Abstract
Spirochaeta smaragdinae Magot et al. 1998 belongs to the family Spirochaetaceae. The species is Gram-negative, motile, obligately halophilic and strictly anaerobic and is of interest because it is able to ferment numerous polysaccharides. S. smaragdinae is the only species of the family Spirochaetaceae known to reduce thiosulfate or element sulfur to sulfide. This is the first complete genome sequence in the family Spirochaetaceae. The 4,653,970 bp long genome with its 4,363 protein-coding and 57 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: spiral shaped, corkscrew-like motility, chemoorganotroph, strictly anaerobe, obligately halophile, rhodanese-like protein, Spirochaetaceae, GEBA
Introduction
Strain SEBR 4228T (= DSM 11293 = JCM 15392) is the type strain of the species Spirochaeta smaragdinae. Currently, there are eighteen species [1] and two subspecies in the genus Spirochaeta [1,2]. The generic name derives from the Greek word ‘speira’ meaning ‘a coil’ and the Greek word ‘chaitê’ meaning ‘hair’, referring to the spiral shape of bacterial cell. The species epithet is derived from the Latin word ‘smaragdinae’ meaning ‘from Emerald’, referring to the name Emerald of an oil field in Congo. Strain SEBR 4228T was isolated from an oil-injection production water sample of a Congo offshore oilfield [3] and described in 1997 by Magot et al. as ‘Spirochaeta smaragdinae’ [3]. Here we present a summary classification and a set of features for S. smaragdinae SEBR 4228T, together with the description of the complete genomic sequencing and annotation.
Classification and features
Strain SEBR 4228T shares 82.2-99.0% 16S rRNA gene sequence identity with the type strains from the other members of genus Spirochaeta [4], with the type strain of S. bajacaliforniensis [5], isolated from a mud sample in Laguna Figueroa (Baja California, Mexico) showing the highest degree of sequence similarity (99%). Notwithstanding the high degree of 16S rRNA gene sequence identity, these two strains are characterized by low genomic similarity (38%) in DNA-DNA hybridization studies and differ by numerous differences in carbon source utilization [3]. Several type strains from the genus Treponema show the highest degree of similarity for non-Spirochaeta strains (82.9-83.6%) [4]. A representative genomic 16S rRNA sequence of strain SEBR 4228T was compared using BLAST with the most recent release of the Greengenes database [6] and the relative frequencies of taxa and keywords, weighted by BLAST scores, were determined. The three most frequent genera were Spirochaeta (76.4%), ‘Sphaerochaeta’ (15.8%) and Cytophaga (7.8%). Within the five most frequent keywords in the labels of environmental samples were 'microbial' (11.7%), 'mat' (10.5%), 'hypersaline' (7.7%), and 'sediment' (1.7%). The environmental samples database (env_nt) contains the marine metagenome genomic clone 1061006082084 (EK988302) that is 92% identical to the 16S rRNA gene sequence of SEBR 4228T. No phylotypes from genomic surveys could be linked to the species S. smaragdinae or even the genus Spirochaeta, indicating a rather rare occurrence of these in the habitats screened so far (as of August 2010).
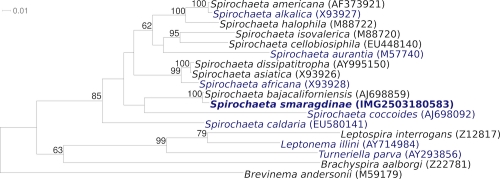
Figure 1 shows the phylogenetic neighborhood of S. smaragdinae SEBR 4228T in a 16S rRNA based tree. The sequences of the two 16S rRNA gene copies differ from each other by up to one nucleotide, and differ by up to five nucleotides from the previously published 16S rRNA sequence generated from DSM 11293 (U80597), which contains two ambiguous base calls.
Figure 1.
Phylogenetic tree highlighting the position of S. smaragdinae SEBR 4228T relative to the type strains of the other species within the genus and of the other genera within the genus Spirochaeta. The tree was inferred from 1,385 aligned characters [7,8] of the 16S rRNA gene sequence under the maximum likelihood criterion [9] and rooted in accordance with the current taxonomy [10]. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 500 bootstrap replicates [11] if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [12] are shown in blue, published genomes in bold.
Strain SEBR 4228T is a Gram-negative, chemoorganotrophic and strictly anaerobic bacterium with spiral shaped, 0.3-0.5 × 5-30 μm long cells (Figure 2 and Table 1). It possesses a multilayer, crenulating, Gram-negative cell envelope, which consists of an outer membrane and an inner membrane adjoining the cytoplasmic membrane [3]. Sillons, which are the contact point between the protoplasmic cylinder, the inner membrane and the outer membrane, are also observed from the cells of S. smaragdinae SEBR 4228T [3]. Strain SEBR 4228T forms translucent colonies with regular edges (0.5 mm of diameter) after two weeks of incubation on SEM agar plates at 37°C [3]. The strain is motile with a corkscrew-like motion, which is characteristic for the typical 1-2-1 periplasmic flagellar arrangement of the members of the genus Spirochaeta [3]. The periplasmic, non-extracellular location of the flagella make the Spirochaeta a valuable candidate for the study of flagella evolution [26]. The enlarged spherical bodies, which are typical for spirochetes, are also observed in strain SEBR 4228T [3]. The temperature range for growth is from 20°C to 40°C, with an optimum temperature at 37°C [3]. The pH range for growth is between 5.5 and 8.0, with an optimum pH of 7.0 [3]. Strain SEBR 4228T is obligately halophilic [3] and is able to grow on media that contains 1-10% of NaCl, with an optimum salinity at 5% NaCl [3]. Under optimum growth conditions, the doubling time is approximately 25 h in the presence of glucose and thiosulfate [3]. Strain SEBR 4228T is able to utilize biotrypcase, fructose, fumarate, galactose, D-glucose, glycerol, mannitol, mannose, ribose, D-xylose and yeast extract, but not acetate, D-arabinose, butyrate, casamino acids, lactate, maltose, propionate, pyruvate, rhamnose, sorbose, sucrose and L-xylose [3]. Yeast extract is required for growth and cannot be replaced by a vitamin mixture [3]. Strain SEBR 4228T ferments fumarate to acetate and succinate [3]. The major end-product of glucose fermentation of strain SEBR 4228T is lactate with traces of H2 and ethanol [3]. S. smaragdinae is the only species of Spirochaeta known to reduce thiosulfate or elemental sulfur to sulfide [3]. Strain SEBR 4228T produces lactate, acetate, CO2 and H2S as the end-products of glucose oxidation when thiosulfate is present in the growth medium [3]. The strain contains a rhodanese-like protein which expresses rhodanese activity [27]. This enzyme is able to reduce thiosulfate to sulfide [28]. Rhodanese is also widely found in other members of the domain Bacteria [29-31].
Figure 2.
Scanning electron micrograph of S. smaragdinae SEBR 4228T
Table 1. Classification and general features of S. smaragdinae SEBR 4228T according to the MIGS recommendations [13].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [14] | |
| Phylum Spirochaetae | TAS [15,16] | ||
| Class Spirochaetes | TAS [16] | ||
| Order Spirochaetales | TAS [17,18] | ||
| Family Spirochaetaceae | TAS [18,19] | ||
| Genus Spirochaeta | TAS [18,20-22] | ||
| Species Spirochaeta smaragdinae | TAS [3,23] | ||
| Type strain SEBR 4228 | TAS [3] | ||
| Gram stain | negative | TAS [3] | |
| Cell shape | spiral | TAS [3] | |
| Motility | yes | TAS [3] | |
| Sporulation | none | NAS | |
| Temperature range | between 20°C and over 40°C | TAS [3] | |
| Optimum temperature | 37°C | TAS [3] | |
| Salinity | 1-10% NaCl (optimum 5%) | TAS [3] | |
| MIGS-22 | Oxygen requirement | obligately anaerobic | TAS [3] |
| Carbon source | polysaccharides | TAS [3] | |
| Energy source | chemoorganotroph | TAS [3] | |
| MIGS-6 | Habitat | oil-fields | TAS [3] |
| MIGS-15 | Biotic relationship | free-living | TAS [3] |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [24] | |
| Isolation | oil-injection water sample in the production system of an oil field |
TAS [3] | |
| MIGS-4 | Geographic location | Emerald oil fields in Congo | TAS [3] |
| MIGS-5 | Sample collection time | 1997 or before | TAS [3] |
| MIGS-4.1 | Latitude | not reported | |
| MIGS-4.2 | Longitude | not reported | |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [25]. If the evidence code is IDA, then the property was directly observed by one of the authors or an expert mentioned in the acknowledgements
Chemotaxonomy
No cellular fatty acids profiles are currently available for S. smaragdinae SEBR 4228T. However, C16:0 dimethyl acetate is the major cellular fatty acids of the type strains of the closely related S. dissipatitropha, S. asiatica and S. americana, and C16:0 fatty acid methyl ester is the major cellular fatty acids of S. africana [20,32].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [33], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [34]. The genome project is deposited in the Genome OnLine Database [12] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Three genomic libraries: 454 pyrosequence standard and PE (12 kb insert size) libraries and one Illumina standard library |
| MIGS-29 | Sequencing platforms | 454 GS FLX Titanium, Illumina GAii |
| MIGS-31.2 | Sequencing coverage | 58.8 × pyrosequence, 6.9 × Illumina |
| MIGS-30 | Assemblers | Newbler version 2.0.0-PostRelease- 11/04/2008, phrap, |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| INSDC ID | CP002116 | |
| Genbank Date of Release | August 6, 2010 | |
| GOLD ID | Gc013354 | |
| NCBI project ID | 32637 | |
| Database: IMG-GEBA | 2503128010 | |
| MIGS-13 | Source material identifier | DSM 11293 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
S. smaragdinae SEBR 4228T, DSM 11293, was grown anaerobically in medium 819 (Spirochaeta smaragdinae medium) [35] at 35°C. DNA was isolated from 0.5-1 g of cell paste using MasterPure Gram Positive DNA Purification Kit (Epicentre MGP04100) following the standard protocol as recommended by the manufacturer, with modification st/LALMice for cell lysis as described in Wu et al. [34].
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website (http://www.jgi.doe.gov/). Pyrosequencing reads were assembled using the Newbler assembler version 2.0.0-PostRelease-11/04/2008 (Roche). The initial Newbler assembly consisted of 51 contigs in one scaffold was converted into a phrap assembly by making fake reads from the consensus, collecting the read pairs in the 454 paired end library. Illumina GAii sequencing data was assembled with Velvet [36] and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. Draft assemblies were based on 273 Mb 454 draft data and all of the 454 paired end data. Newbler parameters are -consed -a 50 -l 350 -g -m -ml 20.
The Phred/Phrap/Consed software package (www.phrap.com) was used for sequence assembly and quality assessment in the following finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolution (http://www.jgi.doe.gov/), Dupfinisher, or sequencing cloned bridging PCR fragments with subcloning or transposon bombing (Epicentre Biotechnologies, Madison, WI) [37]. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR primer walks (J.-F.Chang, unpublished). A total of 147 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. Illumina reads were also used to improve the final consensus quality using an in-house developed tool - the Polisher [38]. The error rate of the completed genome sequence is 0.2 in 100,000. Together, the combination of the Illumina and 454 sequencing platforms provided 65.7× coverage of the genome.
Genome annotation
Genes were identified using Prodigal [39] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [40]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [41].
Genome properties
The genome consists of a 4,653,970 bp long chromosome with a 49.0% GC content (Table 3 and Figure 3). Of the 4,363 genes predicted, 4,306 were protein-coding genes, and 57 RNAs; eighty seven pseudogenes were also identified. The majority of the protein-coding genes (74.2%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 4,653,970 | 100.00% |
| DNA coding region (bp) | 4,315,215 | 92.97% |
| DNA G+C content (bp) | 2,278,823 | 48.97% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 4,363 | 100.00% |
| RNA genes | 57 | 1.31% |
| rRNA operons | 2 | |
| Protein-coding genes | 4306 | 98.69% |
| Pseudo genes | 87 | 1.99% |
| Genes with function prediction | 3,235 | 74.15% |
| Genes in paralog clusters | 818 | 18.75% |
| Genes assigned to COGs | 3,318 | 76.05% |
| Genes assigned Pfam domains | 3,443 | 78.91% |
| Genes with signal peptides | 871 | 26.36% |
| Genes with transmembrane helices | 1,150 | 22.45% |
| CRISPR repeats | 1 |
Figure 3.
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | Value | %age | Description |
|---|---|---|---|
| J | 159 | 4.3 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 328 | 8.8 | Transcription |
| L | 129 | 3.5 | Replication, recombination and repair |
| B | 1 | 0.0 | Chromatin structure and dynamics |
| D | 25 | 0.7 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 58 | 1.6 | Defense mechanisms |
| T | 321 | 8.6 | Signal transduction mechanisms |
| M | 183 | 4.9 | Cell wall/membrane/envelope biogenesis |
| N | 94 | 2.5 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 58 | 1.6 | Intracellular trafficking and secretion, and vesicular transport |
| O | 114 | 3.1 | Posttranslational modification, protein turnover, chaperones |
| C | 223 | 6.0 | Energy production and conversion |
| G | 553 | 14.9 | Carbohydrate transport and metabolism |
| E | 326 | 8.8 | Amino acid transport and metabolism |
| F | 96 | 2.6 | Nucleotide transport and metabolism |
| H | 130 | 3.5 | Coenzyme transport and metabolism |
| I | 61 | 1.6 | Lipid transport and metabolism |
| P | 165 | 4.4 | Inorganic ion transport and metabolism |
| Q | 30 | 0.8 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 450 | 12.1 | General function prediction only |
| S | 212 | 5.7 | Function unknown |
| - | 1,045 | 23.9 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Maren Schröder (DSMZ) for growing cultures of S. smarasgdinae. This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle, and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-1 and Thailand Research Fund Royal Golden Jubilee Ph.D. Program No. PHD/0019/2548 for MY.
References
- 1.Garrity G. NamesforLife. BrowserTool takes expertise out of the database and puts it right in the browser. Microbiol Today 2010; 37:9 [Google Scholar]
- 2.Euzéby JP. List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet. Int J Syst Bacteriol 1997; 47:590-592 10.1099/00207713-47-2-590 [DOI] [PubMed] [Google Scholar]
- 3.Magot M, Fardeau ML, Arnauld O, Lanau C, Ollivier B, Thomas P, Patel BK. Spirochaeta smaragdinae sp. nov., a new mesophilic strictly anaerobic spirochete from an oil field. FEMS Microbiol Lett 1997; 155:185-191 10.1111/j.1574-6968.1997.tb13876.x [DOI] [PubMed] [Google Scholar]
- 4.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 2007; 57:2259-2261 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- 5.Fracek SP, Jr, Stolz JF. Spirochaeta bajacaliforniensis sp. n. from a microbial mat community at Laguna Figueroa, Baja California Norte, Mexico. [RRS] Arch Microbiol 1985; 142:317-325 10.1007/BF00491897 [DOI] [PubMed] [Google Scholar]
- 6.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 8.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 9.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 10.Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Glöckner FO, Rosselló-Móra R. The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 2008; 31:241-250 10.1016/j.syapm.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 11.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [PubMed] [Google Scholar]
- 12.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346-D354 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 16.Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol 2002; 52:7-76 [DOI] [PubMed] [Google Scholar]
- 17.Buchanan RE. Studies in the nomenclature and classification of bacteria. II. The primary subdivisions of the Schizomycetes. J Bacteriol 1917; 2:155-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 19.Swellengrebel NH. Sur la cytologie comparée des spirochètes et des spirilles. Ann Inst Pasteur (Paris) 1907; 21:562-586 [Google Scholar]
- 20.Pikuta EV, Hoover RB, Bej AK, Marsic D, Whitman WB, Krader P. Spirochaeta dissipatitropha sp. nov., an alkaliphilic, obligately anaerobic bacterium, and emended description of the genus Spirochaeta Ehrenberg 1835. Int J Syst Evol Microbiol 2009; 59:1798-1804 [DOI] [PubMed] [Google Scholar]
- 21.Ehrenberg CG. Dritter Beitrag zur Erkenntniss grosser Organisation in der Richtung des kleinsten Raumes Abhandlungen der Preussischen Akademie der Wissenschaften (Berlin) 1835:143-336. [Google Scholar]
- 22.Canale-Parola E. Genus I. Spirochaeta Ehrenberg 1835, 313. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 168-171 [The Williams and Wilkins Co., Baltimore.]. [Google Scholar]
- 23.Validation List No 67. Validation of publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1998; 48:1083-1084 10.1099/00207713-48-4-1083 [DOI] [PubMed] [Google Scholar]
- 24.Classification of bacteria and archaea in risk groups. TRBA 466. [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder LAS, Loman NJ, Fütterer K, Pallen MJ. Bacterial flagellar diversity and evolution: seek simplicity and distrust it? Trends Microbiol 2009; 17:1-5 10.1016/j.tim.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 27.Ravot G, Casalot L, Ollivier B, Loison G, Magot M. rdlA, a new gene encoding a rhodanese-like protein in Halanaerobium congolense and other thiosulfate-reducing anaerobes. Res Microbiol 2005; 156:1031-1038 10.1016/j.resmic.2005.05.009 [DOI] [PubMed] [Google Scholar]
- 28.Singleton DR, Smith DW. Improved Assay for Rhodanese in Thiobacillus spp. Appl Environ Microbiol 1988; 54:2866-2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crolet JL, Magot M. Non-SRB sulfidogenic bacteria in oilfield production facilities. Mater Perform 1996; 35:60-64 [Google Scholar]
- 30.Magot M, Ravot G, Campaignolle X, Ollivier B, Patel BK, Fardeau ML, Thomas P, Crolet JL, Garcia JL. Dethiosulfovibrio peptidovorans gen. nov., sp. nov., a new anaerobic, slightly halophilic, thiosulfate-reducing bacterium from corroding offshore oil wells. Int J Syst Bacteriol 1997; 47:818-824 10.1099/00207713-47-3-818 [DOI] [PubMed] [Google Scholar]
- 31.Ravot G, Magot M. 2000. Detecting sulfate-reducing bacteria utilizing the APS-reductase gene patent GB 2354072 A.
- 32.Hoover RB, Pikuta EV, Bej AK, Marsic D, Whitman WB, Tang J, Krader P. Spirochaeta americana sp. nov., a new haloalkaliphilic, obligately anaerobic spirochaete isolated from soda Mono Lake in California. Int J Syst Evol Microbiol 2003; 53:815-821 10.1099/ijs.0.02535-0 [DOI] [PubMed] [Google Scholar]
- 33.Klenk HP, Goeker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 34.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.List of growth media used at DSMZ: http//www.dsmz.de/microorganisms/media_list.php
- 36.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sims D, Brettin T, Detter JC, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008. [Google Scholar]
- 39.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 41.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]