Patients with tobacco-associated cancer have a higher frequency (26%–71%) of TP53 mutations than patients who never smoked (8%–47%) [72–74]. Until now, several studies have detected hotspots on the TP53 gene, with G:C to T:A (G to T) transversions being a characteristic finding in tobacco-associated lung cancer [44, 58, 74, 75]. Ninety percent of the guanines undergoing these transversion events in lung cancer are located on the nontranscribed DNA strand [42, 76]. Mutational hotspots within the Tp53 gene, for example, codon 157, have been identified for tobacco-related lung cancer whereas these same mutations are rarely found in other cancers. The incidence for G to T transversions in lung cancer tissues from never-smokers is significantly lower than that in those from smokers. The ratio of G to T transversions to G : C to A : T (G to A) transversions was 1.0 in smokers and 0.34 in never-smokers [77]. Another study reported that a screening of 623 genes in 188 lung adenocarcinomas confirmed the higher mutation load and higher fraction of strand-biased G to T transversions in lung tumor of smokers when compared with nonsmokers [78]. The spectrum of G to T transversions is presumably induced by polycyclic aromatic hydrocarbons (PAH) from tobacco smoke. Benzo[α]pyrene is the most studied member of the PAH class, and Benzo[α]pyrene diol epoxide (BPDE) is the main metabolite of Benzo[α]pyrene, one of the most potent carcinogens present in high quantity in tobacco smoke. The BPDE-DNA adduct patterns in the TP53 gene in bronchial epithelial cells coincide with G to T mutational hotspots within codons 157, 248, and 273 in tobacco smoking-associated lung cancers [79, 80]. G to T transversions are typical for bulky adduct-producing mutagens, including the class of PAHs, and BPDE adducts at these codons [81].
3.4. TP53 Mutational Spectra between Never-Smokers and Smokers
Several epidemiological studies have identified lung cancer in people who have never smoked as a distinct entity from tobacco-associated lung cancer, and genetic mutations are regarded to be more common in patients with tobacco-associated lung cancer than in never-smokers. Lung cancer from smokers shows a distinct, unique TP53 mutation spectrum, such as G to T transversions at codons 157, 158, 179, 248, and 273, which is uncommonly observed in lung cancer from nonsmokers [74, 75, 82]. Indeed, in smokers, 43% of the mutations were G to T transversions, but this number dropped to 13% in never-smokers [78]. Another analysis reported that the difference of the TP53 mutational spectra between never-smokers and smokers was detectable only in women [77]. The TP53 mutations in female never-smokers with adenocarcinoma were predominantly transitions (83%). However, in smokers, the TP53 mutations were predominantly transversions (60%) and deletions (20%) [83]. These findings suggest that the TP53 mutations in lung cancer in never-smokers are distinct from those in tobacco-associated lung cancer. In patients with adenocarcinoma, the frequency of TP53 mutations correlated with the amount of tobacco smoked [84]. On the other hand, patients with adenocarcinoma who have never smoked are more likely to have mutations in the epidermal growth factor receptor (EGFR) tyrosine kinase and have a better response to its inhibitors than do patients with tobacco-associated lung cancer [85]. It has been noticed that TP53 mutations are closely related to smokers whereas EGFR mutations are statistically significantly more frequent in females and never-smokers in adenocarcinoma [48, 86]. Furthermore, the prevalence of mutations in k-ras and TP53 differs for patients with lung cancer who never smoked and those with tobacco-associated lung cancer.
4. Therapeutic Strategies for NSCLC with TP53 Mutation
Several studies indicate that TP53 mutations confer chemoresistance to lung cancer cells in vivo and in vitro [87–90]. Determining TP53 status may be of great value in the choice of chemo/radiation therapy. For instance, it is well known that tumors containing the mutant TP53 are more resistant to ionizing radiation than those with the wild-type TP53. The frequent inactivation of TP53 in human tumors suggests that the reconstruction of the TP53 mediated pathway in tumor cells might be an attractive tumor cell-specific strategy for treating cancers. One of the most advanced strategies for therapeutic targeting of the TP53 pathway in cancer is virus-based therapeutic strategies for mutant TP53-carrying tumors. Wild-type TP53 reconstitution in mutant TP53-carrying or TP53-null tumors can be accomplished by gene therapy, that is, the introduction of an intact cDNA copy based on an adenovirus (Adp53). Because of the frequent mutation in TP53 in lung cancer, concomitant treatment with different classes of chemotherapy and TP53 gene replacement strategies has been tested in preclinical as well as in clinical settings [91]. In preclinical studies with lung cancer cell lines expressing mutated TP53, the TP53 replacement strategy has improved chemotherapy and radiotherapy responses [92, 93]. TP53 gene therapy has also been tested in clinical trials in patients with lung cancer, and some clinical studies have in fact shown a beneficial effect of the combination of Adp53 gene therapy and chemotherapeutic drugs and radiotherapy (reviewed in [94]). In a phase I clinical trial, 28 patients with NSCLC whose cancers had progressed on conventional treatments were administered the Adp53 gene into the tumors without any other therapies. Therapeutic activity in 25 evaluable patients included significant reduction in tumor size in two patients (8%) and disease stabilization in 16 patients (64%); the remaining seven patients (28%) exhibited disease progression [95]. In another trial, Adp53 was administered by bronchoscopic intratumoral injection in 12 patients with advanced endobronchial NSCLC; six of the 12 (50%) patients had significant improvement in airway obstruction, and 3 patients met the criteria for partial response [96]. Twenty-four patients with advanced NSCLC and abnormal TP53 function received a total of 83 intratumoral injections with Adp53 with the administration of intravenous cisplatin. As a result, 17 (71%) patients achieved the best clinical response of stable disease, two patients achieved a partial response, four patients had progressive disease, and one patient was not assessable [97]. Nineteen patients with NSCLC were treated with concurrent injection at the tumor site of Adp53 and curative thoracic radiotherapy (60Gy) in a clinical trial. Three months after completion of therapy, pathologic biopsies of the primary tumor revealed no viable tumor in 12 patients (63%), viable tumors in three patients (16%), and no assessment in four patients (21%). Computed tomography and bronchoscopic findings at the primary injected tumor revealed complete response in one patient (5%), partial response in 11 patients (58%), stable disease in three patients (16%), progressive disease in two patients (11%), and no evaluation in two patients (11%) [98]. As reported, reconstitution of the disorganized TP53 tumor suppressor pathway, such as the introduction of Adp53 into tumor cells, is one of the most promising novel concepts for improved cancer therapy as just described. More recently, several approaches for the identification of small molecules that target mutant TP53 have been applied, including rational design and screening of chemical libraries. RITA was originally identified in screening of the diversity set from the National Cancer Institute (NCI) for drugs able to suppress cell proliferation in a wild-type TP53-dependent manner. Upon binding to TP53, RITA reactivates it and induces apoptosis by disrupting the interaction with HDM-2 [99]. Therefore, RITA was proposed as a key molecule to target wild-type TP53 tumors that may prove resistant to drugs that rescue mutant TP53 function, such as PRIMA-1 (p53 reactivation and induction of massive apoptosis) [100]. Zhao et al. showed that the response elicited by RITA is dependent on the presence of mutant TP53 and that it is sustained by reactivation of the apoptotic machinery. They concluded that RITA is a promising lead for the development of anticancer drugs that reactivate the tumor suppressor function of TP53 in cancer cells, irrespective of whether they express the mutant or wild-type TP53 [101]. The compound PRIMA-1 has been identified as a low-molecular-weight compound that selectively inhibits the growth of tumor cells expressing mutant TP53. PRIMA-1 restores wild-type conformation to mutant TP53 by binding to the core and induces apoptosis in human tumor cells [102]. Furthermore, PRIMA-1MET/APR-246, a methylated structural analog of the compound PRIMA-1 that was identified by screening of a chemical library, can function synergistically with cisplatin or other commonly used anticancer drugs to induce tumor cell apoptosis and inhibition of human tumor xenograft growth in vivo in SCID mice [103]. Another study showed that PRIMA-1 alone did not trigger apoptosis but significantly reduced cell viability in human NSCLC cell lines carrying different TP53 proteins: A549 (p53wt), LX1 (p53R273H), and SKMes1 (p53R280K). In combination with adriamycin, PRIMA-1 strengthens the adriamycin-induced apoptosis in A549 and LX1 [104]. Adp53 gene therapy, as well as small molecules, such as PRIMA-1 and PRIMA-1MET, which can restore the transcriptional function of mutant TP53, or RITA, which interferes with MDM2-directed TP53 degradation, has been tested in a preclinical setting, and some of these approaches are currently in clinical development. Furthermore, novel TP53-based therapeutic strategies for NSCLC may be combined with conventional and molecular-targeting cancer therapy, including EGFR-TKI. The study of the somatic genetics of the TP53 pathway in cancer cells and tumors has proven useful in the development of targeted therapies and has shown that the TP53 mutational status can serve as an independent prognostic indicator in some types of cancers [105, 106].
5. Summary
As the molecular basis of lung carcinogenesis must be understood more fully and exploited to enhance survival rates of patients diagnosed with lung cancer, a deeper understanding of the major molecular alterations that occur in lung cancer, including TP53 gene mutation, is needed.
As a genetic alteration in the TP53 pathway is a common denominator to all cancers, an understanding of TP53 tumor suppressor activity has great potential for the development of novel therapeutic treatments of lung cancer. The use of a TP53 gene therapy for cancer cells will be improved with viruses that target only cancer cells or more efficient methods of gene delivery exclusively to the cancer cells.
References
- 1.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40 transformed cells. Nature. 1979;278(5701):261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 2.Linzer DIH, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40 transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17(1):43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 3.Kress M, May E, Cassingena R, May P. Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. Journal of Virology. 1979;31(2):472–483. doi: 10.1128/jvi.31.2.472-483.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melero JA, Stitt DT, Mangel WF, Carroll RB. Identification of new polypeptide species (48-55K) immunoprecipitable by antiserum to purified large T antigen and present in SV40-infected and -transformed cells. Virology. 1979;93(2):466–480. doi: 10.1016/0042-6822(79)90250-2. [DOI] [PubMed] [Google Scholar]
- 5.Munro AJ, Lain S, Lane DP. P53 abnormalities and outcomes in colorectal cancer: a systematic review. British Journal of Cancer. 2005;92(3):434–444. doi: 10.1038/sj.bjc.6602358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;16:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 7.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nature Reviews Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 8.McKinney C, Prives C. Regulation of p53 DNA Binding. 25 Years of p53 Research. Dordrecht, The Netherlands: Springer; 2005. [Google Scholar]
- 9.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 10.Jin S, Levine AJ. The p53 functional circuit. Journal of Cell Science. 2001;114(23):4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 11.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nature Reviews Molecular Cell Biology. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 12.Pomerantz J, Schreiber-Agus N, Liégeois NJ, et al. The Ink4a tumor suppressor gene product, p19(Arf), interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92(6):713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92(6):725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 14.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA:A Cancer Journal for Clinicians. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 15.Bodner SM, Minna JD, Jensen SM, et al. Expression of mutant p53 proteins in lung cancer correlates with the class of p53 gene mutation. Oncogene. 1992;7(4):743–749. [PubMed] [Google Scholar]
- 16.Takahashi T, Nau MM, Chiba I, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246(4929):491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 17.Hainaut P, Soussi T, Shomer B, et al. Database of p53 gene somatic mutations in human tumors and cell lines: updated compilation and future prospects. Nucleic Acids Research. 1997;25(1):151–157. doi: 10.1093/nar/25.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hainaut P, Wiman KG. 30 years and a long way into p53 research. The Lancet Oncology. 2009;10(9):913–919. doi: 10.1016/S1470-2045(09)70198-6. [DOI] [PubMed] [Google Scholar]
- 19.Hofseth LJ, Hussain SP, Harris CC. p53: 25 Years after its discovery. Trends in Pharmacological Sciences. 2004;25(4):177–181. doi: 10.1016/j.tips.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Scientific Publications. 2004;(157):247–270. [PubMed] [Google Scholar]
- 21.Soussi T, Ishioka C, Claustres M, Béroud C. Locus-specific mutation databases: pitfalls and good practice based on the p53 experience. Nature Reviews Cancer. 2006;6(1):83–90. doi: 10.1038/nrc1783. [DOI] [PubMed] [Google Scholar]
- 22.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor Perspectives in Biology. 2010;2(1) doi: 10.1101/cshperspect.a001008. Article ID a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harbor perspectives in biology. 2010;2(3) doi: 10.1101/cshperspect.a001016. Article ID a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soussi T. The p53 pathway and human cancer. British Journal of Surgery. 2005;92(11):1331–1332. doi: 10.1002/bjs.5177. [DOI] [PubMed] [Google Scholar]
- 25.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Human Mutation. 2002;19(6):607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 26.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 Mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 27.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Human Mutation. 2007;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 28.Li FP, Fraumeni JF, Mulvihill JJ, et al. A cancer family syndrome in twenty-four kindreds. Cancer Research. 1988;48(18):5358–5362. [PubMed] [Google Scholar]
- 29.Srivastava S, Zou Z, Pirollo K, Blattner W, Chang EH. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348(6303):747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 30.Olivier M, Goldgar DE, Sodha N, et al. Li-Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Research. 2003;63(20):6643–6650. [PubMed] [Google Scholar]
- 31.Ory K, Legros Y, Auguin C, Soussi T. Analysis of the most representative tumour-derived p53 mutants reveals that changes in protein conformation are not correlated with loss of transactivation or inhibition of cell proliferation. EMBO Journal. 1994;13(15):3496–3504. doi: 10.1002/j.1460-2075.1994.tb06656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Research. 2000;60(24):6788–6793. [PubMed] [Google Scholar]
- 33.Peart MJ, Prives C. Mutant p53 gain of function: the NF-Y connection. Cancer Cell. 2006;10(3):173–174. doi: 10.1016/j.ccr.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Petitjean A, Achatz MIW, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26(15):2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 35.Song H, Xu Y. Gain of function of p53 cancer mutants in disrupting critical DNA damage response pathways. Cell Cycle. 2007;6(13):1570–1573. doi: 10.4161/cc.6.13.4456. [DOI] [PubMed] [Google Scholar]
- 36.Donzelli S, Biagioni F, Fausti F, Strano S, Fontemaggi G, Blandino G. Oncogenomic approaches in exploring gain of function of mutant p53. Current Genomics. 2008;9(3):200–207. doi: 10.2174/138920208784340713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nature Reviews Cancer. 2009;9(10):701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 38.Goldstein I, Marcel V, Olivier M, Oren M, Rotter V, Hainaut P. Understanding wild-type and mutant p53 activities in human cancer: new landmarks on the way to targeted therapies. Cancer Gene Therapy. 2010;18(1):2–11. doi: 10.1038/cgt.2010.63. [DOI] [PubMed] [Google Scholar]
- 39.Pfeifer GP, Besaratinia A. Mutational spectra of human cancer. Human Genetics. 2009;125(5-6):493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinds PW, Finlay CA, Quartin RS, et al. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the "hot spot" mutant phenotypes. Cell Growth & Differentiation. 1990;1(12):571–580. [PubMed] [Google Scholar]
- 41.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Advances in Cancer Research. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 42.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21(48):7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 43.Perwez Hussain S, Hollstein MH, Harris CC. p53 tumor suppressor gene: at the crossroads of molecular carcinogenesis, molecular epidemiology, and human risk assessment. Annals of the New York Academy of Sciences. 2000;919:79–85. doi: 10.1111/j.1749-6632.2000.tb06870.x. [DOI] [PubMed] [Google Scholar]
- 44.Hainaut P, Pfeifer GP. Patterns of p53→T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22(3):367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- 45.Baumann M, Zips D, Appold S. Radiotherapy of lung cancer: technology meets biology meets multidisciplinarity. Radiotherapy and Oncology. 2009;91(3):279–281. doi: 10.1016/j.radonc.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Viktorsson K, De Petris L, Lewensohn R. The role of p53 in treatment responses of lung cancer. Biochemical and Biophysical Research Communications. 2005;331(3):868–880. doi: 10.1016/j.bbrc.2005.03.192. [DOI] [PubMed] [Google Scholar]
- 47.Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. European Respiratory Journal. 2001;18(4):705–719. doi: 10.1183/09031936.01.00062201. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi T, Takahashi T, Suzuki H, et al. The p53 gene is very frequently mutated in small-cell lung cancer with a distinct nucleotide substitution pattern. Oncogene. 1991;6(10):1775–1778. [PubMed] [Google Scholar]
- 49.Hensel CH, Xiang RH, Sagaguchi AY, Naylor SL. Use of the single strand conformation polymorphism technique and PCR to detect p53 gene mutations in small cell lung cancer. Oncogene. 1991;6(6):1067–1071. [PubMed] [Google Scholar]
- 50.Sameshima Y, Matsuno Y, Hirohashi S, et al. Alterations of the p53 gene are common and critical events for the maintenance of malignant phenotypes in small-cell lung carcinoma. Oncogene. 1992;7(3):451–457. [PubMed] [Google Scholar]
- 51.D’Amico D, Carbone D, Mitsudomi T, et al. High frequency of somatically acquired p53 mutations in small-cell lung cancer cell lines and tumors. Oncogene. 1992;7(2):339–346. [PubMed] [Google Scholar]
- 52.Kishimoto Y, Murakami Y, Shiraishi M, Hayashi K, Sekiya T. Aberrations of the p53 tumor suppressor gene in human non-small cell carcinomas of the lung. Cancer Research. 1992;52(17):4799–4804. [PubMed] [Google Scholar]
- 53.Tammemagi MC, McLaughlin JR, Bull SB. Meta-analyses of p53 tumor suppressor gene alterations and clinicopathological features in resected lung cancers. Cancer Epidemiology Biomarkers and Prevention. 1999;8(7):625–634. [PubMed] [Google Scholar]
- 54.Reichel MB, Ohgaki H, Petersen I, Kleihues P. p53 mutations in primary human lung tumors and their metastases. Molecular Carcinogenesis. 1994;9(2):105–109. doi: 10.1002/mc.2940090208. [DOI] [PubMed] [Google Scholar]
- 55.Miller CW, Simon K, Aslo A, et al. p53 Mutations in human lung tumors. Cancer Research. 1992;52(7):1695–1698. [PubMed] [Google Scholar]
- 56.Chiba I, Takahashi T, Nau MM, et al. Mutations in the p53 gene are frequent in primary, resected non-small cell lung cancer. Oncogene. 1990;5(10):1603–1610. [PubMed] [Google Scholar]
- 57.Sozzi G, Miozzo M, Pastorino U, et al. Genetic evidence for an independent origin of multiple preneoplastic and neoplastic lung lesions. Cancer Research. 1995;55(1):135–140. [PubMed] [Google Scholar]
- 58.Harris CC. p53 tumor suppressor gene: from the basic research laboratory to the clinic—an abridged historical perspective. Carcinogenesis. 1996;17(6):1187–1198. doi: 10.1093/carcin/17.6.1187. [DOI] [PubMed] [Google Scholar]
- 59.Chang Y-L, Wu C-T, Shih J-Y, Lee Y-C. Comparison of p53 and epidermal growth factor receptor gene status between primary tumors and lymph node metastases in non-small cell lung cancers. doi: 10.1245/s10434-010-1295-6. Annals of Surgical Oncology. In press. [DOI] [PubMed] [Google Scholar]
- 60.Mitsudomi T, Hamajima N, Ogawa M, Takahashi T. Prognostic significance of p53 alterations in patients with non-small cell lung cancer: a meta-analysis. Clinical Cancer Research. 2000;6(10):4055–4063. [PubMed] [Google Scholar]
- 61.Graziano SL, Gu L, Wang X, et al. Prognostic significance of mucin and P53 expression in stage ib non-small cell lung cancer: a laboratory companion study to CALGB 9633. Journal of Thoracic Oncology. 2010;5(6):810–817. doi: 10.1097/jto.0b013e3181d89f95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsao MS, Aviel-Ronen S, Ding K, et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non-small-cell lung cancer. Journal of Clinical Oncology. 2007;25(33):5240–5247. doi: 10.1200/JCO.2007.12.6953. [DOI] [PubMed] [Google Scholar]
- 63.Custodio AB, González-Larriba JL, Bobokova J, et al. Prognostic and predictive markers of benefit from adjuvant chemotherapy in early-stage non-small cell lung cancer. Journal of Thoracic Oncology. 2009;4(7):891–910. doi: 10.1097/JTO.0b013e3181a4b8fb. [DOI] [PubMed] [Google Scholar]
- 64.Kandioler D, Stamatis G, Eberhardt W, et al. Growing clinical evidence for the interaction of the p53 genotype and response to induction chemotherapy in advanced non-small cell lung cancer. Journal of Thoracic and Cardiovascular Surgery. 2008;135(5):1036–1041. doi: 10.1016/j.jtcvs.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 65.Doll R, Hill AB. Mortality in relation to smoking ten years’ observations of British doctors. British Medical Journal. 1964;1(5396):1460–1467. doi: 10.1136/bmj.1.5396.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chemical Research in Toxicology. 2008;21(1):160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koo LC, Ho JHC. Worldwide epidemiological patterns of lung cancer in nonsmokers. International Journal of Epidemiology. 1990;19(1):S14–S23. doi: 10.1093/ije/19.supplement_1.s14. [DOI] [PubMed] [Google Scholar]
- 68.Tyczynski JE, Bray F, Parkin DM. Lung cancer in Europe in 2000: epidemiology, prevention, and early detection. Lancet Oncology. 2003;4(1):45–55. doi: 10.1016/s1470-2045(03)00960-4. [DOI] [PubMed] [Google Scholar]
- 69.Tyczynski JE, Bray F, Aareleid T, et al. Lung cancer mortality patterns in selected Central, Eastern and Southern European countries. International Journal of Cancer. 2004;109(4):598–610. doi: 10.1002/ijc.20019. [DOI] [PubMed] [Google Scholar]
- 70.Tyczynski JE, Berkel HJ. Mortality from lung cancer and tobacco smoking in Ohio (U.S.): will increasing smoking prevalence reverse current decreases in mortality? Cancer Epidemiology Biomarkers and Prevention. 2005;14(5):1182–1187. doi: 10.1158/1055-9965.EPI-04-0699. [DOI] [PubMed] [Google Scholar]
- 71.Bennett WP, Alavanja MCR, Blomeke B, et al. Environmental tobacco smoke, genetic susceptibility, and risk of lung cancer in never-smoking women. Journal of the National Cancer Institute. 1999;91(23):2009–2014. doi: 10.1093/jnci/91.23.2009. [DOI] [PubMed] [Google Scholar]
- 72.Husgafvel-Pursiainen K, Boffetta P, Kannio A, et al. p53 Mutations and exposure to environmental tobacco smoke in a multicenter study on lung cancer. Cancer Research. 2000;60(11):2906–2911. [PubMed] [Google Scholar]
- 73.Takagi Y, Osada H, Kuroishi T, et al. p53 mutations in non-small-cell lung cancers occurring in individuals without a past history of active smoking. British Journal of Cancer. 1998;77(10):1568–1572. doi: 10.1038/bjc.1998.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vähäkangas KH, Bennett WP, Castrén K, et al. p53 and K-ras mutations in lung cancers from former and never-smoking women. Cancer Research. 2001;61(11):4350–4356. [PubMed] [Google Scholar]
- 75.Hainaut P, Olivier M, Pfeifer GP. TP53 mutation spectrum in lung cancers and mutagenic signature of components of tobacco smoke: lessons from the IARC TP53 mutation database. Mutagenesis. 2001;16:551–553. doi: 10.1093/mutage/16.6.551. [DOI] [PubMed] [Google Scholar]
- 76.Hussain SP, Harris CC. Molecular epidemiology of human cancer. Recent Results in Cancer Research. 1998;154:22–36. doi: 10.1007/978-3-642-46870-4_2. [DOI] [PubMed] [Google Scholar]
- 77.Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Human Mutation. 2003;21(3):229–239. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- 78.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocar. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Denissenko MF, Pao A, Tang MS, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274(5286):430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 80.Smith LE, Denissenko MF, Bennett WP, et al. Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. Journal of the National Cancer Institute. 2000;92(10):803–811. doi: 10.1093/jnci/92.10.803. [DOI] [PubMed] [Google Scholar]
- 81.Denissenko MF, Koudriakova TB, Smith L, O’Connor TR, Riggs AD, Pfeifer GP. The p53 codon 249 mutational hotspot in hepatocellular carcinoma is not related to selective formation or persistence of aflatoxin B adducts. Oncogene. 1998;17(23):3007–3014. doi: 10.1038/sj.onc.1202214. [DOI] [PubMed] [Google Scholar]
- 82.Hernandez-Boussard TM, Hainaut P. A specific spectrum of p53 mutations in lung cancer from smokers: review of mutations compiled in the IARC p53 database. Environmental Health Perspectives. 1998;106(7):385–391. doi: 10.1289/ehp.98106385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gealy R, Zhang L, Siegfried JM, Luketich JD, Keohavong P. Comparison of mutations in the p53 and K-ras genes in lung carcinomas from smoking and nonsmoking women. Cancer Epidemiology Biomarkers and Prevention. 1999;8:297–302. [PubMed] [Google Scholar]
- 84.Kondo K, Tsuzuki H, Sasa M, Sumitomo M, Uyama T, Monden Y. A dose-response relationship between the frequency of p53 mutations and tobacco consumption in lung cancer patients. Journal of Surgical Oncology. 1996;61(1):20–26. doi: 10.1002/(SICI)1096-9098(199601)61:1<20::AID-JSO6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 85.Subramanian J, Govindan R. Molecular genetics of lung cancer in people who have never smoked. The Lancet Oncology. 2008;9(7):676–682. doi: 10.1016/S1470-2045(08)70174-8. [DOI] [PubMed] [Google Scholar]
- 86.Gow CH, Chang YL, Hsu YC, et al. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Annals of Oncology. 2009;20(4):696–702. doi: 10.1093/annonc/mdn679. [DOI] [PubMed] [Google Scholar]
- 87.Fujiwara T, Grimm EA, Mukhopadhyay T, Zhang WW, Owen-Schaub LB, Roth JA. Induction of chemosensitivity in human lung cancer cells in vivo by adenovirus-mediated transfer of the wild-type p53 gene. Cancer Research. 1994;54(9):2287–2291. [PubMed] [Google Scholar]
- 88.Higashiyama M, Kodama K, Yokouchi H, et al. Immunohistochemical p53 protein status in nonsmall cell lung cancer is a promising indicator in determining in vitro chemosensitivity to some anticancer drugs. Journal of Surgical Oncology. 1998;68(1):19–24. doi: 10.1002/(sici)1096-9098(199805)68:1<19::aid-jso5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 89.Rusch V, Klimstra D, Venkatraman E, et al. Aberrant p53 expression predicts clinical resistance to cisplatin-based chemotherapy in locally advanced non-small cell lung cancer. Cancer Research. 1995;55(21):5038–5042. [PubMed] [Google Scholar]
- 90.Vogt U, Zaczek A, Klinke F, Granetzny A, Bielawski K, Falkiewicz B. p53 gene status in relation to ex vivo chemosensitivity of non-small cell lung cancer. Journal of Cancer Research and Clinical Oncology. 2002;128(3):141–147. doi: 10.1007/s00432-001-0305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leslie WT, Bonomi PD. Novel treatments in non-small cell lung cancer. Hematology/Oncology Clinics of North America. 2004;18(1):245–267. doi: 10.1016/s0889-8588(03)00146-1. [DOI] [PubMed] [Google Scholar]
- 92.Fujiwara T, De Wei Cai, Georges RN, Mukhopadhyay T, Crimm EA, Roth JA. Therapeutic effect of a retroviral wild-type p53 expression vector in an orthotopic lung cancer model. Journal of the National Cancer Institute. 1994;86(19):1458–1462. doi: 10.1093/jnci/86.19.1458. [DOI] [PubMed] [Google Scholar]
- 93.Roth JA, Nguyen D, Lawrence DD, et al. Retrovirus-mediated wild-type p53 gene transfer to tumors of patients with lung cancer. Nature Medicine. 1996;2(9):985–991. doi: 10.1038/nm0996-985. [DOI] [PubMed] [Google Scholar]
- 94.Wiman KG. Strategies for therapeutic targeting of the p53 pathway in cancer. Cell Death and Differentiation. 2006;13(6):921–926. doi: 10.1038/sj.cdd.4401921. [DOI] [PubMed] [Google Scholar]
- 95.Swisher SG, Roth JA, Nemunaitis J, et al. Adenovirus-mediated p53 gene transfer in advanced non-small-cell lung cancer. Journal of the National Cancer Institute. 1999;91(9):763–771. doi: 10.1093/jnci/91.9.763. [DOI] [PubMed] [Google Scholar]
- 96.Weill D, Mack M, Roth J, et al. Adenoviral-mediated p53 gene transfer to non-small cell lung cancer through endobronchial injection. Chest. 2000;118(4):966–970. doi: 10.1378/chest.118.4.966. [DOI] [PubMed] [Google Scholar]
- 97.Nemunaitis J, Swisher SG, Timmons T, et al. Adenovirus-mediated p53 gene transfer in sequence with cisplatin to tumors of patients with non-small-cell lung cancer. Journal of Clinical Oncology. 2000;18(3):609–622. doi: 10.1200/JCO.2000.18.3.609. [DOI] [PubMed] [Google Scholar]
- 98.Swisher SG, Roth JA, Komaki R, et al. Induction of p53-regulated genes and tumor regression in lung cancer patients after intratumoral delivery of adenoviral p53 (INGN 201) and radiation therapy. Clinical Cancer Research. 2003;9:93–101. [PubMed] [Google Scholar]
- 99.Issaeva N, Bozko P, Enge M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nature Medicine. 2004;10(12):1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 100.Selivanova G, Wiman KG. Reactivation of mutant p53: molecular mechanisms and therapeutic potential. Oncogene. 2007;26(15):2243–2254. doi: 10.1038/sj.onc.1210295. [DOI] [PubMed] [Google Scholar]
- 101.Zhao CY, Grinkevich VV, Nikulenkov F, Bao W, Selivanova G. Rescue of the apoptotic-inducing function of mutant p53 by small molecule RITA. Cell Cycle. 2010;9(9):1847–1855. doi: 10.4161/cc.9.9.11545. [DOI] [PubMed] [Google Scholar]
- 102.Bykov VJN, Issaeva N, Shilov A, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nature Medicine. 2002;8(3):282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 103.Bykov VJN, Zache N, Stridh H, et al. PRIMA-1 synergizes with cisplatin to induce tumor cell apoptosis. Oncogene. 2005;24(21):3484–3491. doi: 10.1038/sj.onc.1208419. [DOI] [PubMed] [Google Scholar]
- 104.Magrini R, Russo D, Ottaggio L, Fronza G, Inga A, Menichini P. PRIMA-1 synergizes with adriamycin to induce cell death in non-small cell lung cancer cells. Journal of Cellular Biochemistry. 2008;104(6):2363–2373. doi: 10.1002/jcb.21794. [DOI] [PubMed] [Google Scholar]
- 105.Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12(4):303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 106.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nature Reviews Drug Discovery. 2008;7(12):979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]