Abstract
A FRET assembly reports antibiotic affinities to two different RNA targets. A binder was labeled with a fluorophore that acts both as an acceptor for the emissive nucleoside on the bacterial A-site and a donor fluorophore for the terminally-labeled human A-site. Unlabeled drugs were used to dissociate the labeled antibiotic.
The bacterial ribosome is targeted by the majority of diverse and clinically significant antibiotics, of both natural and synthetic origin.1,2 The fundamental role and abundance of this translational ribonucleoprotein machinery in every cell make it an obvious target from evolutionary and functional perspectives, but it presents a formidable challenge for the discovery and development of new antibacterials.2,3 In particular, the similarity between functional rRNA sites in prokaryotes and eukaryotes as well as between naïve and resistant bacteria could significantly limit the therapeutic potential of new agents. While numerous factors influence the efficacy and adverse effects of any drug, its affinity to competing targets is of fundamental significance. The ability to discern the inherent target selectivity of existing and candidate antibiotics could therefore critically impact the discovery and development of new agents.
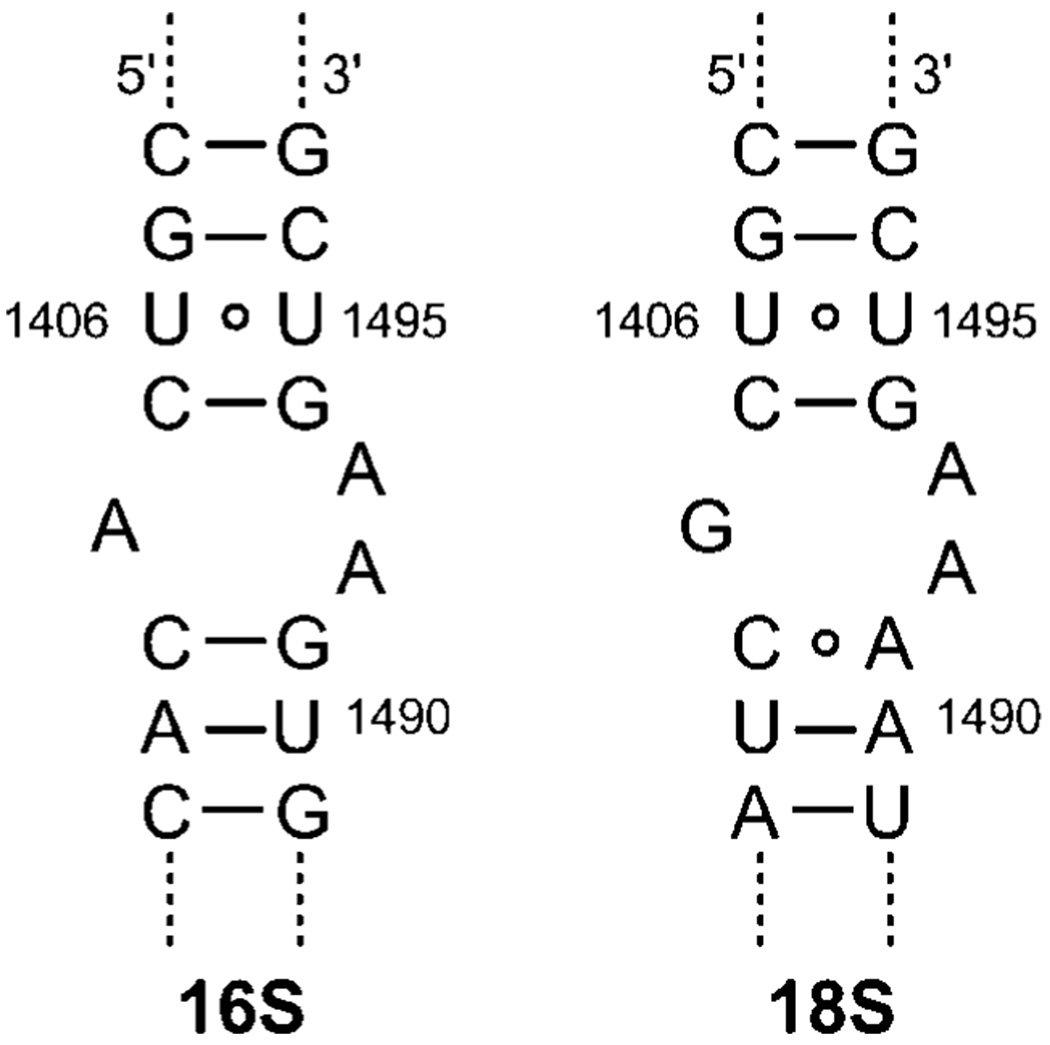
Among the most commonly targeted ribosomal sites, the decoding (or A-site) rRNA is of particular significance.4 It acts as a conformational switch that gauges codon–anticodon recognition.5 Altering its conformational dynamics by bound aminoglycosides, a large family of potent naturally occurring antibiotics, lowers the fidelity of protein synthesis, leading to bacterial cell death.5,6 Although several “mutations” distinguish the prokaryotic 16S decoding site from the corresponding eukaryotic 18S sequence (Fig. 1), biochemical and structural studies illustrate their similarity as well as their ability to bind aminoglycosides.7 It has also been suggested that the clinical value of aminoglycosides could potentially depend on their ability to differentiate between two closely related targets.8,9 We therefore sought out a straightforward approach to determine the selectivity traits of A-site binders. Here we disclose the design and implementation of a FRET-based, three-component assembly that facilitates a rapid determination of the relative affinity of any given binder to the eukaryotic and prokaryotic decoding sites in a single experiment.
Fig. 1.
Sequences of the bacterial (16S) and human (18S) A-sites.
To accomplish this task, we have relied on the following components: (1) an aminoglycoside with modest affinity to both the prokaryotic and eukaryotic A-sites (e.g., kanamycin A), labeled with a small non-perturbing fluorophore (marked F2) at a position that is not essential for rRNA binding; (2) a bacterial 16S A-site RNA construct modified with an isomorphic, emissive nucleoside analog (labeled F1) at a position proximal to the binding site, but not part of it; and (3) a human 18S A-site rRNA construct labeled at its terminus with a third fluorophore (designated F3). To generate unique spectral signatures for each binding event, the following photophysical conditions had to be met: (1) the isomorphic fluorescent probe on the bacterial A-site construct (F1) had to exclusively serve as a FRET donor to the fluorophore placed on the aminoglycoside antibiotic (F2); and (2) the latter, in turn, had to specifically serve as a FRET donor for the terminal fluorophore on the human A-site (F3).
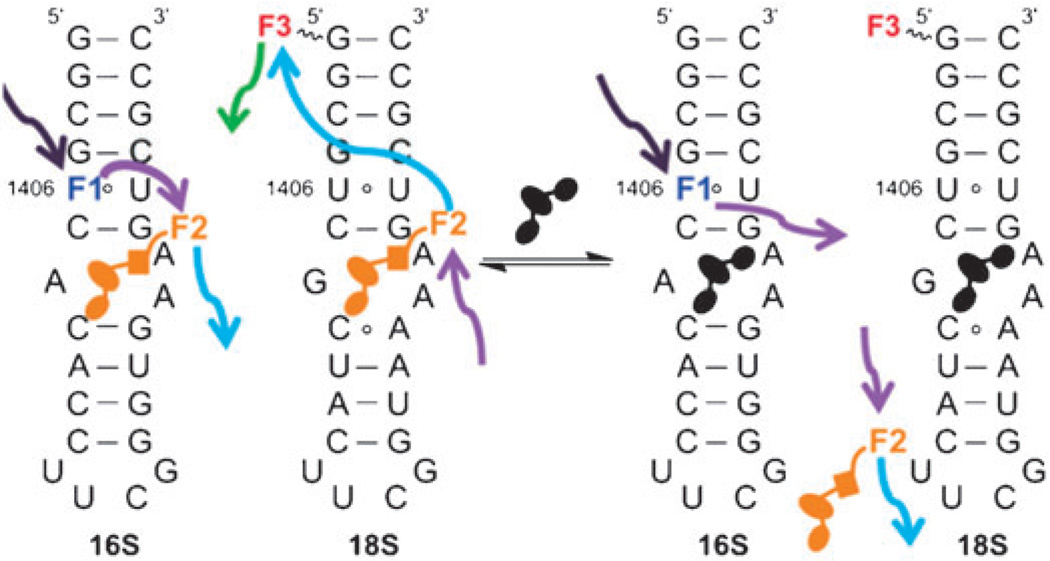
The experiment is conceptually illustrated in Fig. 2. When all three components, the tagged aminoglycoside and the two RNA constructs, are equilibrated together, the presence of the antibiotic on the 16S RNA can be visualized by selectively exciting F1, and monitoring the emission of F2. The fraction of the ligand bound to the 18S A-site can be visualized by selectively exciting F2 and detecting the emission of the acceptor F3. More importantly, when an unlabeled competitor small-molecule is added to the mixture and displaces the tagged antibiotic from the 16S A-site, the acceptor emission F2 is lost, while the fluorescence of the donor nucleoside F1 is recovered. Accordingly, when the “placeholder” tagged antibiotic is displaced from the 18S A-site, the emission of F2 is recovered, and the sensitized emission of F3 is lost. Based on the relative changes in these spectral signatures, measured in one cuvette by following two different emission spectra, the affinity and selectivity of any candidate antibiotic can be determined, as it displaces the tagged ligand on the two related A-sites, according to its inherent selectivity.
Fig. 2.
Secondary structures for the 27-base RNA models of the 16S and 18S A-sites. U1406 of the 16S A-site is replaced with an isosteric emissive nucleoside analogue as a donor (F1); the place-holding molecule is tagged with an appropriate fluorophore (F2); the 18S A-site is tagged with an acceptor (F3) to match the labeled “place-holder” (F2). The affinity and selectivity of unlabeled small-molecules for either A-sites can be accurately monitored using FRET, as the place-holder is displaced.
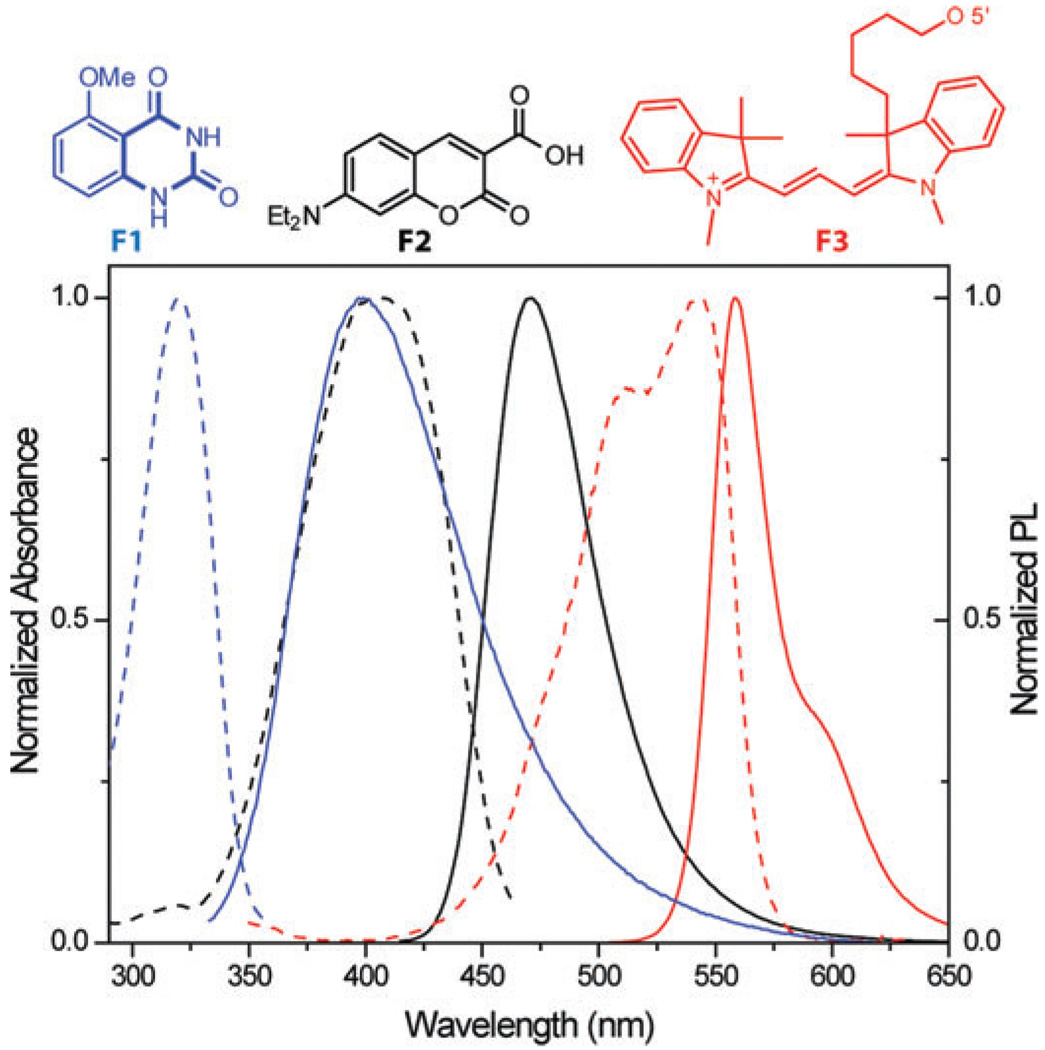
The selection of the two orthogonal, yet matched, FRET pairs is critical to the success of this experiment. We identified 5-methoxyquinazoline-2,4-(1H,3H)-dione F1, an emissive uracil analogue, serving as a suitable donor for 7-diethylaminocoumarin-3-carboxylic acid F2 (Fig. 3).10 We also found that Dy547 F3 is a fitting acceptor for F2. The absorption maximum of F1 at 320 nm corresponds to a wavelength with a minimal absorbance of F2, while the emission of F1, centered at 395 nm (ΦF = 0.16), overlaps perfectly with the absorption band of F2, which emits at 473 nm (ΦF = 0.83) (Fig. 3).10 The emission of F2, in turn, overlaps with the absorption band of F3, which exhibits absorption maxima at 516 and 549 nm and emits at 563 nm (ΦF = 0.27). Importantly, the molar extinction coefficients of F3 are negligible at the absorption maxima of F1 and F2. The critical Förster radii for the F1/F2 (Ro = 27 Å) and F2/F3 (Ro = 45A Å) pairs are apt for the proposed experiments.11 According to previous results12 and control experiments,13 the replacement of U1406 in the 16S A-site by a fluorescent nucleoside does not significantly impact the antibiotic affinity to the model construct.
Fig. 3.
Structures of F1 (blue), F2 (black), and F3 (red) along with their normalized absorption (- - -) and emission spectra (—) in water.
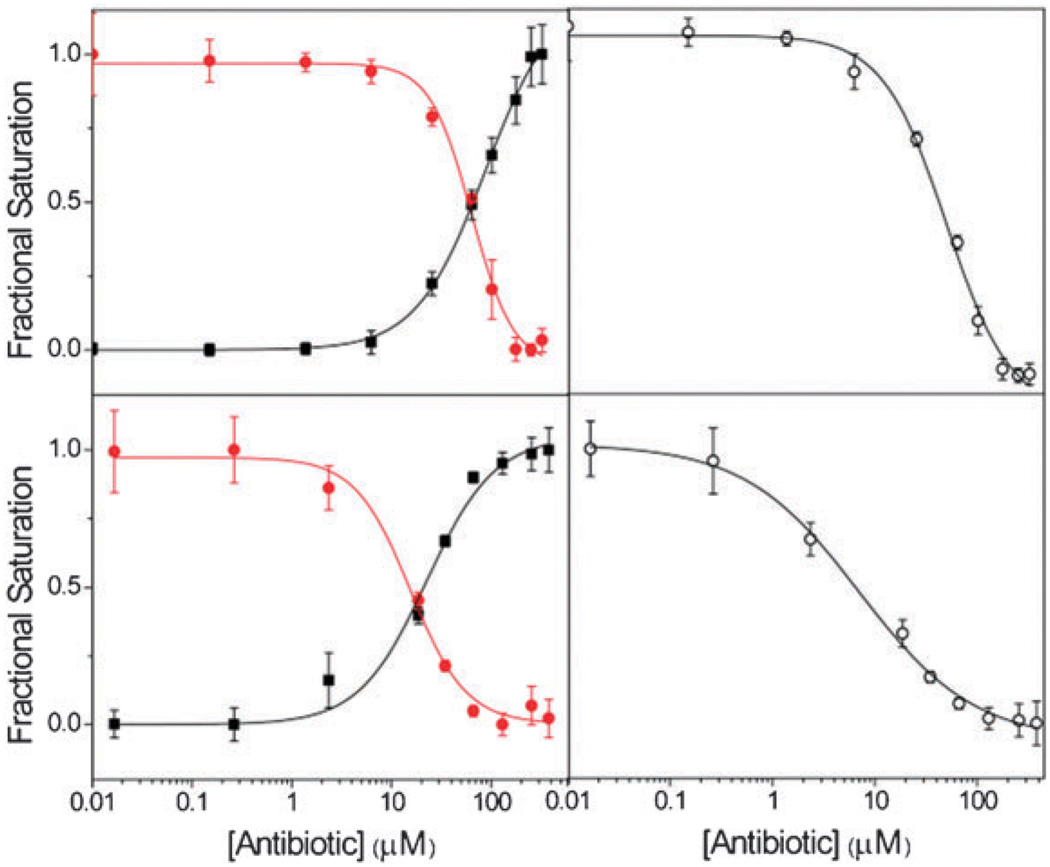
For competition studies, the two A-site constructs were pre-folded separately, mixed together and then treated with a two-mole equivalent of coumarin labeled kanamycin A (Fig. 4).13 Diverse antibiotics were titrated into this mixture of the kanamycin-bound A-sites. These include several aminoglycosides, semisynthetic aminoglycosides, as well as a macrolide, a peptide and an oxazolidinone-based antibiotic (Fig. 4). The relative emissions of all fluorophores were independently recorded. As productive competition on the 16S A-site advances, the emission intensity of the fluorescent nucleobase F1 increased, while the emission of the coumarin F2 was lost (Fig. 5).13 For the 18S A-site, the emission intensity of the Dy547 F3 was reduced. Plotting the fractional fluorescence saturation against the concentration of the competitors yields titration curves (Fig. 5).13 Table 1 provides the IC50 values for both the 16S and 18S A-sites and the selectivity ratio.
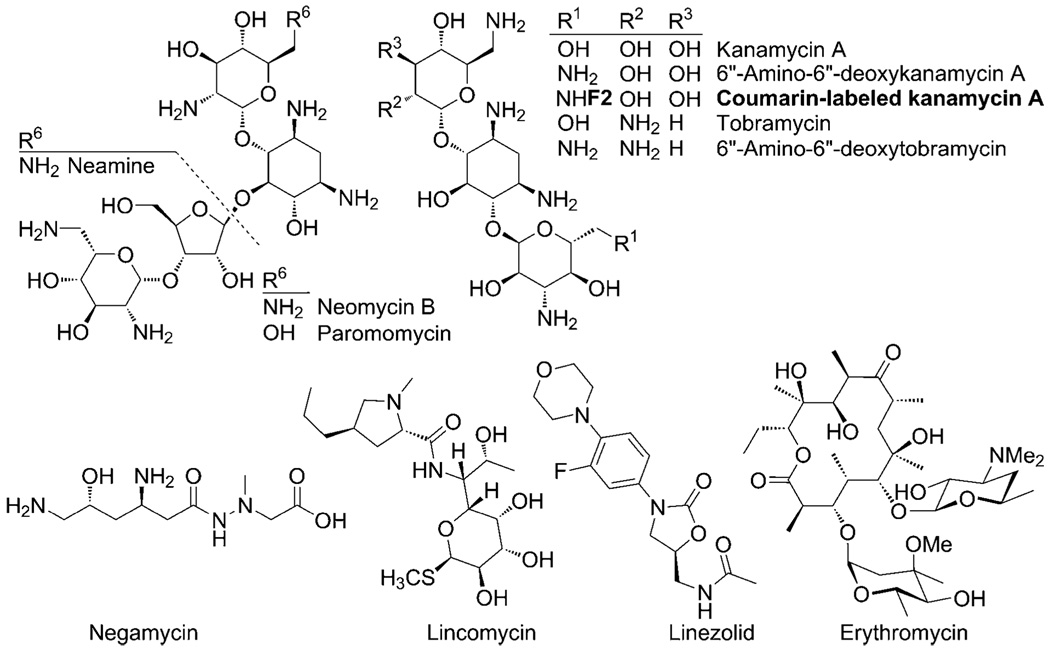
Fig. 4.
rRNA targeting antibiotics studied.
Fig. 5.
Fractional fluorescence saturation of the donor F1 (■) in the labeled 16S A-site, the emissive fluorophore F2 ( ) tagged to kanamycin A, and the emissive acceptor F3 (○) of the 18S A-site in studying the binding of: (top) negamycin; (bottom) neamine. Conditions: 16S RNA (5 × 10−7 M), 18S RNA (5 × 10−7 M), coumarin-labeled-kanamycin A (2.2 × 10−6 M), cacodylate buffer pH 7.0 (2.0 × 10−2 M), NaCl (1.0 × 10−1 M).13
) tagged to kanamycin A, and the emissive acceptor F3 (○) of the 18S A-site in studying the binding of: (top) negamycin; (bottom) neamine. Conditions: 16S RNA (5 × 10−7 M), 18S RNA (5 × 10−7 M), coumarin-labeled-kanamycin A (2.2 × 10−6 M), cacodylate buffer pH 7.0 (2.0 × 10−2 M), NaCl (1.0 × 10−1 M).13
Table 1.
IC50 values of antibiotics for the 16S and 18S A-sitesa
| Antibiotics | 16S A-Site/ 10−6 M |
18S A-Site/ 10−6 M |
Selectivity ratio |
|---|---|---|---|
| Neomycin B | 2.8 (± 0.3) | 4.7 (± 0.2) | 0.60 |
| Tobramycin | 20.2 (± 0.4) | 19.5 (± 0.3) | 1.0 |
| Paromomycin | 9 (± 1) | 8.0 (± 0.6) | 1.1 |
| Kanamycin A | 75 (± 3) | 46 (± 2) | 1.6 |
| Amino-tobramycin | 4.2 (± 0.4) | 3.8 (± 0.4) | 1.1 |
| Amino-kanamycin A | 11.9 (± 0.4) | 4.7 (± 0.3) | 2.5 |
| Negamycin | 62 (± 5) | 42 (± 3) | 1.5 |
| Neamine | 18 (± 2) | 6 (± 1) | 3 |
| Erythromycin | 1880 (± 10) | 1750 (± 10) | 1.1 |
| Lincomycin | > 8.5 × 103 | > 8.5 × 103 | — |
| Linezolid | > 9.6 × 103 | > 9.6 × 103 | — |
Conditions as listed in Fig. 5.
In agreement with reported trends, aminoglycoside antibiotics, including neomycin, tobramycin and paromomycin, do not display a dramatic preference for either the bacterial or human A-sites (Table 1).7c,9,13–15 Among all aminoglycosides tested, neomycin B is the only antibiotic that displays selectivity for the prokaryotic 16S A-site. Neamine, the core aminoglycosidic pharmacophore, binds quite strongly to the eukaryotic A-site. Its affinity to the latter is comparable to that of neomycin, yet its affinity to the prokaryotic A-site is significantly lower than neomycin’s and is comparable to that of tobramycin. Notably, the loss of the neobiosamine moiety from neomycin lowers the preference for the prokaryotic A-site. Two semi-synthetic amino-aminoglycoside derivatives were also tested. Amino-tobramycin and amino-kanamycin A,16 while displaying higher affinity for both A-sites compared to the parent antibiotics, still prefer the 18S RNA, with amino-kanamycin A displaying a higher 16S/18S selectivity ratio.
Negamycin, a dipeptide antibiotic, is an active bactericidal compound discovered in the 1970s.17 Interestingly, little is certain about its mode of action. While originally identified as a protein synthesis inhibitor with enhanced miscoding activity, suggestive of A-site binding,18 a recent crystal structure implied that its bactericidal potency could result from its binding to the wall of the peptide exit tunnel of the large ribosomal subunit.19 Our data show that negamycin indeed binds both A-sites with affinities similar to that of kanamycin A. Finally, a macrolide (erythromycin), a lincosamide (lincomycin) and an oxazolidinone-based antibiotic (linezolid) showed, as expected, little or no binding to both ribosomal RNA targets (Table 1).
While it is tempting to compare selectivity ratios and apparent antibiotic toxicities, the nature of the latter complicates such correlations. In particular, the diverse organisms, conditions and antibiotics used hinder the development of firm relationships.20 There is, however, a qualitative correlation between the trends observed for aminoglycoside antibiotics (Table 1) and their nephrotoxicity as evaluated in rat models. Paromomycin, while displaying a lower affinity for the 18S A-site compared to neomycin, has a higher histopathology score than neomycin (Fig. S1†).13,21 This could potentially be explained by its higher preference for the human A-site. Similarly, tobramycin has a five-fold lower affinity for the 18S A-site compared to neomycin, but is more selective for the eukaryotic A-site. These opposing factors could contribute to its low histopathology score, which is similar to that of neomycin.
In summary, we have developed a three-component assembly that facilitates the real-time evaluation of the affinity and selectivity of small-molecules to the bacterial and human ribosomal decoding sites in a single experiment. It relies on two orthogonal FRET pairs that act in concert to generate unique spectral signatures for each binding and displacement event. The wide spectral window spanned by the absorption and emission of the selected chromophores (ca. 300–600 nm) and their sequential overlap are facilitated by the use of an isomorphic nucleoside analogue, which provides a short wavelength trigger while maintaining the bacterial RNA fold. In addition to assessing the selectivity traits of known antibiotics, we were able to gather affinity and selectivity data for compounds that are not generally considered to be A-site binders. While we note, naturally, that a multitude of factors contribute to the apparent toxicity of any drug, where target selectivity is just one of them, having a simple tool to screen derivatives prior to advancing them into preclinical evaluation could prove highly valuable.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM 069773) for support and the National Science Foundation (instrumentation grants CHE-9709183 and CHE-0741968).We are grateful to Drs Yuzuru Akamatsu and Yoshikazu Takahashi for a generous gift of negamycin and to Dr Keiichi Ajito for helpful discussions.
Footnotes
Electronic supplementary information (ESI) available: Experimental details, photophysical data.
Notes and references
- 1.Gale EF, Cundliffe E, Renolds PE, Richmond MH, Waring MJ. The Molecular Basis of Antibiotic Action. London: John Wiley & Sons; 1981. [Google Scholar]; Green R, Noller HF. Annu. Rev. Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]; Puglisi JD, Blanchard SC, Dahlquist KD, Eason RG, Fourmy D, Lynch SR, Recht MI, Yoshizawa S. In: The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. Garrett RA, Douthwaite SR, Liljas A, Matheson AT, Moore PB, Noller HF, editors. Washington D.C: ASM Press; 2000. pp. 419–429. [Google Scholar]
- 2.Yonath A. Annu. Rev. Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 3.Wimberly BT. Curr. Opin. Investig. Drugs. 2009;10:750–765. [PubMed] [Google Scholar]
- 4.Gallego J, Varani G. Acc. Chem. Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]; Tor Y. ChemBioChem. 2003;4:998–1007. doi: 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]; Sutcliffe JA. Curr. Opin. Microbiol. 2005;8:534–542. doi: 10.1016/j.mib.2005.08.004. [DOI] [PubMed] [Google Scholar]; Hermann T. Biochimie. 2006;88:1021–1026. doi: 10.1016/j.biochi.2006.04.020. [DOI] [PubMed] [Google Scholar]; Tor Y. Biochimie. 2006;88:1045–1051. doi: 10.1016/j.biochi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]; Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]; Vicens Q, Westhof E. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- 6.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]; Vicens Q, Westhof E. ChemBioChem. 2003;4:1018–1023. doi: 10.1002/cbic.200300684. [DOI] [PubMed] [Google Scholar]; Francois B, Russell RJM, Murray JB, Aboul N, Masquida B, Vicens Q, Westhof E. Nucleic Acids Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kaul M, Barbieri CM, Pilch DS. J. Am. Chem. Soc. 2006;128:1261–1271. doi: 10.1021/ja056159z. [DOI] [PubMed] [Google Scholar]
- 7.(a) Recht MI, Douthwaite S, Puglisi JD. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lynch SR, Puglisi JD. J. Mol. Biol. 2001;306:1037–1058. doi: 10.1006/jmbi.2000.4420. [DOI] [PubMed] [Google Scholar]; (c) Kaul M, Barbieri CM, Pilch DS. J. Mol. Biol. 2005;346:119–134. doi: 10.1016/j.jmb.2004.11.041. [DOI] [PubMed] [Google Scholar]; (d) Hobbie SN, Kalapala SK, Akshay S, Bruell C, Schmidt S, Dabow S, Vasella A, Sander P, Bottger EC. Nucleic Acids Res. 2007;35:6086–6093. doi: 10.1093/nar/gkm658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobbie SN, Akshay S, Kalapala SK, Bruell CM, Shcherbakov D, Böttger EC. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20888–20893. doi: 10.1073/pnas.0811258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Ryu DH, Rando RR. Bioorg. Med. Chem. 2001;9:2601–2608. doi: 10.1016/s0968-0896(01)00034-7. [DOI] [PubMed] [Google Scholar]; (b) Wong C, Hendrix M, Priestley ES, Greenberg WA. Chem. Biol. 1998;5:397–406. doi: 10.1016/s1074-5521(98)90073-4. [DOI] [PubMed] [Google Scholar]
- 10.Xie Y, Dix AV, Tor Y. J. Am. Chem. Soc. 2009;131:17605–17614. doi: 10.1021/ja905767g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Based on crystal structures (PDB 2ESI, 1FYO, and 1FYP), the estimated distance between F1 and F2 is less than 10 Å, while the distance between F2 and F3 is less than 20 Å.
- 12.Srivatsan SG, Tor Y. J. Am. Chem. Soc. 2007;129:2044–2053. doi: 10.1021/ja066455r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.See supporting information for additional details.
- 14.Griffey RH, Hofstalder SA, Sannes-Lowery KA, Ecker DJ, Crooke ST. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10129–10133. doi: 10.1073/pnas.96.18.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Few studies have examined aminoglycoside affinity to both A-sites under identical experimental conditions.
- 16.Wang H, Tor Y. Angew. Chem., Int. Ed. 1998;37:109–111. [Google Scholar]
- 17.Mizuno S, Nitta K, Umezawa H. J. Antibiot. 1970;23:581–588. [PubMed] [Google Scholar]; Kondo S, Shibahara S, Takahashi S, Maeda K, Umezawa H, Ohno M. J. Am. Chem. Soc. 1971;93:6305–6306. doi: 10.1021/ja00752a072. [DOI] [PubMed] [Google Scholar]
- 18.Arakawa M, Shiozuka M, Nakayama Y, Hara T, Hamada M, Kondo SI, Ikeda D, Takahashi Y, Sawa R, Nonomura Y, Sheykholeslami K, Kondo K, Kaga K, Kitamura T, Suzuki-Miyagoe Y, Takeda SI, Matsuda R. J. Biochem. 2003;134:751–758. doi: 10.1093/jb/mvg203. [DOI] [PubMed] [Google Scholar]
- 19.Schroeder SJ, Blaha G, Moore PB. Antimicrob. Agents Chemother. 2007;51:4462–4465. doi: 10.1128/AAC.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilhelm JM, Jessop JJ, Pettitt SE. Biochemistry. 1978;17:1149–1153. doi: 10.1021/bi00600a002. [DOI] [PubMed] [Google Scholar]; Wilhelm JM, Pettitt SE, Jessop JJ. Biochemistry. 1978;17:1143–1149. doi: 10.1021/bi00600a001. [DOI] [PubMed] [Google Scholar]; Böttger EC, Springer B, Prammananan T, Kidan Y, Sander P. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kostrub CF, Diokno R, Aggen JB, Miller GH, Judice JK, Tulkens PM. 19th European Congress of Clinical Microbiology and Infectious Diseases; Helsinki, Finland: Blackwell Publishing; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.