Abstract
Hundreds of small nuclear non-coding RNAs, including small nucleolar RNAs (snoRNAs), have been identified in different organisms, with important implications in regulating gene expression and in human diseases. However, functionalizing these nuclear RNAs in mammalian cells remains challenging, due to methodological difficulties in depleting these RNAs, especially snoRNAs. Here we report a convenient and efficient approach to deplete snoRNA, small Cajal body RNA (scaRNA) and small nuclear RNA in human and mouse cells by conventional transfection of chemically modified antisense oligonucleotides (ASOs) that promote RNaseH-mediated cleavage of target RNAs. The levels of all seven tested snoRNA/scaRNAs and four snRNAs were reduced by 80–95%, accompanied by impaired endogenous functions of the target RNAs. ASO-targeting is highly specific, without affecting expression of the host genes where snoRNAs are embedded in the introns, nor affecting the levels of snoRNA isoforms with high sequence similarities. At least five snoRNAs could be depleted simultaneously. Importantly, snoRNAs could be dramatically depleted in mice by systematic administration of the ASOs. Together, our findings provide a convenient and efficient approach to characterize nuclear non-coding RNAs in mammalian cells, and to develop antisense drugs against disease-causing non-coding RNAs.
INTRODUCTION
It has been shown that the majority of the mammalian genome is transcribed largely as non-coding RNAs (ncRNAs), whereas only ∼2% encodes mRNAs (1–3). These ncRNAs, including miRNAs, small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), Piwi-interacting RNAs and long ncRNAs, are involved in multiple biological processes, such as DNA and RNA production, translation and protein translocation, e.g. (4–5). However, functionalizing individual ncRNAs in mammals has lagged in time, mainly due to lack of convenient knockout or knockdown approaches. This is especially the case for snoRNAs.
In eukaryotes, several hundred snoRNAs have been identified that belong to two major groups: C/D box and H/ACA box snoRNAs. Most snoRNAs guide nucleotide modifications in rRNAs, whereas a subset of similar RNAs are present in Cajal bodies (scaRNAs) and direct modifications in snRNAs. In both cases, the C/D box RNAs guide 2′-O-methylation (Nm) and H/ACA RNAs for pseudouridylation (Ψ). The site specificity of modifications is provided by base-pairing of a snoRNA/scaRNA with the substrate RNA(s) (6–8). The mechanisms of RNA-guided RNA modification are conserved in eukaryotes, and similar machineries also exist in archaea (6,9). Although yeast snoRNAs have been better characterized through genetic knockout, the functions of most snoRNAs in other organisms have not been verified. This is particularly problematic since many snoRNAs are species specific.
The majority of human snoRNAs were predicted to guide modifications in rRNAs and snRNAs, but the predictions are not experimentally verified (10). In addition, even bioinformatics prediction was shown to be difficult, since no obvious target sites could be found in rRNAs and snRNAs for many snoRNAs, which are called orphan snoRNAs. For example, ∼110 in ∼360 human snoRNAs have no potential target sites in rRNAs or snRNAs (10). These observations raise an interesting possibility that snoRNAs may have other roles, e.g. in modulating expression of protein-coding genes. Indeed, a mammalian brain-specific snoRNA (HBII52) has been shown to regulate alternative splicing and/or mRNA editing (11–13), and miRNAs derived from snoRNAs have been identified in human and Giardia lamblia (14–16). Finally, the snoRNA-related machinery has already been implicated in human diseases, such as Prader–Willi Syndrome, Dyskeratosis congenita as well as cancer (16–19). Thus it is important to functionalize these ncRNAs, and to be able to manipulate their expression for therapeutical purpose.
Several features make it difficult to knockout or knockdown snoRNAs in mammals. First, these RNAs are highly structured and exist in stable snoRNP complexes. Each snoRNP contains a unique snoRNA, and two sets of four core proteins, with Nop56, Nop58, snu13/15.5K and Nop1/fibrillarin for C/D box snoRNPs and Nop10, Gar1, Nhp2 and Cbf5/dyskerin for H/ACA box snoRNPs (6). Second, the gene organizations of snoRNAs are highly diverged in different organisms (20). For example, most yeast snoRNAs are independently transcribed from single copy genes, making it easier for genetic knockout. In contrast, most mammalian snoRNAs are encoded in introns of host genes (21), whereas almost all trypanosome snoRNAs are encoded in reiterated polycistronic snoRNA clusters (22,23), and many plant snoRNAs are encoded in intronic or polycistronic gene clusters (24). Thus, maturation of mammalian (and trypanosome and plant) snoRNAs involves post-transcriptional processing from the host precursor RNAs (8,25). Additionally, many mammalian snoRNA genes have multi-copies or isoforms (10), adding more obstacles for genetic manipulation of snoRNAs.
Several attempts have been made to knock down snoRNAs. In trypanosomes, it has been shown that over-expression of antisense RNA or double-stranded RNA was able to degrade snoRNAs, yet not all tested snoRNAs were depleted (26,27). The working system for studying vertebrate snoRNAs is Xenopus oocyte, where antisense oligonucleotides (ASOs) were microinjected into nucleus to knockdown snoRNA (28). However, this approach cannot fulfill the functional study for mammalian snoRNAs, since many snoRNAs are species specific. In addition, other approaches have been attempted to knockdown mammalian snoRNAs, including siRNA, ribozyme, locked nucleic acid (LNA) and peptide–nucleic-acid oligonucleotides. In no case was the function of the targeted snoRNA disrupted, albeit one tested snoRNA (U81) was reduced by ∼60% using LNA or ribozyme (29), suggesting that a more efficient depletion approach is required for functional analysis. During our study, a recent work was published showing that snoRNAs could be depleted in human cells by 2′-methyloxylethyl modified (2′-MOE), phosphorothioate backbone-containing RNA–DNA chimeric ASOs (30). However, delivery of ASOs was performed in that study using electroporation with a specialized device (nucleofection), making it difficult to judge if ASOs could be delivered to the nucleus in animals by systematic administration and used as drugs to target disease-causing nuclear ncRNAs. In addition, it is also not clear if such an approach is suitable for functionalization of snoRNAs, since most mammalian snoRNAs are encoded in introns of host RNAs, which, in principle, can also be targeted to degradation by the ASOs.
In this study, we determined if snoRNAs could be efficiently depleted by our 2′-MOE/chimeric ASOs using conventional transfection in human and mouse cells. These ASOs can efficiently and specifically deplete mRNAs through an RNaseH1-based mechanism and exhibit high affinity to RNA (31). Our strategy includes full-length screening of ncRNAs to map RNA accessible sites with these ASOs, and using selective reduction of ncRNAs to investigate functional changes of targeted RNAs. We found that all tested snoRNAs and snRNAs could be dramatically depleted with high specificity, and that depletion of intronic snoRNAs does not affect expression of the host genes. We used this method to evaluate the proposed functions of several snoRNAs. Importantly, we also demonstrated that snoRNAs can be dramatically depleted in mice by administration of ASOs.
MATERIALS AND METHODS
Oligonucleotides
Synthesis of RNA/DNA chimeric phosphorothioate/2′-O-MOE oligonucleotides was carried out as described previously (32). Unless indicated, all ASOs used for targeting snoRNAs were 5–10–5 RNA/DNA chimeric oligonucleotides linked with phosphorothioate backbone, with 10 deoxyribonucleotides flanked by 5 2′-O-MOE modified ribonucleotides at both sides. DNA oligonucleotide probes used for northern hybridization, primer extension and quantitative real time PCR (qRT-PCR) are listed in Supplementary Data.
Cell growth and ASO transfection
HeLa cells were grown on plates in DMEM medium supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin at 37°C in 5% CO2 incubator. For knock-down of targeted snoRNA genes, sub-confluent cells (∼30–50%) were treated with ASOs at a 50 nM final concentration (or as indicated) in Opti-MEM medium containing 4 µg/ml Lipofectamine 2000 (Invitrogen) for 4 h, based on the manufacturer’s instruction. Four hours after transfection, medium was replaced with DMEM medium supplemented with 10% FCS and 1% penicillin/streptomycin. For ASO screening, transfection was performed in six-well dishes. For other experiments, transfection was carried out in 75 cm2 dishes. Cells were harvested 48 h after transfection, or at different time points as indicated in figure legends. For multiple snoRNA depletion, ASOs targeting different snoRNAs were co-transfected, at final concentrations described in figure legends.
Northern hybridization and qRT-PCR
Total RNA was prepared using Tri-Reagent (Sigma), based on the manufacturer’s instructions. Five microgram total RNA was separated in 8% polyacrylamide–7 M urea gels and was transferred onto membrane, using semi-dry transfer apparatus. Northern hybridization was performed using 5′-end-labeled oligonucleotide probes, as described previously (33). Real-time PCR (qRT-PCR) was performed using gene-specific primers and probes essentially as described (32). A forward primer T7F2 (5′ tacttaatacgactcactataggctagcctcg 3′) that is specific to a mini-reporter gene derived from exons 6–8 of SMN2 pre-mRNA and a reverse primer SMN-E6/7R (5′ TTTTGTCTAAAACCCATATAATAGCC 3′) were used to detect spliced mRNA, using SMN-E6P (6FAM-CAGATTCTCTTGATGATGCTGATGCTTTGG-IABkFQ) as a probe. Another reverse primer SMN-I6R (5′ TGTCAGGAAAAGATGCTGAGTG 3′) was used in combination with T7F2 and SMN-E6P to detect un-spliced pre-mRNA (T. A. Vickers et al., manuscript in preparation).
Detection of 2′-O-methylations
Methylation was determined by primer extension assay using two different dNTP concentrations (0.05 and 5 mM), as described previously (34). Primer extension sequencing reaction was performed as in (35). Extension products were separated in 8% polyacrylamide–7 M urea gels. For quantitative analysis of 2′-O-methylation, RNase H-cleavage assay was performed (36). Briefly, 2 µl (4 µg) total RNA was hybridized with 3 µl (300 pM) chimeric oligonucleotide (XL086) by heating at 90°C for 5 min, incubated at 37°C for 10 min and cooled on ice for 2 min. Next, 5 µl 2× RNase H buffer (40 M Tris.Cl pH 7.5; 20 mM MgCl2; 200 mM KCl; 50 mM DTT; 10% sucrose) containing 3 units of RNase H (NEB) and 2 units of RNase Inhibitor (NEB) were added. The RNA was digested at 37°C for 60 min, separated in a 1.2% agarose gel and transferred onto a membrane. 18 S rRNA and its 5′ cleaved product were detected by northern hybridization using a 5′-end-labeled probe.
Analysis of pseudouridines
Total RNA was treated with CMC, and pseudouridines were detected by primer extension assay using 5′-end-labeled primers downstream to the pseudouridine sites, as described previously (37).
Chemical probing of snoRNA structure
Preparation of HeLa cell nucleus and chemical modification with dimethylsulfate (DMS) were performed essentially as described (38), except that nuclei were incubated with DMS at a 3% final concentration for 5 min at room temperature (RT), and total nuclear RNA was prepared using Tri-Reagent. Primer extension was performed using 10 µg nuclear RNA and a 5′-end-labeled oligonucleotide specific to U16 snoRNA.
Western blotting
Whole cell lysate was separated in 4–12% gradient SDS–PAGE gels. Proteins were transferred to PVDF membrane for 1 h using a semi-dry transfer apparatus at 25 V constant. The membranes were blocked for 1 h with block buffer (5% dry milk in 1× TBS), and incubated with primary antibodies against RPL4 (11302-1-AP, Proteintech, 1:1500), alpha-tubulin (T-5168, Sigma, 1:8000) or Nucleolin (ab22758, Abcam, 1:2000) at RT for 3–4 h. After three washes with wash buffer (1× TBS, 0.1% Tween-20), membranes were incubated with anti-rabbit or anti-mouse secondary antibody at RT for 1 h. After three washes, proteins were detected using ECL (Abcam).
Immunofluorescence
HeLa cells grown in glass-bottom dishes (MEK) were transfected using Lipofectamine 2000 with 50 nM of different oligonucleotides and incubated for 6 h at 37°C in a 5% CO2 incubator. Cells were washed three times with 1× PBS, fixed at RT for 30 min with 4% formaldehyde and permeabilized for 5 min with 0.1% Triton-100 in 1× PBS. After 30 min blocking with block buffer (1 mg/ml BSA in 1× PBS), cells were incubated for 4 h with block buffer containing an rabbit polyclonal antibody (6653) that recognizes phosphorothioate backbone containing oligonucleotides (1:200, a gift from Erich Koller) and a mouse monoclonal antibody for nucleolin (1:400, ab13541, Abcam). After three washes (5 min each) with wash buffer (0.1% tween-20 in 1× PBS), cells were incubated for 1 h with block buffer containing secondary antibodies against rabbit (ab6717, Abcam, conjugated with FITC; 1:400) and mouse (ab6563, Abcam, conjugated with Cy5; 1:400) IgG. After three washes, anti-fade solution was added and the slides were covered. Cells were imaged with a confocal microscope (Olympus, Fluoview 1000) and images were processed using software FV10-ASW 2.1.
RESULTS
U16 snoRNA can be dramatically depleted by 2′-MOE/chimeric ASOs using conventional transfection in different human cells
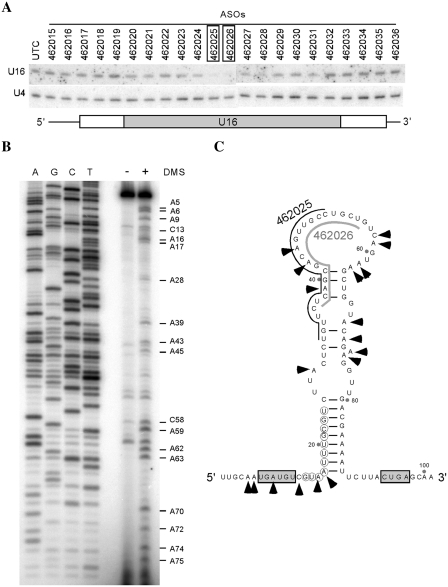
To determine if the 2′-MOE/chimeric ASOs are able to reduce snoRNAs through conventional transfection, we initially targeted U16, a C/D box snoRNA. Twenty seven ASOs were synthesized that cover the entire coding region and adjacent flanking sequences of U16 snoRNA (Figure 1A and Supplementary Figure S1A). Each ASO contains 20 nts linked by Phosphorothioate backbone, with 10 deoxyribonucleotides in the middle flanked at both ends by 5 ribonucleotides containing 2′-MOE modification (Supplementary Figure S1B). HeLa cells were transfected using Lipofectamine 2000 (Invitrogen) with individual ASOs at 50 nM final concentration. The effects of ASO treatment on U16 snoRNA level were determined by northern hybridization. Two ASOs (462025 and 462026) reduced U16 snoRNA by >90%, and moderate reduction (∼40–50%) was found with two neighboring ASOs (462027–462028) (Figure 1A). However, most other ASOs, including those targeting the flanking sequences, had no or little effects.
Figure 1.
A C/D box snoRNA U16 can be specifically depleted by 2′-MOE/chimeric ASOs. (A) Identification of the active ASOs by full RNA screening. HeLa cells were treated with 50 nM ASOs targeting different regions of the U16 snoRNA for 48 h, and total RNA was prepared and subjected to northern hybridization using a 5′-end-labeled oligonucleotide probe specific to U16 snoRNA. U4 snRNA was detected and served as a loading control. The ASOs causing highest reduction in U16 snoRNA levels are boxed. UTC, untreated cells. Lower panel depicts U16 snoRNA regions targeted by the ASOs. The gray box, white box and thin line represent the coding region, the junctions between the coding region and flanking sequence and flanking sequence, respectively. (B) Structure probing for U16 snoRNA. Nuclei were prepared from HeLa cells and incubated with (+) or without (–) 3% DMS at RT for 5 min. RNA was prepared and subjected to primer extension using a 5′-labeled primer specific to U16 snoRNA. Extension products were analyzed in an 8% polyacrylamide, 7 M urea gel, next to a DNA sequencing ladder generated with the same primer. The nucleotides accessible to DMS are identified. (C) Proposed secondary structure of U16 snoRNA. The structure was predicted using program mFold (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi), and refined based on the probing data in (B). The accessible nucleotides are marked with arrowheads. The C and D motifs are boxed. The nucleotides complementary to rRNA for guiding modification are circled. The sequences targeted by the active ASOs (462025 and 462026) are marked by lines.
Since snoRNAs are structured and assembled in RNP complexes, the different effects of ASOs on snoRNA reduction may be related to the accessibility of the snoRNA regions to the ASOs. Indeed, we found that the U16 snoRNA regions targeted by the two most active and three moderately active ASOs exhibit an open structure in vivo, as determined by chemical structure probing using DMS, which modifies accessible A and C nucleotides. The snoRNA regions (U32–U66) targeted by ASO462025–ASO462028 are clearly modified by DMS (Figure 1B and C), suggesting that, as expected, the knockdown efficiency of ASOs correlates with the accessibility of the binding sites in the snoRNP.
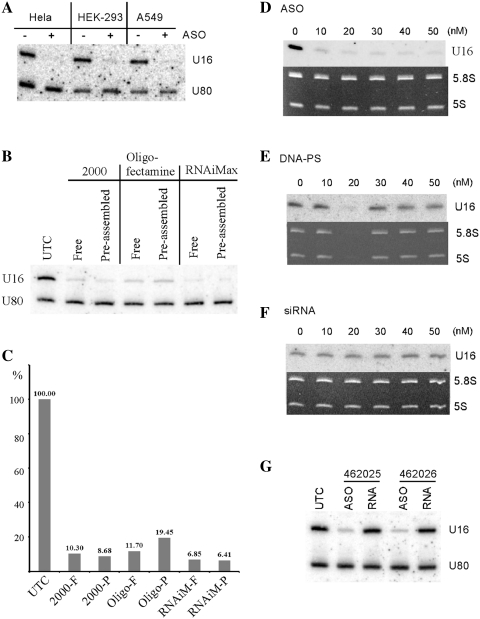
In addition to HeLa cells, U16 snoRNA could also be dramatically depleted in HEK-293 or A549 cells, by transfection of ASO462026 (50 nM) using Lipofectamine 2000 (Figure 2A), indicating that ASOs can knock down U16 RNA in different cell types, by conventional transfection. In fact, dramatic reduction of U16 snoRNA could also be achieved using different transfection reagents. More than 90% reduction was achieved using RNAiMax, similar to Lipofectamine 2000, and ∼85% reduction was obtained with Oligofectamine (Figure 2B and C). Interestingly, pre-assembly of the ASOs with the transfection reagent did not increase the level of snoRNA reduction, as compared with transfection of non-assembled ASOs. The following studies were performed in HeLa cells using Lipofectamine 2000 as transfection reagent, unless indicated.
Figure 2.
2′-MOE/chimeric ASOs operates in different human cells to deplete U16 snoRNA and are more effective than other types of oligonucleotides. (A) Three different types of human cells were treated with (+) or without (–) 50 nM of ASO462026 for 48 h, and levels of U16 snoRNA were determined by northern hybridization. U80 snoRNA was detected and served as a loading control. (B) U16 snoRNA was depleted in HeLa cells by ASO462026 treatment using different transfection reagents. Total RNA was prepared 24 h after transfection, and analyzed by northern hybridization using probes specific to U16 and U80 snoRNAs. 2000, Lipofectamine 2000; Oligo, Oligofectamine; RNAiMax, Lipofectamine RNAiMax (Invitrogen). Free: transfection was performed by adding 4 µg/ml (Lipofectamine 2000 and Oligofectamine) or 6 µg/ml (Lipofectamine RNAiMax) reagents into culturing medium (Opti-MEM, Invitrogen), followed by addition of the U16 ASO to a final concentration of 50 nM. Pre: ASOs were pre-mixed with transfection reagent, and the mixture was incubated at room temperature for 10 min, followed by addition to the culturing medium (Opti-MEM). UTC, untreated cells. (C) The U16 levels detected in (B) were quantified using ImageJ and normalized to the level of U80 snoRNA. The relative levels were plotted and indicated. The transfection reagents were as described in (B). F, free transfection; P, pre-assembled. (D) HeLa cells were treated for 48 h with different concentrations of ASO462026, or a DNA oligonucleotide linked with phosphorothioate backbone that has the same sequence as ASO462026 (E), or siRNA with the same sequence (F). The level of U16 snoRNA was determined by northern hybridization. 5.8 S and 5 S rRNAs are shown as ethidium bromide staining of the gel. (G) HeLa cells were treated for 48 h with 50 nM of either 2′-MOE/chimeric ASOs (ASO) or 2′-MOE modified, phosphorothioate linked RNA oligonucleotides with the same sequences (RNA) as ASOs. Total RNA was prepared and subjected to northern hybridization, as in (A). U80 snoRNA served as a loading control.
While the 2′-MOE/chimeric ASOs are effective in depleting U16 snoRNA, a DNA oligonucleotide linked with phosphorothioate backbone with the same sequence as ASO462026 has little effect. For example, at 50 nM concentration, >90% reduction of U61 snoRNA was obtained with the 2′-MOE/chimeric ASO (Figure 2D), yet only ∼30% reduction was found for the DNA oligonucleotide transfected in the same way (Figure 2E). In addition, treatment with siRNA containing the ASO462026 sequence did not decrease the level of U16 snoRNA (Figure 2F), supporting the previous observation (29). Consistent with the possibility that the ASOs deplete U16 snoRNA through an RNaseH-based mechanism, uniform 2′-MOE/RNA oligonucleotides with the same sequences as ASO462025 or ASO462026 could not reduce U16 snoRNA (Figure 2G). Together, these results indicate that the 2′-MOE/chimeric ASOs can efficiently reduce U16 snoRNA using conventional transfection, in various cell types.
Depletion of U16 snoRNA impaired its function in guiding rRNA modification
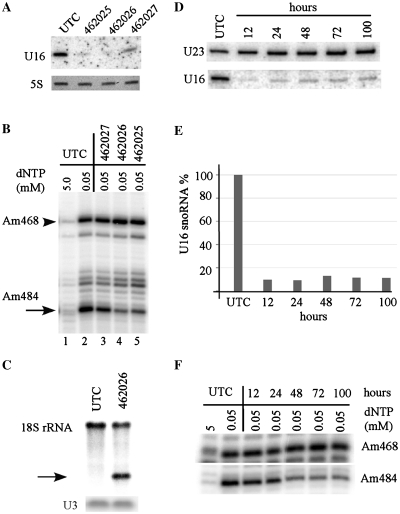
As U16 snoRNA was postulated to guide 2′-O-methylation at position A484 of 18 S rRNA (39), we determined if depletion of U16 snoRNA reduced the methylation level at the predicted site. HeLa cells were treated with different ASOs, and total RNA was prepared 48 h after transfection. As expected, ASO462025 and ASO462026 reduced U16 snoRNA by ∼90%, whereas ASO462027 led to a ∼65% decrease (Figure 3A). The 2′-O-methylation was determined using a primer extension assay, in which methylated nucleotides cause extension to stop one nucleotide before the methylation sites at low but not high dNTP concentration (34). Indeed, dramatic depletion of U16 snoRNA by ASO462025 and 462026 caused ∼70% reduction in the signal strength of primer extension stop at Am484 site (Figure 3B), but not at a neighboring methylation site (Am468). In contrast, moderate reduction of U16 snoRNA by ASO462027 only led to a slight decrease (∼15%) in the methylation signal, consistent with the previous finding that a 60% reduction of a snoRNA (U81) did not impair its guide function (29).
Figure 3.
Depletion of U16 snoRNA impairs its function in guiding 2′-O-methylation. (A) U16 snoRNA was depleted to different extents by treatment with different ASOs (462025, 462026 or 462027) in HeLa cells for 48 h, as determined by northern analysis. Ethidium bromide staining of 5 S rRNA is shown in the lower panel. (B) The same total RNA as used in (A) was subjected to primer extension analysis using a primer specific to 18 S rRNA, to determine the methylation status at the predicted site (A484). Extension products were separated in an 8% polyacrylamide gel. The targeted Am484 is indicated with an arrow, whereas an adjacent methylation (Am468) is marked with an arrowhead. m, 2′-O-methylation. (C) Methylation reduction at site A484 was confirmed by an RNase H-cleavage assay. Total RNA was annealed with a DNA–RNA chimeric oligonucleotide complementary to A484 region of 18 S rRNA, followed by digestion using E. coli RNaseH. The digested products were analyzed by northern hybridization using a probe specific to 18 S rRNA, upstream to A484 site. The cleaved fragment representing unmodified 18 S rRNA is marked with an arrow. U3 snoRNA served as a loading control. (D) Time course of U16 snoRNA depletion by ASO treatment. HeLa cells were transfected with 50 nM ASO462026 for different times, as indicated above lanes. Total RNA was prepared and the level of U16 was analyzed by northern hybridization. U23 was probed and used as a loading control. (E) Quantification of U16 snoRNA as detected in (D), and normalized to U23 snoRNA. (F) Methylation reduction at A484 was apparent at later time points. Total RNA as used in (D) was subjected to primer extension analysis to detect the levels of methylation, as in (B). Methylation at site A468 was shown as a control.
To further confirm the methylation reduction at position A484 of 18 S rRNA, we employed a more quantitative approach, an oligonucleotide directed, site-specific RNaseH cleavage assay (36), in which only the 18 S rRNA lacking A484 methylation could be cleaved. Total RNA was incubated with a DNA/RNA chimeric oligonucleotide specific to the A484 region, digested by RNase H, and 18 S rRNA was probed by northern hybridization. Approximately 55% of 18 S rRNA from U16 depleted cells was cleaved, whereas no rRNA cleavage was found for control cells (Figure 3C), indicating that depletion of U16 indeed reduced the level of A484 methylation. The incomplete loss of A484 modification may stem either from the long half-life of mammalian rRNAs (∼3–7 days) (40), or from the remaining enzymatic activity of the residues U16 snoRNP. Nevertheless, these results indicate that ASO-mediated depletion of U16 snoRNA specifically reduced the methylation guided by this snoRNA, and that the predicted function of this RNA could be confirmed.
Since rRNAs have a long half-life, significant reduction of methylation is expected to appear at a time point later than depletion of the snoRNA. To determine the kinetics of snoRNA and methylation reduction, HeLa cells were treated with ASO462026 for different times, and the levels of U16 snoRNA and methylation at A484 of 18 S rRNA were analyzed. Although U16 snoRNA was reduced to a maximum extent within 12 h (as early as 4 h) after transfection (Figure 3D and E, and data not shown), strong reduction in the methylation level was only detected at 48 h and after (Figure 3F). Note that reduction of U16 snoRNA maintained for at least 100 h after transfection, suggesting the high stability and long-lasting activity of the ASOs. Together, these results suggest that the suitable time window for functional study of snoRNAs could be 48–100 h after transfection.
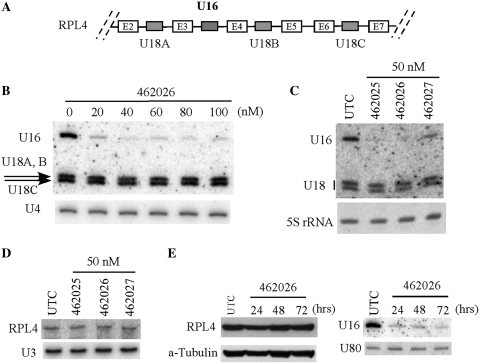
ASOs targeting U16 snoRNA do not impair the expression of the host gene
Like most human snoRNAs, U16 is embedded in an intron of its host gene encoding a ribosomal protein RPL4 (10). The same pre-mRNA also contains another intronic snoRNA, U18, which has three isoforms in different introns (Figure 4A). To determine whether the ASOs targeting U16 snoRNA act on the U16 coding region of the host pre-mRNA, or on the mature U16 snoRNA, we detected the expression of the host gene and the levels of the neighboring U18 snoRNAs. No difference in the levels of U18 snoRNAs was observed in cells treated with different concentrations of ASO462026 (Figure 4B), or with different ASOs targeting different regions of U16 snoRNA (Figure 4C), although U16 snoRNA was dramatically depleted in these cases. This observation suggests that U16 ASOs may not act on pre-mRNA. Indeed, the level of mature RPL4 mRNA was not affected by treatment of ASOs targeting different regions of U16 snoRNA (Figure 4D). Importantly, the level of RPL4 protein also remained unchanged even 72 h after transfection, as detected by western analysis (Figure 4E, left panel), although in the same cells U16 snoRNA was dramatically depleted by ASO treatment (Figure 4E, right panel). Together, these results indicate that the ASOs act on mature U16 snoRNA or on pre-snoRNA after pre-mRNA splicing, or both. This observation is consistent with a previous study, in which degradation of trypanosome snoRNAs by over-expression of antisense RNA also occurred at the mature snoRNA level (26), although the RNA-mediated snoRNA degradation in trypanosomes should utilize a mechanism different from RNaseH. In sum, our results indicate that ASO-mediated U16 depletion does not affect expression of the host gene.
Figure 4.
Depletion of U16 snoRNA by ASO treatment does not impair the expression of the host gene. (A) Schematic depiction of the U16 snoRNA gene locus. The partial host pre-mRNA (RPL4) is shown. The gray boxes indicate intronic snoRNAs (U16 and three isoforms of U18 snoRNA) in the host gene. E2–E7, exons of RPL4 pre-mRNA. (B) ASOs targeting U16 did not affect the levels of U18 snoRNAs encoded in the same host pre-mRNA. HeLa cells were treated for 48 h with different concentrations of ASO462026. Total RNA was prepared and subjected to northern hybridization with probes specific to U16 and U18 snoRNAs. U4 snRNA was detected as a loading control. (C) HeLa cells were treated for 48 h with 50 nM of three different ASOs targeting U16 snoRNA. Northern hybridization was performed as in (B). Ethidium bromide staining of 5 S rRNA is shown in lower panel. (D) The mRNA level of the host gene (RPL4) was not changed by U16 ASOs. Aliquots of RNAs as in (C) were separated in a 6% polyacrylamide gel, and RPL4 mRNA was detected by northern hybridization. U3 snoRNA served as a loading control. (E) Depletion of U16 by ASO did not alter the protein level of RPL4. HeLa cells were treated with U16 ASO (462026) for different times, as indicated above lanes. Whole cell extracts were prepared and subjected to western analysis using an anti-RPL4 antibody (left panel). As a control for loading, the same membrane was re-probed with an alpha-tubulin antibody. In addition, total RNA was prepared from an aliquot of test cells and subjected to northern hybridization, using probes specific to U16. The same membrane was re-probed for U80 snoRNA, which served as a loading control (right panel).
Other snoRNAs can also be depleted by ASO treatment using conventional transfection
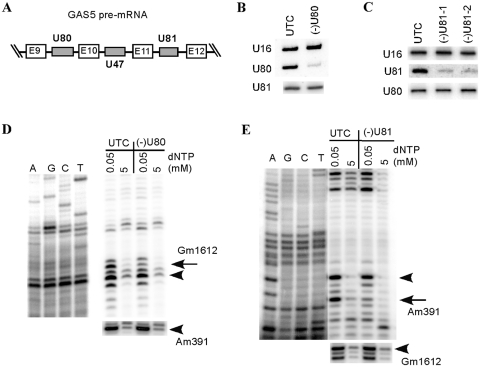
As described above, ASO treatment through conventional transfection efficiently and specifically depleted U16 snoRNA in human cells. To determine if this approach can also be used to reduce other snoRNAs, we targeted two additional snoRNAs, U80 and U81. These two C/D box snoRNAs are encoded in different introns of the same host non-protein-coding RNA, GAS5 (41) (Figure 5A). Nine ASOs were synthesized for each snoRNA to cover the entire coding regions of these RNAs (Supplementary Figure S2A and C). Individual ASOs were transfected into HeLa cells using Lipofectamine 2000, in the same way as for U16 snoRNA depletion. Total RNA was prepared 48 h after transfection, and subjected to northern hybridization. One active ASO for U80 and two active ASOs for U81 were identified that caused >85% reduction of these snoRNAs, respectively (Supplementary Figure S2B and D), indicating that other C/D snoRNAs can also be depleted by ASO treatment. Interestingly, the active ASOs for both U80 and U81 snoRNAs are identified to target snoRNA guide sequences (Supplementary Figure S2). This observation is in agreement with the view that snoRNA guide regions have a relatively open structure and are ready to base-pair with the substrate RNAs. Note that the same region of U81 snoRNA has been targeted in a previous study using LNA or ribozyme, yet only ∼60% reduction was obtained in that study (29), suggesting the high efficiency of our current approach.
Figure 5.
2′-MOE/chimeric ASOs can also be used to deplete other C/D box snoRNAs. (A) Gene organization of U80 and U81 snoRNAs in the introns of GAS5 gene. Gray boxes indicate the intronic snoRNAs. The E9–E12 exons of GAS5 gene are shown in white boxes. (B) Northern hybridization for U80 snoRNA in HeLa cells treated [(–)U80] or not treated (UTC) with 50 nM of an active ASO (477457). U16 snoRNA was detected and served as a loading control. The same blot was re-hybridized with a probe specific to U81 snoRNA. (C) U81 snoRNA was dramatically depleted by U81-specific ASOs. HeLa cells were treated with 50 nM of different active ASOs targeting U81 snoRNA [477443 for (–)U81-1, and 477444 for (–)U81-2]. Total RNA was prepared and subjected to northern hybridization, as in (B). (D) Depletion of U80 snoRNA specifically reduced the methylation level at the predicted site in 28 S rRNA (G1612). The modification was mapped by a primer extension assay using a primer specific to 28S rRNA. Extension products were analyzed in an 8% polyacrylamide gel, next to primer extension sequencing reactions performed with the same primer. The Gm1612 site is indicated with an arrow, whereas a neighboring methylation site or the site guided by U81 (Am391) are marked with arrowheads. Note that the methylation level of A391 guided by U81 snoRNA was not affected by depletion of U80 snoRNA, as determined by primer extension using a different primer (lower panel). (E) Depletion of U81 snoRNA specifically impaired its function in guiding methylation at site A391 of 28 S rRNA. Primer extensions were performed as in (D), using different primers specific to different regions of 28 S rRNA. The arrow indicates the Am391 site guided by U81, whereas arrowheads mark a neighboring methylation or Gm1612 methylation guided by U80 snoRNA (lower panel).
Although U80 and U81 are encoded in different introns of the same pre-mRNA, depletion of U80 by its active ASO did not affect the level of U81 snoRNA, and vice versa (Figure 5B and C). This result is consistent with the observation that ASOs targeting U16 snoRNA do not act on pre-mRNA. Again, depletion of U80 snoRNA specifically reduced the methylation level by ∼65% at the predicted position (G1612) of 28 S rRNA, yet no level change was found for methylation at site A391 of 28 S rRNA that was predicted to be guided by U81 snoRNA (Figure 5D). Similarly, depletion of U81 specifically reduced methylation level (∼70%) at A391, but not G1612 of 28 S rRNA (Figure 5E). Together, these data indicate that ASO-mediated snoRNA depletion can also be used to target other snoRNAs, and that a ∼80–85% reduction of snoRNAs is sufficient to cause significant loss of function.
ASOs are highly specific to the target snoRNAs
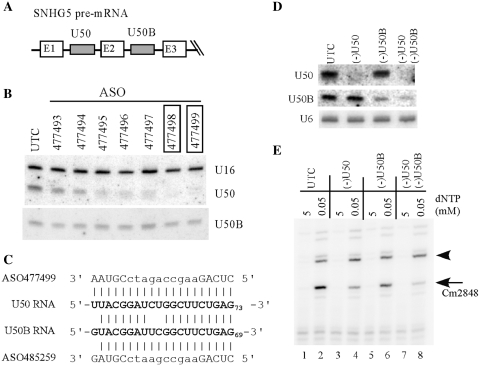
Many mammalian snoRNAs have isoforms with significant sequence similarities (10), thus ASOs targeting a particular snoRNA may act on other isoforms or even other RNAs through imperfect base-pairing. To demonstrate the specificity of the ASOs, two isoforms of a C/D box snoRNA were chosen: U50 and U50B, which are encoded in different introns of the same host gene (SNHG5) and share >75% identity (Figure 6A and Supplementary Figure S3). ASOs were screened for U50, and two active ASOs were identified that caused >90% reduction of the snoRNA (Figure 6B). However, no significant change on the level of U50B snoRNA was observed in cells treated with these ASOs, suggesting that the ASOs are specific. Indeed, one active ASO that causes ∼90% reduction of U50 snoRNA targets the guide sequence of the snoRNA (ASO477499, Supplementary Figure S3A), however, no significant reduction of U50B was observed, despite that the guide sequence is conserved between U50 and U50B snoRNAs, except two continuous mis-matches in the middle and one at the 5′-end (Figure 6C).
Figure 6.
ASOs are highly specific to the target snoRNAs. (A) The U50 snoRNA and its isoform U50B are encoded in different introns of SNHG5, a non-protein coding gene. (B) Northern hybridization of U50 and U50B snoRNAs in cells treated for 48 h with 50 nM of different ASOs targeting U50 snoRNA. The active ASOs are boxed. U16 snoRNA was detected and served as a loading control. (C) Sequence comparison of ASOs targeting the guide region of U50 and U50B snoRNAs. The snoRNA sequences are in bold. The uppercase letters in ASO sequences indicate 2′-MOE/RNA sequence, whereas lowercase letters represent DNA sequence. (D) ASOs specific to U50 (ASO477499) or U50B snoRNA (ASO485259) act specifically on the targeted snoRNAs. HeLa cells were treated for 48 h with ASOs targeting either U50 or U50B alone (50 nM), or with two ASOs targeting both snoRNAs (30 nM each). Total RNA was prepared and subjected to northern hybridization using 5′-end-labeled oligonucleotides specific to U50 or U50B, respectively. U6 snRNA was detected and served as a loading control. (E) Depletion of U50 or U50B snoRNA impairs the methylation guided by the snoRNA. Modification at C2848 in 28S rRNA was detected by primer extension assay using a primer specific to the C2848 region. The targeted modification site is indicated with an arrow, and a neighboring potential modification site is marked with an arrowhead.
To further confirm the specificity, a U50B specific ASO was synthesized (ASO485259) that contains one mis-match at the 5′-end of the U50B snoRNA guide region. This ASO has two mis-matches in the middle of the duplex formed with U50 snoRNA (Figure 6C). Consistently, transfection of the U50B ASO specifically depleted U50B, but not U50 snoRNA, and co-transfection of U50 and U50B ASOs depleted both snoRNAs (Figure 6D). This result indicates that two mis-matches in the middle of the duplex are sufficient to inactivate the ASOs. In addition, it appears that one mis-match at the end of the duplex does not affect the ASO activity, as shown for the U50B ASO. Interestingly, depletion of either U50 or U50B snoRNA reduced the methylation level at the predicted site (Cm2848 in 28 S rRNA) by ∼70 or ∼30%, respectively, however, simultaneous depletion of both snoRNAs caused greater reduction (∼85%) in methylation, indicating that both isoforms are functional in cells (Figure 6E). Together, these results show that 2′-MOE/chimeric ASOs can deplete snoRNAs and inactivate their functions in a highly specific manner.
ASO treatment can also deplete H/ACA type of snoRNAs
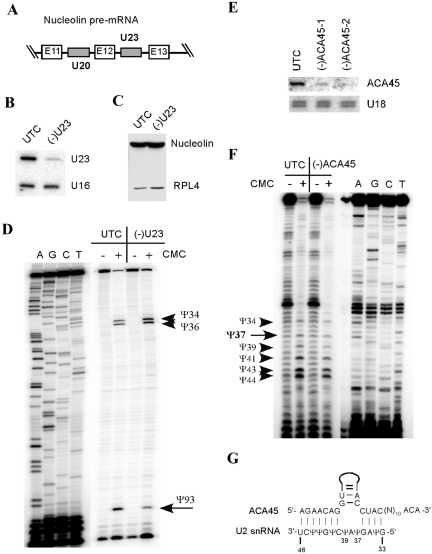
All the five snoRNAs targeted above by 2′-MOE/chimeric ASOs were successfully depleted, indicating that this approach is not unique to individual RNAs. Since these snoRNAs are all C/D box RNAs, next, we investigated if another group of snoRNAs, H/ACA box RNAs that typically contain two hairpin structures, could also be depleted in this way. Twenty five ASOs were synthesized for U23, an H/ACA snoRNA encoded in an intron of Nucleolin pre-mRNA (Figure 7A and Supplementary Figure S4A). Two active ASOs were identified that reduced the level of U23 snoRNA by 80–85% (Figure 7B and Supplementary Figure S4B), indicating that H/ACA type of snoRNAs can also be depleted by conventional transfection of the ASOs. The two active ASOs target the terminal or internal loop regions in the 3′-hairpin of U23 snoRNA, suggesting a higher accessibility of these regions (Supplementary Figure S4C).
Figure 7.
H/ACA box RNAs can be depleted by ASO treatment. (A) Schematic representation of partial pre-mRNA of Nucleolin. The intronic snoRNAs U23 (H/ACA type) and U20 (C/D type) are shown in gray boxes. (B) Northern hybridization for U23 snoRNA in cells treated [(–)U23] or not treated (UTC) with 50 nM of an active ASO targeting this snoRNA (ASO483788). U16 snoRNA served as a loading control. (C) The protein level of Nucleolin was not affected by the U23 ASO, as determined by western analysis. RPL4 protein was also probed and served as a loading control. (D) Depletion of U23 H/ACA snoRNA impaired its function in guiding pseudouridylation at site U93 of 18S rRNA. Total RNA was treated (+) or not treated (–) with CMC (N-cyclohexyl-N′-(2-morpholinoethyl)-carbodiimide methyl-p-toluolsulfonate), and subjected to primer extension analysis using a primer specific to U93 region of 18S rRNA. CMC treatment causes extension to stop one nucleotide before the pseudouridylated sites. Extension products were separated in an 8% polyacrylamide gel, next to primer extension sequencing reactions preformed with the same primer. The targeted pseudouridylation (Ψ93) is indicated. Two neighboring pseudouridines (Ψ34 and Ψ36) are marked with arrowheads. (E) An H/ACA type of scaRNA can also be degraded by ASOs. HeLa cells were treated for 48 h with two different ASOs targeting ACA45 scaRNA [462037 for (–)ACA45-1 and 462038 for (–)ACA45-2]. Total RNA was prepared and subjected to northern hybridization using a probe specific to ACA45 scaRNA. U18 snoRNA was detected and served as a loading control. (F) Dramatic depletion of ACA45 scaRNA did not disrupt pseudouridylation at the predicted site in U2 snRNA (Ψ37). Pseudouridines were detected as in (D), using a primer specific to U2 snRNA. Extension products were separated in an 8% polyacrylamide gel, next to primer extension sequencing ladders generated using the same primer. The predicted target site (Ψ37) is indicated, and other neighboring pseudouridines are marked with arrowheads. (G) Base pairing potential between ACA45 scaRNA and U2 snRNA. U2 snRNA sequence is numbered and the pseudouridines (Ψ) in U2 snRNA are shown.
As was the case for depleting C/D snoRNAs, reduction of U23 H/ACA snoRNA by ASO treatment occurred at the mature U23 snoRNA, since the level of another snoRNA (U20) encoded in the same precursor was not altered (Supplementary Figure S4B), and the protein level of the host gene (Nucleolin) remained unchanged (Figure 7C). These results further support the view that ASO-mediated snoRNA depletion does not affect expression of the host genes. Next, we analyzed if a ∼80% reduction of U23 snoRNA could impair its predicted function in guiding pseudouridylation at position U93 of 18 S rRNA (10). Pseudouridylation was determined by a primer extension assay following CMC treatment of total RNA (37). As expected, depletion of U23 snoRNA significantly reduced the level of pseudouridylation at the predicted position (U93) in 18 S rRNA, as compared with the level of neighboring pseudouridylation sites (Figure 7D). Thus, despite of their highly structured nature, H/ACA snoRNAs can also be depleted by 2′-MOE/chimeric ASOs via conventional transfection, and an 80% reduction of U23 snoRNA is sufficient to inactivate its function.
ASOs act on scaRNAs
In addition to snoRNAs that are accumulated in the nucleoli, similar RNAs (scaRNAs) are also present in the Cajal bodies where these RNAs guide nucleotide modification in snRNAs (42). To determine if scaRNAs can also be targeted by ASO treatment, 16 ASOs were synthesized for ACA45 (Supplementary Figure S5A), a scaRNA predicted to direct pseudouridylation at site U37 of U2 snRNA (43). Two active ASOs were identified that target the terminal loop region of the 5′-hairpin (Supplementary Figure S5B and C). The level of ACA45 scaRNA was reduced by ∼85% with these two ASOs (Figure 7E), indicating that ASOs can also be used to deplete scaRNAs. Interestingly, no change was observed for the level of pseudouridylation at the predicted position (U37) in U2 snRNA (Figure 7F). This result suggests that either the small amount of ACA45 left is sufficient for full function, or that this RNA does not have the predicted role. Consistent with the latter possibility, we noticed that three nucleotides (U37A38U39) of U2 snRNA are unpaired in between the two intermolecular duplexes formed between U2 snRNA and ACA45 RNA (Figure 7G). This is different from the guide rule in which two nucleotides are unpaired in the middle (44), suggesting that ACA45 may not have the predicted function.
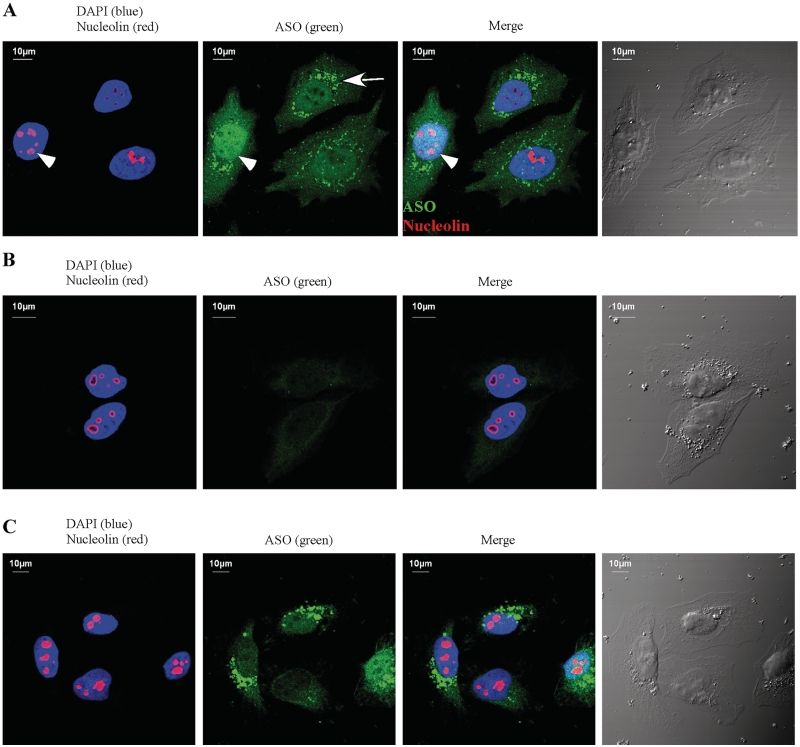
The ASOs targeting a snoRNA can localize to the nucleus
As demonstrated above, ASO-mediated snoRNA depletion does not occur at pre-mRNA level, rather, it takes place most likely at mature snoRNA level. Since snoRNAs are produced in the nucleoplasm, followed by import into the nucleolus where they accumulate (6), snoRNA degradation could occur either in the nucleoplasm or in the nucleolus, or both. To determine if ASOs accumulate in the nucleolus, ASO462026 targeting U16 snoRNA was transfected into HeLa cells, and the ASOs were detected through indirect immunofluorescence using a rabbit polyclonal antibody that recognizes phosphorothioate backbone containing oligonucleotides in a sequence independent manner (a gift kindly provided by Dr Erich Koller). As shown in Figure 8A, the 2′-MOE chimeric ASO is enriched and evenly distributed in the nucleus, indicating that the ASO can be delivered to this compartment by conventional transfection. Most nucleoli lack ASO staining, however, nucleolar signal of ASOs can be detected in certain cases (Figure 8A, indicated by an arrowhead). This observation suggests that ASOs can localize in the nucleolus as well, although to a lesser extent than in the nucleoplasm. The large cytoplasmic speckles most likely reflect the ASOs that are still present in the endosomal particles. Note that the staining is specific to the phosphorothioate linked oligonucleotides, since no significant signal was detected especially in the nucleus of cells transfected in the same way with a conventional, unmodified oligodeoxynucleotide (Figure 8B). Despite the fact that a phosphorothioate linked oligodeoxynucleotide that contains the same sequence as ASO462026 can also be delivered to the nucleus in this way (Figure 8C), this oligodeoxynucleotide failed to deplete U16 snoRNA (Figure 2E), suggesting that higher affinity of ASOs to target RNA is required for efficient reduction of snoRNAs. Together, these data imply that most likely ASOs act on snoRNAs in the nucleoplasm. However, we cannot completely exclude the possibility that ASOs can trigger snoRNA degradation in the nucleolus.
Figure 8.
ASOs are enriched in the nucleus after conventional transfection. HeLa cells were transfected with either ASO462026 (A), or a conventional DNA oligonucleotide [XL011, (B)], or the DNA oligonucleotide linked with phosphorothioate backbone, as used in Figure 2E (C). After incubation at 37°C for 6 h, cells were fixed, stained with antibodies against the phosphorothioate linked oligonucleotides (green) and Nucleolin (red), a nucleolar marker protein, as described in ‘Materials and Methods’ section. The nucleus was stained with DAPI (blue). The arrow indicates cytoplasmic ASO particles, whereas the arrowhead marks the nucleolus with ASO staining. The scale bars: 10 µm.
snRNAs can also be degraded by ASO treatment
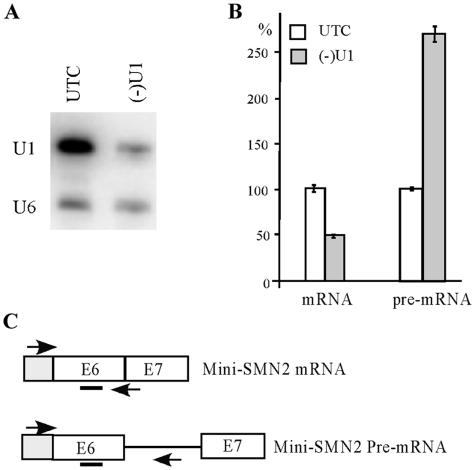
Next, we tested if snRNAs that are present in nuclear speckles can also be depleted in the same way. ASOs were synthesized for four spliceosomal U snRNAs (U1, U2, U4 and U6) and transfected into HeLa cells. All the tested snRNAs were successfully reduced by >85% (T. A. Vickers et al., manuscript in preparation). As exemplified for U1 snRNA, ASO treatment reduced the level of this snRNA by ∼90%, as determined using northern hybridization (Figure 9A). As expected, depletion of U1 snRNA impaired pre-mRNA splicing, resulting in reduction of mature mRNA and accumulation of pre-mRNA, as determined by qRT-PCR for a reporter minigene, using different probe-primer sets for mature or pre-mRNA (Figure 9B and C). This result indicates that ASOs can also successfully target ncRNAs in the nucleoplasm.
Figure 9.
Spliceosomal snRNAs can be depleted by ASO treatment using conventional transfection. (A) HeLa cells were treated for 48 h with 50 nM of a U1 specific ASO (46,9508). Total RNA was prepared and U1 snRNA was detected by northern hybridization. U6 snRNA was determined and served as a loading control. UTC, untreated cells. (B) ASO-mediated U1 depletion impaired pre-mRNA splicing. The levels of mature mRNA and un-spliced pre-mRNA of a mini-reporter gene (derived from SMN2 gene, exon 6–8) were determined by qRT-PCR, using primer probe sets specific to spliced or un-spliced RNAs. (C) Positions of primer probe sets used in (B). Exons (E) are shown in boxes. Primer positions are indicated using arrows. The probe position in exon 6 is marked with a bar.
Multiple snoRNAs can be simultaneously depleted by ASO treatment
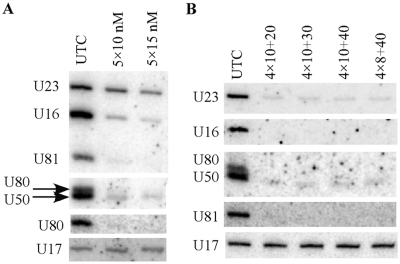
Based on the study performed in yeast, genetic deletion of most guide snoRNAs individually has no obvious effects on cell growth, whereas combined knockout of several snoRNAs targeting modifications in a particular domain of the rRNA can significantly impair ribosome biogenesis and function, e.g. (33,45,46). These findings suggest that several snoRNAs need to be depleted at the same time in order to study the function of modifications. Thus, we determined if multiple snoRNAs could be depleted simultaneously in human cells by co-transfection of a set of ASOs targeting different RNAs. Five snoRNAs, including four C/D snoRNAs (U16, U50, U80, U81) and one H/ACA snoRNA (U23), were targeted by co-transfection of five active ASOs specific to these RNAs, with 10 nM or 15 nM final concentrations of each ASO. The four C/D snoRNAs were all dramatically depleted (>85% reduction), however, the H/ACA snoRNA (U23) was only moderately reduced at these ASO concentrations (Figure 10A), consistent with the fact the H/ACA RNAs have a more structured feature. This result suggests that ASO concentration for the H/ACA snoRNA should be higher than that for C/D snoRNAs, in order to obtain significant reduction.
Figure 10.
Multiple snoRNAs can be depleted simultaneously. (A) HeLa cells (∼50% confluency) were transfected with five different ASOs targeting C/D snoRNAs U16, U50, U80, U81 and H/ACA snoRNA U23 (ASOs 462026, 477499, 477457, 477443 and 483788, respectively), at 10 nM (5 × 10 nM) or 15 nM (5 × 15 nM) final concentration of each ASO. Total RNA was prepared 24 h after transfection and subjected to northern hybridization for the levels of these snoRNAs. U17 snoRNA was detected and served as a loading control. Note that U80 snoRNA was detected twice to separate from U50 snoRNA. (B) HeLa cells (∼30% confluency) were transfected with the five ASOs as indicated in (A), but with different ASO concentrations. 4 × 10 or 4 × 8, the final concentrations of the four ASOs targeting the four C/D snoRNAs was either 10 or 8 nM, respectively. The concentrations of the U23 ASO were 20, 30 or 40 nM (+20, +30 and +40). Northern hybridization was performed as in (A).
Next, we tested the effects of different ASO concentrations on depleting U23 snoRNA, in combination with 10 or 8 nM ASOs for C/D snoRNAs. Indeed, U23 snoRNA was reduced by >85% at 20–40 nM ASO concentrations, and the four C/D snoRNAs were also dramatically depleted (>90%) at 10 or 8 nM concentration of ASOs (Figure 10B). These observations indicate that for multiple depletions, different snoRNAs may require different ASO concentrations, with higher concentration for H/ACA snoRNAs. We note that cells grew normally even at the highest ASO concentration (total 80 nM), suggesting that there was no toxicity under these conditions. In addition, we also noticed that lower cell confluency for transfection could lead to a greater reduction. For example, at 10 nM ASO concentration, ∼85% reduction was obtained for U16 snoRNA with ∼50% confluency of starting cells (Figure 10A), whereas >95% reduction was achieved with ∼30% confluency (Figure 10B), suggesting that cell confluency affects transfection efficiency, which in turn affects depletion effects. All together, these results indicate that multiple snoRNAs can be simultaneously depleted in human cells by ASOs using conventional transfection.
snoRNAs can be depleted in vivo by systematic administration of the ASOs
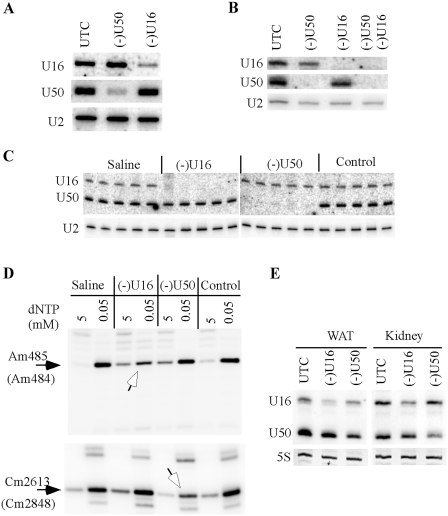
The results described above indicate that the 2′-MOE/Chimeric ASOs are highly suitable and convenient for functional study of the nuclear ncRNAs, by conventional transfection. These findings suggest that the ASOs may also operate in animals, such as mice, to manipulate the level of snoRNAs. To demonstrate this possibility, we first targeted snoRNAs in different mouse cells in vitro, to ensure the activity of the ASOs. Mouse U16 and U50 snoRNAs were targeted, since a human U16 snoRNA ASO (462026) and a U50 snoRNA ASO (477499) have perfect complementarities to the mouse snoRNAs, respectively. The two mouse snoRNAs were reduced by >80% in b.ENDs cells (Figure 11A) or by >95% in primary hepatocytes (Figure 11B), using conventional transfection of the corresponding ASOs. Like the case in human cells, the two snoRNAs can also be simultaneously depleted in mouse cells by co-transfection of the ASOs, indicating that ASO-mediated snoRNA depletion also operates in mouse cells.
Figure 11.
snoRNAs can be depleted in mice by systematic administration of ASOs. ASO477499 (for U50) and ASO462026 (for U16) were transfected individually (50 nM) or together (35 nM each) into bEND.3 cells (A) or mouse primary hepatocytes (B) using Lipofectamine 2000, and total RNA was prepared 24 h after transfection. The levels of U16 and U50 were determined by northern hybridization. U2 snRNA was detected and served as a loading control. (C) 2′-MOE/chimeric ASOs can deplete snoRNAs in vivo. Seven-week-old mice (n = 5) were subcutaneously injected with 100 mg/kg of ASO462026 (U16), ASO477499 (U50) or ASO141923 (Control), and sacrificed 72 h later. Total RNA was prepared from liver and subjected to northern hybridization to determine the levels of U16 and U50 snoRNAs. U2 snRNA was used as a loading control. Different lanes represent total liver RNA prepared from different mouse. Saline, total liver RNA prepared from mouse injected with saline. (D) Reduction of snoRNAs in vivo decreased the levels of targeted rRNA methylation. Total RNA used in (C) was pooled for each group, and subjected to primer extension analysis to detect rRNA methylation at positions targeted by U16 snoRNA (Am485 in 18 S rRNA) or U50 snoRNA (Cm2613 in 28 S rRNA). Open arrows indicate the reduced methylation level that is reflected by decreased signal strength at 0.05 mM dNTP concentration, as compared with control samples. The modified positions are numbered with mouse rRNA. The equivalent positions in human rRNAs are shown in parentheses. (E) Moderate reduction of snoRNAs can also be observed in other tissues. Total RNA was prepared from different tissues of the animals used in (C), pooled for each group, and analyzed by northern hybridization, as in (C). Ethidium bromide staining of 5 S rRNA is shown. WAT, white adipose tissue.
Next, ASO effects on snoRNA depletion in vivo were determined through systematic administration of a single dose of 100 mg/kg ASOs in 7-week-old male BALB/c mice (n = 5). Animals were sacrificed 72 h after subcutaneous injection and total RNA was prepared from mouse liver. As detected by northern hybridization, U16 or U50 snoRNA was almost completely depleted in liver of mice administrated with the corresponding ASOs (Figure 11C). No significant difference in depletion efficiency was observed among the five animals within each group. As controls, injection of either saline or a control ASO (ISIS141923) has no effect on the levels of these snoRNAs, indicating the specificity of the ASOs and that ASOs can be delivered into liver and act in the nucleus. Importantly, depletion of these snoRNAs also caused specific reduction (∼50%) in the levels of corresponding rRNA methylation guided by these snoRNAs, as determined by a primer extension assay (Figure 11D), consistent with the in vitro results. In addition, moderate reduction (∼40–60%) was also observed for U16 and U50 snoRNAs in kidney and white fat (WAT), but no significant depletion was found in heart, spleen and muscle (Figure 11E and data not shown), suggesting that ASOs can also be delivered to tissues other than liver. Together, these results indicate that snoRNAs can be dramatically depleted in mice by systematic administration of the 2′-MOE/chimeric ASOs.
DISCUSSION
Using conventional transfection, here we showed that the 2′-MOE/chimeric ASOs can efficiently degrade all tested ncRNAs that are present in different sub-nuclear structures, including nucleolus, Cajal bodies and nuclear speckles. Depletion was achieved in different human and mouse cells, using different transfection reagents. Reduction of snoRNAs significantly reduced the level of nucleotide modification in endogenous rRNAs guided by the snoRNAs. Surprisingly and attractively, ASOs targeting intronic snoRNAs do not affect expression of the host genes, thus minimizing unexpected effects for functional studies. Importantly, snoRNAs can also be depleted in vivo by subcutaneous injection of the 2′-MOE/chimeric ASOs, making ASOs a powerful approach to functionalize small ncRNAs, and potentially as drugs to target disease-causing ncRNAs.
In our study, the most active ASOs were identified using a full RNA screening approach. The candidate target regions of snoRNAs can be proposed based on our data and published information. For four of the five tested C/D snoRNAs, the most potent ASOs were found to target the guide regions (ASO 477457 for U80 snoRNA, ASO477444 for U81 snoRNA, ASO477499 for U50 snoRNA and ASO485259 for U50B RNA). This observation is consistent with the fact that C/D snoRNAs contain long, continuous antisense guide sequence (12–24 nt) that base-pair with the substrate RNAs (47), therefore the guide sequences are more likely to be accessible to the ASOs. However, this is not always the case, as shown for the U16 snoRNA, for which the most potent ASOs target snoRNA regions outside the guide sequence. Thus, it is reasonable to target the guide region(s) of C/D snoRNAs as a first attempt. However, screening for active sites may be required if no success with the guide region(s). For H/ACA RNAs, which typically contain two hairpin structures, the most active sites for ASO target were found to be the terminal loop regions, as well as the internal loop where the guide sequence may be present.
The observation that at least five snoRNAs could be depleted simultaneously is interesting. Co-transfection of five different ASOs caused dramatic reduction of all the five targeted snoRNAs. This finding suggests that these ASOs do not strongly compete for the recruitment of RNaseH, or that RNaseH is not a limiting factor under these experimental conditions. This feature makes the ASOs a very convenient and efficient approach to analyze the biological roles of multiple modifications in a particular domain of rRNAs or snRNAs, since in many cases deletion of a single modification had no effect, yet cumulative or synergistic effects could be observed when several modifications were co-deleted from rRNA, e.g. (33,45–46).
During this work, a report published recently that showed that ASO-mediated snoRNA reduction (>80%) in human cells was achieved using a specialized device, but not by conventional transfection (30). Here we show clearly that 2′-MOE/chimeric ASOs can efficiently deplete small ncRNAs present in different sub-nuclear structures by conventional transfection in different human and mouse cells, even with different transfection reagents. The different observations for conventional transfection may step from the fact that, in our study, the most active ASOs were identified by full RNA screening, whereas in the previous study the sequences of ASOs were not optimized. Nevertheless, our results indicate that different nuclear ncRNAs can be depleted by convenient, conventional transfection to a level that is sufficient to cause functional loss.
Consistent with previous studies (29,30), here we also found that siRNA does not act on snoRNAs, inferring that RISC is not active in the nucleus of human cells. To support an RNaseH-based mechanism, transfection of MOE/RNA oligoes did not reduce the level of target snoRNA. Additionally, a DNA oligonucleotide with the same sequence as a most potent ASO only caused a moderate reduction of the target snoRNA, indicating the advantages of 2′-MOE/chimeric ASOs in depleting snoRNAs. Indeed, we showed that U81 snoRNA could be dramatically depleted by MOE/chimeric ASOs (∼85% reduction), accompanied by a clear reduction in the methylation level at the targeted site of rRNA (Figure 5). This same snoRNA has been targeted by different approaches in a previous study (29), including a LNA that has a sequence almost identical to an active ASO identified in this study (477444). The snoRNA level could only be reduced by ∼60% using the LNA, and no functional loss of the U81 snoRNA has been observed in the previous study (29). The difference in knockdown efficiency of 2′-MOE/chimeric ASO and LNA likely stems from the higher affinity of MOE chimeric ASOs and the more thorough screening we performed. Different transfection efficiencies may also affect the activity. Indeed, lower depletion efficiency of snoRNAs was observed when higher confluency of starting cells was used (Figure 10 and data not shown), suggesting that transfection efficiency significantly affects snoRNA depletion. Thus we optimized the transfection by starting at ∼30% confluency, with which >85% reduction of snoRNAs could often be achieved.
ASOs targeting snoRNAs have the ability to base-pair with snoRNA precursors, thus it is possible to trigger degradation of pre-snoRNAs. However, we found that the 2′-MOE/chimeric ASOs are capable to deplete the intronic snoRNAs without affecting the expression of the host genes. These observations indicate that the ASOs targeting snoRNAs do not act on pre-mRNA. The different effects of ASOs on depleting mature snoRNAs and pre-mRNAs may stem from different accessibilities of these molecules, due to differences in the structures and compositions of pre-mRNA–protein complex and the mature snoRNP complex, thus leading to differential accessibility to ASOs. Indeed, mature snoRNPs contain only the four core proteins, whereas additional proteins are required for snoRNA maturation and snoRNP assembly. For example, Naf1 and Shq1 are required for H/ACA snoRNP assembly but are absent in mature snoRNP (48–50). It has been shown that Naf1 interacts with pre-snoRNP co-transcriptionally, and after splicing, this protein is replaced by a H/ACA snoRNP core protein, Gar1 (51–53). Thus, compositional exchange of the RNPs may lead to structural changes between pre-snoRNP and mature snoRNP complexes. To support this view, re-structuring of snoRNP complex or core proteins has been found for C/D snoRNPs, and it was proposed that this restructuring could reflect the conversion from pre-snoRNP to mature snoRNP complex (54). These findings may explain why pre-mRNA cannot be targeted by the snoRNA ASOs. However, other factors can also contribute to the difference, such as different cellular localizations and half-lives between pre-mRNA and mature snoRNAs. Despite of the uncertainty of the mechanisms, it is clear that ASOs can specifically target intronic snoRNAs, without affecting expression of the host genes. This feature makes ASOs highly suitable for functional studies of mammalian nuclear ncRNAs.
Small ncRNAs have already been implicated in human diseases, and more links between impaired expression/function of ncRNAs and diseases are to be identified. However, the ways to manipulate the expression of nuclear ncRNAs are limited, even in vitro. In this study, we showed that snoRNAs can be dramatically depleted in mice by systematic administration of ASOs. Greatest depletion was achieved in mouse liver, and moderate depletion was found in kidney and white fat. Since only a single dose was used in this experiment, it is conceivable to achieve higher depletion in other tissues, by multi-dose treatment or by different delivery approaches. Thus, the 2′-MOE/chimeric ASOs have great potential to develop drugs against disease-causing ncRNAs that are difficult to be depleted by other means, such as siRNAs.
SUPPLEMENTARY DATA
Supplementary data are available at NAR online.
FUNDING
Funding for open access charge: ISIS Pharmaceuticals.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Sheri Booten and Sue Murray for help in animal manipulation, Sue Freier and Huynh-Hoa Bui for designing the ASOs; Walt Lima and Hongjiang Wu for stimulating discussions; Erich Koller for kindly providing the ASO antibody; and Sue Freier, Brett Monia, Frank Bennet and Frank Rigo for critical reading of the manuscript.
REFERENCES
- 1.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 2.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J. Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 3.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 5.Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 6.Bertrand E, Fournier MJ. In: The Nucleolus. Olson MOJ, editor. Georgetown, TX: Landes Bioscience Publishing; 2004. pp. 225–261. [Google Scholar]
- 7.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 8.Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- 9.Omer AD, Ziesche S, Decatur WA, Fournier MJ, Dennis PP. RNA-modifying machines in archaea. Mol. Microbiol. 2003;48:617–629. doi: 10.1046/j.1365-2958.2003.03483.x. [DOI] [PubMed] [Google Scholar]
- 10.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 12.Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin. Genet. 2008;73:103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 18.Rogelj B. Brain-specific small nucleolar RNAs. J. Mol. Neurosci. 2006;28:103–109. doi: 10.1385/JMN:28:2:103. [DOI] [PubMed] [Google Scholar]
- 19.Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, Dong JT. Implication of snoRNA U50 in human breast cancer. J. Genet. Genomics. 2009;36:447–454. doi: 10.1016/S1673-8527(08)60134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 22.Liang XH, Uliel S, Hury A, Barth S, Doniger T, Unger R, Michaeli S. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA. 2005;11:619–645. doi: 10.1261/rna.7174805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uliel S, Liang XH, Unger R, Michaeli S. Small nucleolar RNAs that guide modification in trypanosomatids: repertoire, targets, genome organisation, and unique functions. Int. J. Parasitol. 2004;34:445–454. doi: 10.1016/j.ijpara.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 24.Brown JW, Echeverria M, Qu LH. Plant snoRNAs: functional evolution and new modes of gene expression. Trends Plant Sci. 2003;8:42–49. doi: 10.1016/s1360-1385(02)00007-9. [DOI] [PubMed] [Google Scholar]
- 25.Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 26.Liang XH, Liu Q, Michaeli S. Small nucleolar RNA interference induced by antisense or double-stranded RNA in trypanosomatids. Proc. Natl Acad. Sci. USA. 2003;100:7521–7526. doi: 10.1073/pnas.1332001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta SK, Hury A, Ziporen Y, Shi H, Ullu E, Michaeli S. Small nucleolar RNA interference in Trypanosoma brucei: mechanism and utilization for elucidating the function of snoRNAs. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq599. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tycowski KT, Smith CM, Shu MD, Steitz JA. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl Acad. Sci. USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ploner A, Ploner C, Lukasser M, Niederegger H, Huttenhofer A. Methodological obstacles in knocking down small noncoding RNAs. RNA. 2009;15:1797–1804. doi: 10.1261/rna.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ideue T, Hino K, Kitao S, Yokoi T, Hirose T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15:1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crooke ST, Vickers T, Lima W, Wu H-J. In: Antisense Drug Technology—Principles, Strategies, and Application. Crooke ST, editor. Boca Raton, FL. USA: CRC Press; 2006. [Google Scholar]
- 32.Vickers TA, Lima WF, Wu H, Nichols JG, Linsley PS, Crooke ST. Off-target and a portion of target-specific siRNA mediated mRNA degradation is Ago2 ‘Slicer’ independent and can be mediated by Ago1. Nucleic Acids Res. 2009;37:6927–6941. doi: 10.1093/nar/gkp735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 34.Maden BE, Corbett ME, Heeney PA, Pugh K, Ajuh PM. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995;77:22–29. doi: 10.1016/0300-9084(96)88100-4. [DOI] [PubMed] [Google Scholar]
- 35.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 36.Yu YT, Shu MD, Steitz JA. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- 37.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23 S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 38.Antal M, Boros E, Solymosy F, Kiss T. Analysis of the structure of human telomerase RNA in vivo. Nucleic Acids Res. 2002;30:912–920. doi: 10.1093/nar/30.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 40.Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol. 1999;19:3989–3997. doi: 10.1128/mcb.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 45.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 46.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 48.Fatica A, Dlakic M, Tollervey D. Naf1 p is a box H/ACA snoRNP assembly factor. RNA. 2002;8:1502–1514. [PMC free article] [PubMed] [Google Scholar]
- 49.Dez C, Noaillac-Depeyre J, Caizergues-Ferrer M, Henry Y. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 2002;22:7053–7065. doi: 10.1128/MCB.22.20.7053-7065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang PK, Rotondo G, Porras T, Legrain P, Chanfreau G. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 2002;277:45235–45242. doi: 10.1074/jbc.M207669200. [DOI] [PubMed] [Google Scholar]
- 51.Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell. Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richard P, Kiss T. Integrating snoRNP assembly with mRNA biogenesis. EMBO Rep. 2006;7:590–592. doi: 10.1038/sj.embor.7400715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richard P, Kiss AM, Darzacq X, Kiss T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol. Cell. Biol. 2006;26:2540–2549. doi: 10.1128/MCB.26.7.2540-2549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 2007;27:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.