Abstract
The development of genetic switches and their integrated forms (genetic circuits) with desired specifications/functions is key for success in synthetic biology. Due to the difficulty in rational design, genetic switches and circuits with desirable specifications are mostly obtained by directed evolution. Based on a virus-derived nucleotide kinase as a single-gene dual selector, we constructed a robust, efficient and stringent selection system for genetic switches. This method exhibited unprecedented enrichment efficacy (>30 000-fold) of functional switches from non-functional ones in a single selection cycle. In addition, negative (OFF) selection was exceptionally stringent, allowing the rapid and efficient selection of non-leaky from leaky circuits.
INTRODUCTION
Forward engineering of genetic circuits in living cells has various biotechnological applications (1–3) and provides valuable insights into the design principles of natural gene networks (4,5). Significant progress has been made in the rational design of genetic switches (6–9) and circuits (10–14). However, it is still a challenge to construct such circuits at will; the physics of each device component is not satisfactory for the reliable integration into complex genetic circuits in cells. Due to the leakiness, cross-talk, improper switching thresholds and context dependency (15) of genetic switches and their assemblies, implementation of functional circuits, especially those containing toxic genes, remains a major challenge in synthetic biology (16,17).
Directed evolution has been used to efficiently construct genetic switches and circuitry (11,18–22). All genetic circuits ultimately turn on/off gene expression under defined conditions, irrespective of the types and molecular mechanisms of gene regulation. Thus, functional selection of genetic circuits can be established simply by coupling the output of switches (or circuits) with the survival of the host cells. Here, the library of switches/circuits is placed in an environment where the output should be ON, and all variants with the OFF phenotype are removed (ON selection). The survivors are then placed under the OFF condition, followed by selection against variants that are inappropriately in the ON state (OFF selection). Iterative rounds of these ON/OFF selections yield various switches or circuits with desirable behaviors or specifications (17,19,23,24).
Several selection systems have been developed for the evolutionary engineering of genetic switches and circuits. In early systems, the selection was achieved by coupling the circuit output with the expression of two independent genes: a conditional rescuer (ON-selector) and a conditional killer (OFF-selector) (24,25). However, the use of two independent selector genes weakens the robustness of the screening process, resulting in the frequent emergence of false positives. To address this problem, single genes that function as both the ON- and the OFF-selector have been developed (26,27). TetA, a tetracycline/H+ antiporter, can be used as an ON-selector by adding tetracycline, while it functions as an OFF-selector in the presence of toxic metal salts such as NiCl2.
In this article, we report another type of single-gene dual selector for genetic switches/circuits. Herpes simplex virus thymidine kinase (hsvTK), frequently used as a suicide gene in gene therapy (28), was adapted for the OFF selection of a genetic circuit. In the presence of mutagenic nucleoside dP (29), hsvTK works as the OFF selection system with excellent speed and power. Conversely, thymidine kinases (TKs) are known to rescue tdk- strains from death by thymidine deficiency (30). We adapted this system for the ON selection of a genetic circuit. Used in conjunction, we established an efficient, robust and rapid selection platform for selecting genetic circuits with desired specifications from the large pool of non-functional ones. We also demonstrate how this system would be especially useful for the selection of genetic circuits with high stringency.
MATERIALS AND METHODS
Materials
The 6-(β-D-2-deoxyribofuranosyl)-3,4-dihydro-8H-pyrimido[4,5-c][1,2]oxazin-7-one (dP) and 5-fluoro-2′-deoxy-uridine (5FdU) were purchased from Berry and Associates (MI) and Sigma and Aldrich (MO, USA), respectively. Oligonucleotides were synthesized by FASMAC Co., Ltd (Kanagawa, Japan) and BEX Co., Ltd (Tokyo, Japan). All other chemicals and media were of the highest grade available.
Strains and plasmids
Escherichia coli JW1226 [from the KEIO collection (31)] was used throughout this study except for the GFPUV assay. Unless otherwise noted, cells were grown at 37°C in LB broth (2.0% w/v, Invitrogen).
Plasmid pTrc-hsvtk was constructed by subcloning of PCR-amplified reading frame of hsvTK [from pET-hsvtk (32)] into pTrc-99A. The reading frame of hsvtk was amplified by PCR using primers containing the additional sequence coding for PL promoter/ribosome-binding sites (rbs). The PCR products were subcloned into pACYC184 (33), yielding selector plasmid pCI-hsvtk. Similarly, screening plasmid pCI-gfpUV, was constructed by the insertion of the gfpUV (from pGFPUV, Clontech) attached with the PL promoter/rbs.
For the construction of Plasmid-A, the following components were assembled in series: trc promoter, luxR, a transcription terminator, Plux promoter and the CI repressor gene. The entire assembly was inserted into the multi-cloning site of pTrc99A (34), resulting in Plasmid-A. Plasmid-B was identical to Plasmid-A except that the CI was truncated at its C-terminus. Plasmid-C was constructed by the deletion of the rbs sequence for the CI from Plasmid-B. Plasmid-D was constructed by the deletion of the portion containing the transcriptional terminator and PL promoter from Plasmid-B.
Positive (ON) selection
For selection on an agar plate, an overnight culture of JW1226 harboring appropriate plasmids was diluted and plated onto an ‘ON selection plate’ [2.0% (w/v) tryptone, 0.5% (w/v) NaCl, 1.5% (w/v) agar, 20 μg/ml 5FdU, 10 μg/ml dT and 10 μg/ml uridine]. After a 20-h incubation at 37°C, colonies from the plates were isolated for further analysis.
For selection in liquid media, approximately 106 cells were taken from the overnight culture and diluted in 1 ml of ‘ON selection medium’ [2.0% (w/v) tryptone, 0.5% (w/v) NaCl, 10 μg/ml thymidine, 1 μg/ml adenosine and 0–25 μg/ml 5FdU]. After shaking for 12–15 h at 37°C, the culture was plated on LB agar to isolate survivors. To determine selection efficiency, the optical density (OD600) and colony-forming units (c.f.u./20 μl) of the culture were compared with the culture grown in non-selective media (i.e. lacking 5FdU).
Negative (OFF) selection
For selection on an agar plate, an overnight culture of JW1226 [or BL21Δtdk (32)] harboring appropriate plasmids was plated onto an ‘OFF selection plate’ containing 1.5% (w/v) agar, 100 nM dP and 1 μM AHL and incubated for 12–15 h at 37°C.
For OFF selection in liquid media, the cell culture was diluted to a final OD600 of 0.002 (∼106 cells/ml) in 1 ml of the ‘OFF selection media’ containing varying concentrations (10–1000 nM) of dP. After shaking for 5 min–12 h at 37°C, the culture was plated on LB agar to isolate the survivors.
Mock selection
Plasmid-A, -B, -C or -D was transformed into JW1226 harboring the selector plasmid pCI-hsvtk and cultured overnight at 37°C. The transformants were mixed together in different ratios and then subjected to OFF/ON selection in series. First, approximately 106 cells from the mixed culture was inoculated in the LB medium containing AHL (1.0 µM) and dP (10–1000 nM) for 1 h (OFF selection). The cell culture mixture was washed twice with 2 ml LB medium and resuspended in 1 ml LB medium. A portion (10 µl) was inoculated into 2 ml ‘ON selection media’ without AHL and cultured for 12–20 h at 37°C (ON selection). Plasmids were collected by miniprep of the culture before and after each selection process.
Determination of the abundance ratio of plasmids
Plasmids prepared from each selection culture were treated with BamHI to digest the selector plasmid pCI-hsvtk. The reaction was column-purified and transformed into E. coli (JW1226 or XL10 Gold) harboring pCI-gfpUV and plated on LB-agar plates (supplemented with 100 μg/ml ampicillin and 30 μg/ml chloramphenicol) with or without AHL (1.0 µM) for 12 h at 37°C.
The enrichment factor of Plasmid-B over the non-functional plasmids (-C and -D) was obtained from the ratio of fluorescent/non-fluorescent colonies on AHL (+)/(–) plates. To determine the existing ratio of the two functional plasmids (-A and -B), a portion of the culture was subjected to PCR using primer sets that amplify part of the circuitry from both plasmids: one was designed to anneal to the N-terminal region of cI (CIf: 5′-TAGCGTTGAAGAATTTAGCCCTTCAATCGC-3′) and another was targeted to the region downstream of the cI (CIr: 5′-ACATCATAACGGTTCTGGCAAATATTCTG-3′). Due to the difference in size of the Cl genes, this primer set yields distinguishable bands for Plasmid-A (639 bp) and Plasmid-B (367 bp). We confirmed that both plasmids were amplified with roughly the same efficiency. The ratio of their band intensity from the PCR products was used to estimate the relative abundance of Plasmid-A and -B.
RESULTS AND DISCUSSION
Positive (ON) selection using hsvTK
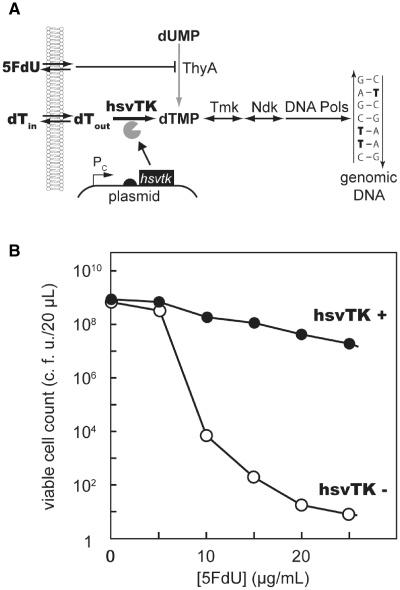
In E. coli, the nucleotide dTMP is synthesized via two routes: the de novo pathway and the salvage pathway (Figure 1A). When 2′-deoxy-5-fluorouridine (5FdU) is added, it is phosphorylated to yield 5F-dUMP, a potent inhibitor of thymidylate synthase (ThyA) (35). When the de novo supply of dTMP is blocked, E. coli growth becomes dependent on exogenously supplemented dT. Thus, strains deficient in thymidine kinase (Tdk), the first enzyme in this pathway, fail to grow in 5FdU-containing media. Plasmid-encoded TKs have been shown to support cell growth when supplemented with dT by complementing tdk (36,37).
Figure 1.
TK as an ON-selector. (A) Mechanism of action. In the presence of 5FdU, the expression of hsvTK rescues tdk strains by complementing the thymidine salvage pathway (37), while cells lacking hsvTK die of thymidine depletion. (B) Viable JW1226 cell count with (filled circles) or without (open circles) expression of hsvTK after a 12-h incubation in ON selection media with various concentration of 5FdU.
Escherichia coli JW1226 (tdk strain) was incubated in the presence of ‘ON selection medium’ (‘Materials and Methods’ section) with various concentrations of 5FdU. The number of viable cells in the medium precipitously dropped with increasing concentrations of 5FdU. After 12 h of incubation in the presence of 25 µg/ml 5FdU, the cell count was approximately 10−8 that of the culture without 5FdU (Figure 1B) due to the cell death from thymidine deficiency (30). We observed growth of JW1226 expressing hsvTK under the same conditions, indicating that hsvTK rescues the thymidine salvage pathway of E. coli. We observed a slight decline in cell count with increasing concentrations of 5FdU, even for cells expressing hsvTK. This may be due to other factors causing cytotoxicity, such as misincorporation of metabolites into DNA/RNA. Overall, the survival rate of JW1226 was 2.1 × 106-fold higher upon hsvTK expression under this selection condition. In the future, even higher selection efficiency may be achieved simply by extending the growth time in this selection medium.
Negative (OFF) selection using hsvTK
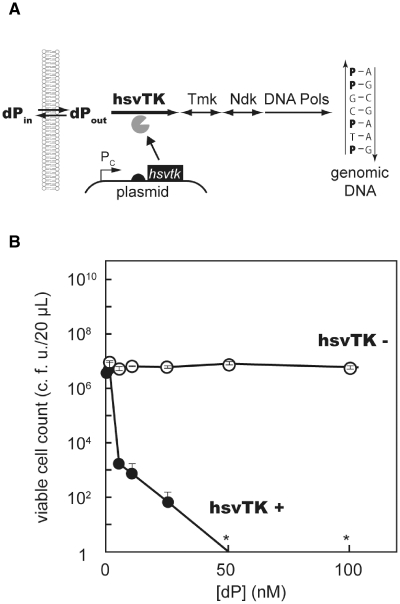
Many synthetic nucleosides are known to become toxic upon phosphorylation. Due to their promiscuous nature, viral TKs and their variants have been used to activate the killer effect of synthetic nucleosides such as ganciclovir (GCV), acyclovir (ACV) and bromovinyldeoxyuridine (BVdU) (28). In the search for other nucleoside candidates for killing agents upon phosphorylation, we encountered a synthetic nucleoside called dP (38). Unlike other nucleoside analogs that act as chain terminators or as inhibitors of nucleotide metabolism, dP is a potent mutagenic agent (29). When added to the media, it passes through the cellular membrane, is phosphorylated by the nucleoside kinases, and is incorporated into the genomic DNA (Figure 2A). In duplex, nucleobase P forms base pairs with A and G, thereby elevating the mutation frequency of the cell. Due to the random damage in genomic information, ∼80% of E. coli cells lost their viability (c.f.u.) after overnight incubation in media containing 37 µM dP (29).
Figure 2.
TK as an OFF-selector. (A) Mechanism of action. dP travels through the cell membrane, is phosphorylated by hsvTK, and is incorporated into genomic DNA. P can pair with both A and G, elevating the mutation rate within the cell. Having their genetic information damaged rapidly, cells quickly die of lethal mutagenesis. (B) The number of viable cells after 12 h incubation in the presence of dP (0–100 nM). Filled circles represent BL21 (tdk) expressing hsvTK; open circles represent BL21 (tdk) without hsvTK expression.
In the course of searching for a better dP kinase, we found that hsvTK was by far the best sensitizer of the cells to dP. Viability of E. coli strains expressing hsvTK sharply decreased with increasing concentrations of dP. In media containing 100 nM dP, the cell count reproducibly dropped to zero (Figure 2B). In sharp contrast, cells not expressing hsvTK showed no detectable loss of viability nor increase in mutation frequency in the same media. Due to the complete loss of cell viability, we were unable to precisely determine the selection efficiency, but the selection guaranteed an at least 107-fold enrichment of cells not expressing hsvTK. Interestingly, we obtained the same result for the tdk+ strains (data not shown). dP does not have an effect on cells via the aid of endogenous TK when its concentration is submicromolar.
We found that only 5 min of dP treatment was necessary for reliably eliminating hsvTK-expressing cells (Supplementary Figure S1). Phosphorylation of dP proceeds only in cells expressing hsvTK. Once formed, the toxic dP-nucleotides cannot travel through the cell membrane, ensuring the safety of neighboring cells. Given the high killing efficiency, quickness, and selectivity, the dP kinase activity of hsvTK provides an ideal OFF-selector for genetic switches and circuits.
Model circuits
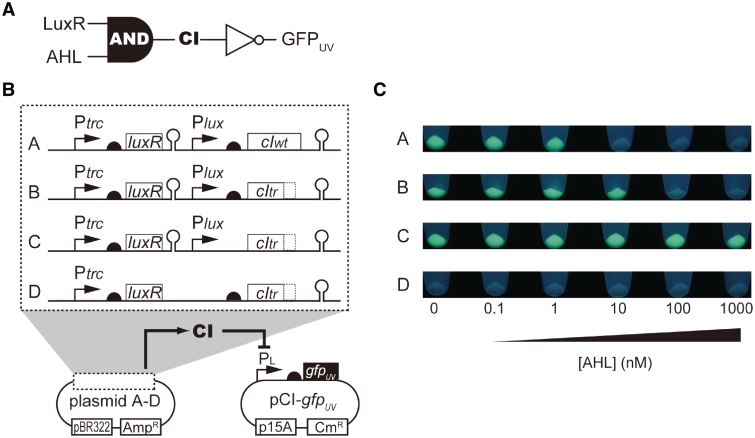
We constructed the model circuit illustrated in Figure 3A. This circuit is implemented with two compatible plasmids. Plasmid-A constitutively expresses LuxR (39), a quorum sensor protein, under the trc promoter (34). In the presence of acyl-homoserine lactones (AHLs; i.e. the quorum-sensing signaling molecule of gram-negative bacteria), LuxR activates the expression of the Lambda CI repressor (40) placed under the Plux promoter. The CI protein represses the expression of the reporter gene (gfpUV in this work) under the PL promoter (40) on a separate plasmid, pCI-gfpUV (Figure 3B). Altogether, the output of the entire circuit (i.e. the fluorescent signal of GFPUV) is inversely related to the concentration of the AHL (Figure 3C). Plasmid-B is similar to Plasmid-A except that a portion (91 amino acids) from the C-terminal dimerization domain (40) of Cl is deleted. The function of this truncated CI (CItr) is weaker than the wild-type, thereby requiring a higher concentration of AHL for the complete switching OFF of the output signal (fluorescence) from pCI-gfpUV. Plasmid-C was constructed by removing the ribosome-binding site (rbs) from Plasmid-B. Lack of translation of the CI repressor results in a circuit that is ‘always ON’ when combined with pCI-gfpUV. Plasmid-D was made by the deletion of the transcription terminator downstream of LuxR from Plasmid-B (together with Plux promoter). Read-through from the trc promoter constitutively expresses CI, resulting in the always OFF behavior of the circuit as a whole.
Figure 3.
Plasmid-born model circuits function as AHL/GFPUV inverters. Logic diagram (A), plasmid constructs (B), and the actual behaviors (C) of the AHL/GFPUV inverter circuits. Plasmid-A constitutively expresses LuxR, which activates the CI repressor in the presence of AHL. CI represses GFPUV on pCI-gfpUV, which is under the control of the PL promoter. Overall, the level of GFPUV output is the inverse of the input signal, AHL. Plasmid-B also produces an AHL/GFPUV inverter, but it has a partial truncation of the CI repressor, requiring a higher concentration of AHL to fully switch off the output (GFPUV fluorescence). Plasmid-C lacks translational signals for CI, resulting in an always ON phenotype. Plasmid-D lacks the transcriptional terminator downstream of the LuxR, which results in the constitutive repression of GFPUV (always OFF). The semicircles and the hairpins indicate the ribosome-binding sites and transcription terminators, respectively. (C) The cell pellets of Strains containing Plasmid-B, -C, or -D together with pCI-gfpUV after 12 h of incubation in the presence of various concentrations of AHL.
Dual genetic selection on solid plates
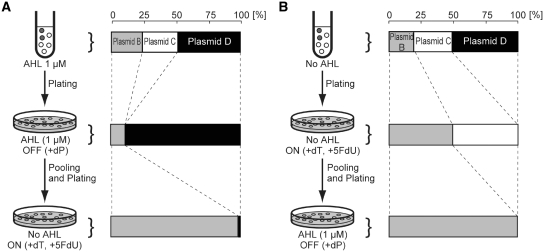
A functional inverter Strain-B was selected from non-functional strains (-C and -D) by a single round of OFF and ON selections (Figure 4A). Plasmid-B, -C and -D were transformed into JW1226 harboring the selector plasmid pCI-hsvtk. The resulting Strain-B, -C and -D were mixed and plated on an ‘OFF selection plate’ (100 nM dP) containing no AHL. Colonies were collected and pooled into LB media containing 1 µM AHL to set the circuit output to ON. The culture was then plated on an ‘ON selection plate’ containing 1 µM AHL.
Figure 4.
Schematic illustration of dual selection of genetic circuits. (A) OFF→ON selection. A mixture of E. coli JW1226 harboring selector plasmid pCI-hsvtk and Plasmid-B, -C, or -D was plated on an ‘OFF selection plate’ containing AHL. The colonies were pooled and re-plated on an ‘ON selection plate’ without AHL. (B) ON→OFF selection. The same cell mixture was first grown on ON selection plates in the absence of AHL, followed by growth on an OFF selection plate containing AHL. The bar graphs represent the abundance ratios of Plasmid-B, -C and -D found in the media taken from each selection step.
Before selection, the ratio of Strains-B, -C and -D was 0.2, 0.3 and 0.5, respectively (Figure 4A). After the first selection (OFF selection), Strains-C (always ON) was completely eradicated, giving a mixture of the two Strains -B and -D. The following ON selection efficiently removed Strain-D (always OFF), resulting in a culture dominated by Strain-B. Thus, a single round of OFF/ON selection effectively enriched the strain harboring functional genetic circuits from the background of non-functional ones. Similar results were obtained by dual selection in the reversed order (ON selection followed by OFF selection, Figure 4B). When OFF selection was conducted immediately after ON selection, however, the majority of Strain-B died off, likely due to the remaining activity of hsvTK that was expressed in the previous ON selection process. This inappropriate death of Strain-B did not occur when 1–2 h of pre-incubation in the OFF condition (1 µM AHL in this case) was conducted before OFF selection.
Dual selection in liquid media
To automate circuit construction, it is important to conduct the entire selection processes with only liquid handling. The mixture of Strain-B, -C and -D was subjected to dual selection in liquid media (Figure 5A). First, the strains were incubated in ‘OFF selection media’ containing 1 µM AHL. Then, they were directly shifted into ‘ON selection media’ free from AHL and were incubated for 12 h at 37°C. A portion of the culture was removed after each step, and the plasmid (mixture) was isolated. The relative abundance of Plasmid-B, -C and -D was determined by re-transformation of the isolated plasmid mixtures into E. coli harboring pCI-gfpUV. Before selection (fraction #1, Figure 5B), both fluorescent/non-fluorescent colonies were observed on both AHL (+)/(–) plates. In the sample taken after OFF selection (fraction #2), no colonies fluoresced on the AHL (+) plate, indicating the complete loss of Plasmid-C. We observed both fluorescent/non-fluorescent colonies on the AHL (–) plate from the same sample, indicating the presence of Plasmid-B and -D. After ON selection, almost all colonies fluoresced on the AHL (–) plates (fraction #3, Figure 5B). These results demonstrate that Strain-B (the strain harboring Plasmid-B) was efficiently enriched solely by liquid-based processes.
Figure 5.
Liquid-based dual selection of AHL/GFPUV inverter circuits. (A) Schematic illustration of the selection procedure. The mixture of Strain-B, -C and -D was incubated in the OFF selection media for 1 h in the presence of 1 µM AHL. After AHL removal by centrifugation/washing, the cells were grown in the ON selection media without AHL. (B) The switching properties of the mixed culture in each step of the dual selection. The plasmids collected from each step were re-transformed into cells harboring pCI-gfpUV and grown on AHL (+)/(–) plates.
Mimicking the situation encountered under actual selection conditions, where one desirable circuit plasmid must be selected from tens of thousands of non-functional ones, we conducted liquid-based dual selection starting from various mix ratios of Plasmid-B:-C:-D. Plasmids isolated from the cells that survived both the OFF/ON selections were transformed into cells harboring pCI-gfpUV to determine the relative abundance of the three starting plasmids after selection. In all cases, we obtained excellent selection efficiency (up to 3.2 × 104; Table 1) with a single round of OFF/ON selection. To the best of our knowledge, this selection efficiency is higher than any other reported value. When starting ratios of Plasmid-B, -C and -D was 1:100 000:100 000, the ratio of Plasmid-B remained ∼16% after dual selection. It should be noted, however, that this does not suggest the upper limit of the selection efficiency of this method. We found that under high concentrations of AHL, Strain-B had a very small but certain level of genes expression under the PL promoter due to the incomplete repression capability of the CItr (not shown). More stringent circuits (such as circuit A) should be selected for with higher efficiency.
Table 1.
Selection efficiency of functional inverters from non-functional ones
| Original ratios (Plasmid-B:-C:-D) | The final ratios of Plasmid-B (%) | Enrichment factor |
|---|---|---|
| 1:103:103 | 84.0 | 1.7 × 103 |
| 1:104:104 | 82.0 | 1.6 × 104 |
| 1:105:105 | 16.0 | 3.3 × 104 |
Competition for stringency
Plasmid-A and -B were subjected to competition through dual selection. Both -A and -B behave as AHL/PL inverters. They repress gene expression under the PL promoter when AHL levels are high (>100 nM), and they turn on the output when AHL levels are low (<10 nM). The difference is that the CItr in Plasmid-B is less efficient in repressing the PL promoter than the wild-type CI in Plasmid-A. Thus, the PL promoter is expected to be leakier when combined with Plasmid-B.
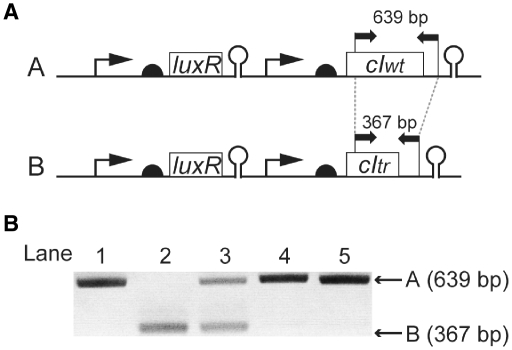
The mixed culture of the Strain-A and -B was first subjected to OFF selection in the presence of 100 nM AHL. The survivors of the OFF selection were then grown in non-selective media for 12 h and subjected to ON selection in the absence of AHL. PCR of the starting mixture yielded two bands: one (639 bp in size) is the amplification product from Plasmid-A, while the other (367 bp) is from Plasmid-B (Figure 6).
Figure 6.
Competition of two inverter circuits. The mixture of Strain-A and -B was subjected to OFF selection in the presence of 100 nM AHL, followed by ON selection in its absence. The existing ratio of the Plasmids-A and -B was tracked by PCR of the culture after overnight growth in selection media. (A) Annealing sites of the primers used for PCR analysis to the Plasmid-A and -B. (B) Gel electrophoresis of the PCR products taken from each step of the selection procedure. Lane: 1, culture containing Strain-A; 2, culture containing Strain-B; 3, mixed culture of Strain-A and -B before selection; 4, mixed culture after OFF selection; and 5, mixed culture after the subsequent ON selection.
Based on the relative intensity of these bands, we estimated that they existed in roughly the same amount as in the starting mixture (lane 3 in Figure 6). After dP selection, bands for Plasmid-B disappeared (lane 4). The band pattern was unchanged after subsequent ON selection (lane 5). It is assumed that Plasmid-B allowed trace expression from the PL promoter due to the compromised function of CItr. Note that both circuits exhibited the ‘OFF’ phenotype (non-fluorescent) when GFPUV was used as the output with this AHL concentration (Figure 3C). Perhaps even the leakiness that was undetectable with the GFPUV reporter was fatal under OFF selection due to its exceptionally high killing efficiency of the hsvTK dP kinase activity (Figure 1B).
Tuning Circuit B into NAND gate and AHL inverter
Even simple networks built out of a few, well-characterized components could behave in various ways (15). In theory, the circuit-B (Figure 3A) should behave as NAND gate with two chemical inputs, IPTG and AHL: it should fluoresce in all conditions except where both IPTG and AHL are present. Or, the circuit could be insensitive to IPTG due to the leakiness of the trc promoter, thereby becoming a simple AHL inverter. We tried to convert circuit B into both circuit functionalities, simply by tuning the translational efficiency of LuxR gene.
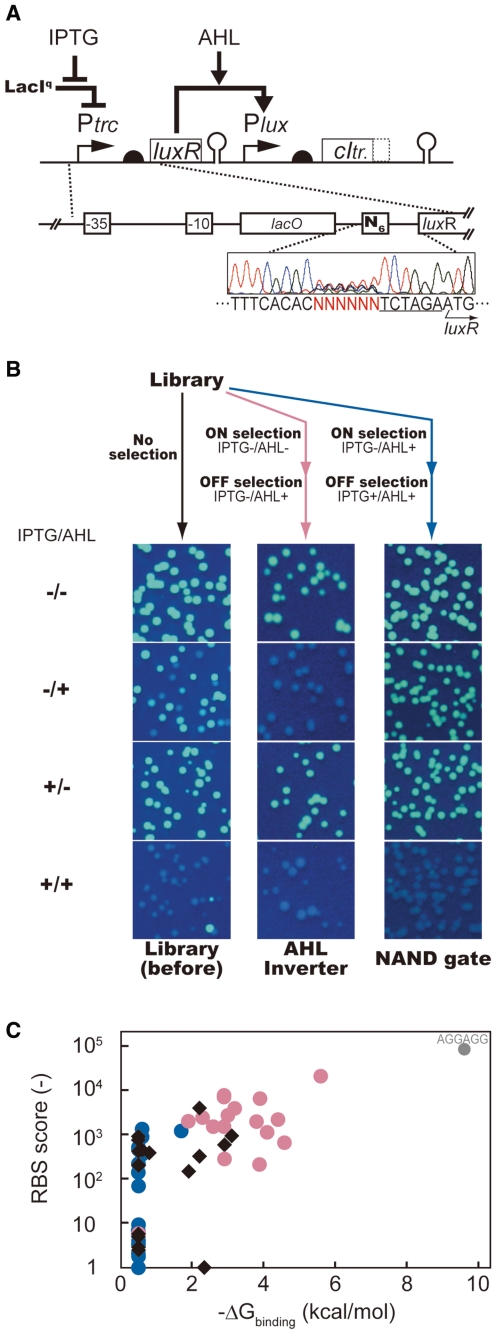
Using the primer containing N6 (N stands for equimolar mixture of A, G, C and T), we randomized 6 nt corresponding to the rbs for LuxR gene on Plasmid-B. The resultant library (Figure 7A, library size, 2.2 × 105) was transformed into the cell harboring selector plasmid pCI-hsvtk. The culture was split into two. One culture was subjected to the ‘AHL inverter selection’: ON selection in IPTG(–)/AHL(–) media, followed by OFF selection in IPTG(–)/AHL(+) media. The other culture was subjected to ‘NAND-gate selection’ where ON selection was conducted in IPTG(–)/AHL(+) media, followed by OFF selection in IPTG(+)/AHL(+) media. Plasmids were minipreped from each culture and re-transformed into the cell harboring pCI-gfpUV to analyze the distribution of the switching properties of the variants (Figure 7B).
Figure 7.
Isolating AHL Inverters and NAND-gates from a single rbs library. (A) Library design. The six bases (N6 in figure) corresponding to rbs of luxR gene were randomized. (B) The phenotype variation of the circuit library before (left), after the ‘AHL-inverter selection’ (middle), and after the ‘NAND-gate selection’ (right). (C) Distribution of the rbs scores in the variants from original (black filled diamonds), AHL inverter-selected (red filled circles), and NAND gate-selected (blue filled circles) libraries. X-axis represents the free binding energy of these rbs sequences (ΔGbinding) and anti-rbs sequence, calculated using the RNA duplex software component of the Vienna RNA Package (45). Rbs scores for Y-axis were calculated using RBS Calculator (14). The sequence AGGAGG (perfect match to the anti-rbs) was independently constructed by site-directed mutagenesis. It was found to behave as an AHL inverter.
The initial library (Figure 7B, left column) included three types of variants: IPTG/AHL NAND-Gates (NAND-variants, 84%), simple AHL inverters that are insensitive to IPTG (AHL inverter variants, 13%) and the variants that always fluoresce (3%, cloning artifacts). After ‘AHL inverter selection’, 96% of the library was found to be AHL-inverter variants (middle column in Figure 7B). After ‘NAND-gate selection’, the library was exclusively consisted of the NAND-gate variants (right column in Figure 7B). Thus, two types of genetic circuits were quickly constructed in parallel from a single circuit library.
From the library before and after the selections, we randomly picked about 20 clones, and analyzed their rbs sequences (Supplementary Table S1). Each of the sequence obtained was scored in its translational efficiency using two independent ways. One is the binding energy (ΔGbinding) with anti-rbs, the last twelve nucleotides of the E. coli 16S rRNA (3′-AUUCCUCCACUA-5′). The other is given by RBS Calculator (14), the freeware package to estimate the translation initiation rate based on various molecular interactions involved in the translation initiation event besides the binding energy with anti-rbs.
There was a significant variation in rbs scores in the original library (Figure 7C). These scores were shifted toward much lower values after the NAND-gate selection (Figure 7C). This suggests the level of expression from trc promoter is non-negligible even in the absence of IPTG. Thus, down tuning of the rbs was necessary to make the LuxR’s expression IPTG-dependent. In contrast, the ‘AHL inverter selection’ enriched the variants with higher rbs scores (Figure 7C): elevation in the translational efficiency magnifies the effect of leakiness of trc promoter, thereby making LuxR expressed constitutively. On top of such qualitative interpretation, Figure 7C adds some quantitative insights: in this particular architecture, threshold score separating these two circuit functions (IPTG/AHL NAND gates and Simple AHL inverters) is about −1.0 kcal/mol in binding energy with anti-rbs and about 1000 for the values given by the RBS Calculator (14).
CONCLUSIONS
Based on the discovery that hsvTK kills cells with exceptional efficiency upon dP addition, we established a new single-gene dual selector for the construction of genetic switches and circuits. There are several reported single-gene dual selectors for genetic switches (27,41), all of which are compatible with the system described in this work. The combined use of hsvTK with other selector systems will allow us to select for more complex circuits with multiple outputs.
One of the important features of selection systems is their selection efficiency. The selection efficiency of our ON selection was more than 106, which may be increased simply by extending the duration of the selection process. Our system offers the most powerful OFF selection by far. Due to its exceptional killing power, cells expressing even a minimal level of hsvTK were quickly removed with remarkable efficiency (selection efficiency more than 107). The successive round of ON/OFF selection could also be reproducibly conducted in liquid media with unprecedented selection efficiency (more than 104 in a single round).
We have shown that our selection even removes circuits with the proper logic configuration if they are leaky (Figure 6). To construct integrated circuits by assembling multiple genetic switches, each component must be as stringent as possible in its OFF state. The exceptionally high killing efficiency of hsvTK as an OFF-selector should be valuable in the construction of stringent genetic circuits or evolutionary engineering of circuitry components (switches/sensors).
Despite the remarkable killing power of the OFF selection process, hsvTK remains completely non-toxic without the addition of dP. This is in sharp contrast to natural-born killing enzymes, such as cell-lysing, DNA/RNA degrading, or addiction modules (42,43). Conditional killer genes can be stably retained in cells for generations due to the lack of counter-selection against their existence, enabling their use in situ (on duty). Thus, the system described in this article would be useful not only for constructing genetic circuits but also for the maintenance or as life-extension technology of cell populations/consortiums implemented with various genetic circuits.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JFE 21st Century Foundation; Ministry of Education, Culture, Sports, Science and Technology, Japan (Grant-in-Aid for Young Scientists, A). Funding for open access charge: Futaba Electronics Memorial Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Yoichi Mashimo (Department of Public Health, Chiba University) for sequence analysis and Maiko Furubayashi for helpful discussion. The authors thank the National BioResource Project (NIG, Japan) for E. coli KEIO strains. The plasmids constructed in this work partially consist of Biobricks (44).
REFERENCES
- 1.Carothers JM, Goler JA, Keasling JD. Chemical synthesis using synthetic biology. Curr. Opin. Biotechnol. 2009;20:498–503. doi: 10.1016/j.copbio.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell. Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 3.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 5.Mukherji S, van Oudenaarden A. Synthetic biology: understanding biological design from synthetic circuits. Nat. Rev. Genet. 2009;10:859–871. doi: 10.1038/nrg2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Looger LL, Dwyer MA, Smith JJ, Hellinga HW. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 7.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinkhabwala A, Guet CC. Uncovering cis regulatory codes using synthetic promoter shuffling. PLoS One. 2008;3:e2030. doi: 10.1371/journal.pone.0002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl Acad. Sci. USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 16.Voigt CA. Genetic parts to program bacteria. Curr. Opin. Biotechnol. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Haseltine EL, Arnold FH. Synthetic gene circuits: design with directed evolution. Annu. Rev. Biophys. Biomol. Struct. 2007;36:1–19. doi: 10.1146/annurev.biophys.36.040306.132600. [DOI] [PubMed] [Google Scholar]
- 18.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins CH, Leadbetter JR, Arnold FH. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat. Biotechnol. 2006;24:708–712. doi: 10.1038/nbt1209. [DOI] [PubMed] [Google Scholar]
- 20.Sayut DJ, Niu Y, Sun L. Construction and enhancement of a minimal genetic and logic gate. Appl. Environ. Microbiol. 2009;75:637–642. doi: 10.1128/AEM.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SK, Chou HH, Pfleger BF, Newman JD, Yoshikuni Y, Keasling JD. Directed evolution of AraC for improved compatibility of arabinose- and lactose-inducible promoters. Appl. Environ. Microbiol. 2007;73:5711–5715. doi: 10.1128/AEM.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daber R, Lewis M. A novel molecular switch. J. Mol. Biol. 2009;391:661–670. doi: 10.1016/j.jmb.2009.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nomura Y, Yokobayashi Y. Reengineering a natural riboswitch by dual genetic selection. J. Am. Chem. Soc. 2007;129:13814–13815. doi: 10.1021/ja076298b. [DOI] [PubMed] [Google Scholar]
- 24.Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat. Chem. Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 25.Yokobayashi Y, Arnold FH. A dual selecion module for directed evolution of genetic circuits. Nat. Comput. 2005;4:245–254. [Google Scholar]
- 26.Muranaka N, Sharma V, Nomura Y, Yokobayashi Y. An efficient platform for genetic selection and screening of gene switches in Escherichia coli. Nucleic Acids Res. 2009;37:e39. doi: 10.1093/nar/gkp039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomura Y, Yokobayashi Y. Dual selection of a genetic switch by a single selection marker. Biosystems. 2007;90:115–120. doi: 10.1016/j.biosystems.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Patterson AV, Saunders MP, Greco O. Prodrugs in genetic chemoradiotherapy. Curr. Pharm. Des. 2003;9:2131–2154. doi: 10.2174/1381612033454117. [DOI] [PubMed] [Google Scholar]
- 29.Negishi K, Loakes D, Schaaper RM. Saturation of DNA mismatch repair and error catastrophe by a base analogue in Escherichia coli. Genetics. 2002;161:1363–1371. doi: 10.1093/genetics/161.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 31.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Black ME, Newcomb TG, Wilson HM, Loeb LA. Creation of drug-specific herpes simplex virus type 1 thymidine kinase mutants for gene therapy. Proc. Natl Acad. Sci. USA. 1996;93:3525–3529. doi: 10.1073/pnas.93.8.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose RE. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988;16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 35.Yagil E, Rosner A. Phosphorolysis of 5-fluoro-2'-deoxyuridine in Escherichia coli and its inhibition by nucleosides. J. Bacteriol. 1971;108:760–764. doi: 10.1128/jb.108.2.760-764.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dube DK, Black ME, Munir KM, Loeb LA. Selection of new biologically active molecules from random nucleotide sequences. Gene. 1993;137:41–47. doi: 10.1016/0378-1119(93)90249-3. [DOI] [PubMed] [Google Scholar]
- 37.Summers WC, Raksin P. A method for selection of mutations at the tdk locus in Escherichia coli. J. Bacteriol. 1993;175:6049–6051. doi: 10.1128/jb.175.18.6049-6051.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin PK, Brown DM. Synthesis and duplex stability of oligonucleotides containing cytosine-thymine analogues. Nucleic Acids Res. 1989;17:10373–10383. doi: 10.1093/nar/17.24.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ptashne M. A Genetic Switch, Third Edition: Phage Lambda Revisited. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 41.Topp S, Gallivan JP. Random walks to synthetic riboswitches–a high-throughput selection based on cell motility. Chembiochem. 2008;9:210–213. doi: 10.1002/cbic.200700546. [DOI] [PubMed] [Google Scholar]
- 42.Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 43.Paul D, Pandey G, Jain RK. Suicidal genetically engineered microorganisms for bioremediation: need and perspectives. Bioessays. 2005;27:563–573. doi: 10.1002/bies.20220. [DOI] [PubMed] [Google Scholar]
- 44.Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.