Abstract
We present evidence that the reverse transcriptase (RT) of human immunodeficiency virus type-1 stabilizes in vitro very short (2-nt) duplexes of 3′-overhangs of the primer strand that are annealed to complementary dinucleotides tails of DNA or RNA template strands, provided that these sequences contain at least one C or G. This RT-induced strand ‘clamping’ activity promotes RT-directed DNA synthesis. This function is achieved only when the functional template strand is adjacent to a second DNA or RNA segment, annealed upstream to most of the primer (without gaps). The combined clamp/polymerase activity is typical to RTs, as it was found in different RTs from diverse retroviral groups, whereas cellular DNA-polymerases (devoid of 3′→5′ exonucleolytic activity) showed no clamp activity. The clamp-associated DNA-binding activity is markedly stabilized by dGTP, even when dGTP is not incorporated into the nascent DNA strand. The hereby-described function can help RTs in bridging over nicks in the copied RNA or DNA templates, encountered during reverse transcription. Moreover, the template-independent blunt-end synthesis of RTs can allow strand transfers onto compatible acceptor strands while synthesizing DNA. These RT properties can shed light on potentially-new roles of RTs in the reverse-transcription process and define new targets for anti-retroviral drugs.
INTRODUCTION
Reverse transcription (RTN) is a critical step in the life cycle of retroviruses and the related long terminal repeat (LTR) retrotransposons. This complex process is performed by the retroviral reverse transcriptase (RT) that converts the viral single-stranded RNA into integration competent double-stranded viral DNA (1–3). To perform RTN, RTs possess a DNA polymerase activity, which is both DNA-dependent and RNA-dependent (DDDP and RDDP, respectively), and a ribonuclease H (RNase H) activity that concurrently with DNA synthesis cleaves the RNA template in the generated RNA/DNA duplex (4). DNA synthesis produces both (−) and (+) DNA strands, while RNase H removes the tRNA-primer and the viral genomic RNA template (1,3–5).
During RTN, there are two strand transfers, or template switches events, where the 3′-end of the elongated DNA primer switches to a second template (1–3,5). In the first one, designated (−) strand transfer, the DNA is translocated onto the 3′-end of the genomic RNA. In the second one, (+) strand transfer, the 3′-end of the (+) strand with the primer-binding site (PBS) sequence switches to a complementary sequence in the (−) DNA strand. These transfers depend on stable sequence complementarities between the ends of the growing (donor) DNA and the acceptor RNA or DNA strands. These complementary sequences are relatively long. In human immunodeficiency virus type-1 (HIV-1), the terminal repeat (R), which promotes (−) strand transfer, is 98-nt long and in murine leukemia virus (MLV), it is 68-nt long, whereas the PBS, is usually 18-nt long in all retroviruses that use a tRNA primer (including HIV-1 and MLV) (1,6).
We present here in vitro evidence that RTs can perform template switches even with a very short (2-nt) complementarity between the 3′-ends of the primer donor strand and the DNA or RNA template acceptor strands. These dinucleotide duplexes are markedly stabilized by RT that ‘clamps’ together these otherwise unstable duplexes. This stabilization of sequence micro-homology efficiently promotes DNA synthesis. The apparently-new RT clamp function is performed by all RTs tested but not by cellular DNA polymerases. Thus, this activity can potentially allow the RT-associated DNA synthesis to bridge over nicks in the copied RNA or DNA templates that are encountered during RTN. Alternatively, the combined clamp activity and the capacity of some RTs to perform template-independent blunt-end DNA synthesis can promote strand transfers onto compatible acceptor strands while synthesizing the viral DNA.
MATERIALS AND METHODS
RTs and DNA polymerases
All purified recombinant RTs carry six-histidine tags, as previously described by us. These RTs were of the BH-10 strain of HIV-1 and of the HIV-2 Rod strain (7) as well as the RTs of Moloney MLV (8), the LTR retrotransposon Tf1 (9) and bovine immunodeficiency virus (BIV) (10). Viral avian myeloblastosis virus (AMV) RT and recombinant Klenow fragment of Escherichia coli Pol I (KF) (3′→5′ exo-) polymerase were purchased from NEB. Recombinant DNA polymerase β (pol-β) was a generous gift from Drs W. Beard and S. Wilson from National Institutes of Health.
Oligonucleotides
All oligonucleotides were custom synthesized and HPLC purified by Metabion (Supplementary Table S1).
Combined clamp/polymerase assay reactions
Primers were 5′-end-labeled by T4 polynucleotide kinase and [γ32P]ATP, followed by heat inactivation and annealing to the appropriate templates (Supplementary Table S2), as described in detail (11). The molar ratios between the templates to primers were 4:1, unless otherwise stated. Reactions were performed (unless otherwise stated) in final volumes of 12.5 µl with 0.33 pmol of template/primer (T/P), 150 ng (128 nM) HIV-1 RT (or equal DDDP activities of the other enzymes used, according to Supplementary Figure S1) in 30 mM Tris–HCl, pH 7.5, 43 mM NaCl, 5 mM MgCl2, 4 mM dithiothreitol, 24 µg/ml bovine serum albumin and 4% glycerol (clamp/polymerase reaction buffer). The contributions to the final NaCl concentration in the reaction mixtures by each enzyme preparation were up to 3 mM (depending on the enzyme dilutions performed). Reactions were initiated by adding all four dNTPs, each at 50 µM (unless otherwise indicated) for 30 min at 37°C, and then stopped by adding formamide loading buffer. The reactions were analyzed by urea–polyacrylamide gel electrophoresis (urea–PAGE), followed by autoradiography, as described earlier (8,11).
ELISA-based DNA-binding assay
For a schematic description see Supplementary Figure S2. Standard binding assays were performed with 3 pmol of the appropriate T/P and 50 µM dNTPs, as described above for the combined clamp/polymerase assay. After incubation at 37°C for 1 h, some of the reactions were heat-denatured for 5 min at 95°C, followed by annealing, while the others were kept on ice. Next, 50 µl of the clamp/polymerase reaction buffer were added to each reaction. In parallel, microtiter wells that were pre-coated with NeutrAvidin [2 µg per well, in phosphate buffered saline (PBS), incubated overnight at room temperature] were blocked with 5% BSA in PBS for 1 h at room temperature, followed by three washes with PBS. Then, each diluted binding reaction mixture was added to a separate well and incubated for 1 h at room temperature. Unbound DNA was removed by three washes with PBS and then the bound DNA was detected by adding 200 µl of anti-digoxigenin antibody (anti-dig), conjugated to horse reddish peroxidase (HRP) (from Roche, diluted 1:1000 in 100 mM Tris–HCl, pH 7.5 and 150 mM NaCl). After a 1 h incubation at room temperature, the wells were washed five times with PBS and a further incubation was carried out with 100 µl of 0.5 mg/ml 2,2′-azino-bis(3-ethylbenz-thiazoline-6-sulfonic acid), ABTS, in 12.5 mM Tri-sodium citrate, 12.5 mM citric acid and 0.02% H2O2 .The resulting green color was quantified at 405 nm using a SpectraFluorPlus plate reader and the Magellan4 software.
RESULTS AND DISCUSSION
The HIV-1 RT clamp phenomenon
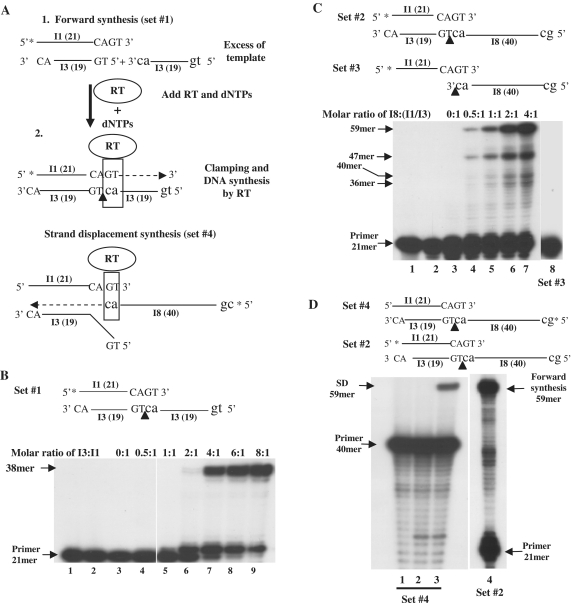
While studying the in vitro HIV-1 RT DNA synthesis, we have noticed that when the DNA primer has a 2-nt 3′-end overhang over the template strand, and this template strand is in a molar excess over the primer, there was a strong extension of the 21-nt primer to 38-nt long products (e.g. Figure 1B, lane 7). Since there is a two base complementarity between the 3′-terminal sequences of the primer (5′-GT-3′) and of the template strand (5′-AC-3′, Supplementary Table S2, set #1), the extension by RT can be explained as follows: after, or along with the annealing of one template strand (designated I3) to the primer (oligo I1), a second identical I3 molecule is juxtaposed 5′ to the first I3 via the 2-bp complementarity between the 3′-ends of the primer and template strands, thus RT-directed DNA synthesis is initiated (for a schematic description, see Figure 1A, steps 1–2). The 38-nt long product is in agreement with this model, as the expected length is: 21-nt (of the primer, I1) plus 17-nt (19-nt of I3, minus 2). To support the proposed hypothesis, we have tested DNA synthesis as a function of varying the molecular ratios of oligo I3 over I1 in set #1 (Figure 1B). Indeed, substantial DNA synthesis is apparent when this ratio is above 2:1 (lane 6) up to 8:1 (lane 9), suggesting that two template molecules are annealed to a single primer molecule. Without RT, the duplex of the two strands is likely to be unstable, with a predicted basic melting temperature (Tm) value of merely 6°C. Therefore, stabilizing these duplexes by the RT-associated clamping activity is imperative for primer extension.
Figure 1.
The combined clamp/polymerase activities of HIV-1 RT. Reactions were conducted with the indicated template/primer sets, as described in the ‘Materials and Methods’ section. Each oligonucleotide is indicated by the designation that appears in Supplementary Table S1 along with its length in nucleotides (in parenthesis). Asterisks indicate the 5′-end 32P label. The ends of the primer (I1) and template (I3) are indicated by capital letters and those of the functional templates (either I3 or I8) are indicated by lower case letters. Nicks between two closely-adjacent templates are indicated by arrowheads. (A) A schematic description of the clamp/polymerase activity. This scheme is drawn for the forward synthesis with set #1 (steps 1 and 2) and for the strand displacement synthesis with set #4. See the text for further explanations. (B) Effects of template/primers (T/Ps) molar ratios on HIV-1 RT activity with substrate set #1. 1, T/P only; 2, T/P+RT; 3–9, T/P+RT+dNTPs, at the indicated molar ratios of template over primer. (C) Effects of T/Ps molar ratios on HIV-1 RT activity with substrate set #2. 1, T/P (I3/I1) only; 2, T/P+RT; 3–8, RT+dNTPs+T/P+I8. In all lanes, the molar ratios of I3 to I1 were 1:1 and the ratios of I8 over (I3/I1) are indicated. In lane 8, I8 was 4-fold higher than I1, with no I3 present. (D) The strand displacement synthesis (SD) with set #4. The molar ratios of I1:I3:I8 were 4:4:1, respectively. 1, T/P only; 2, T/P+RT; 3, T/P+RT+dNTPs; 4, a control reaction of RT+dNTPs+set #2 [as shown above in (C)].
To test whether the combined clamp/polymerase activity can be achieved also with another functional template, we have used a third 40-nt long oligo (I8, Supplementary Table S1) with a terminal AC sequence, matching the primer’s GT overhang (while the rest of the sequence is not complementary), that served as the acceptor second template (Figure 1C and Supplementary Table S2, set #2). Here, the ratio of I3:I1 was equimolar, while I8 concentrations were varied. The primer was elongated to 59-nt products, agreeing with the anticipated length of 21-nt (of the primer) plus 38-nt (40 of the template I8, minus 2). As expected, extension was apparent even when the ratio of I8:(I3/I1) was only 0.5:1 (Figure 1C, lane 4). However, when I3 was omitted (set #3, Figure 1C and Supplementary Table S2), there was no apparent extension (lane 8), indicating that the first complementary strand is obligatory for stabilizing the RT/DNA complex. In many experiments shown here, not all extended primers reach their maximal lengths, as the processivity of DNA synthesis by RTs is not very high. Therefore, despite their ability to polymerize up to a few hundred nucleotides in a single synthesis round, RTs tend to fall off in vitro at specific sequences or structures, defined as ‘strong stops’, leading to premature terminations of synthesis, see for example, (1,3,7,12). Thus, in both cases, with sets #1 and #2 (Figure 1B and C, respectively), there is also a significant clamp-dependent addition of a single nucleotide (forming a 22-nt products). In the case of set #2 (Figure 1C), premature terminations are apparent (with product lengths of mostly 47, 40 and 36-nt), whereas with set #1 these longer premature terminations are less evident (see also below, Figures 2A and 4A, regarding set #1). These results support the concept that the extent of RT-dependent processivity and, consequently pausing, can depend, for a given RT, on the sequences of the copied templates, for example, (12,13).
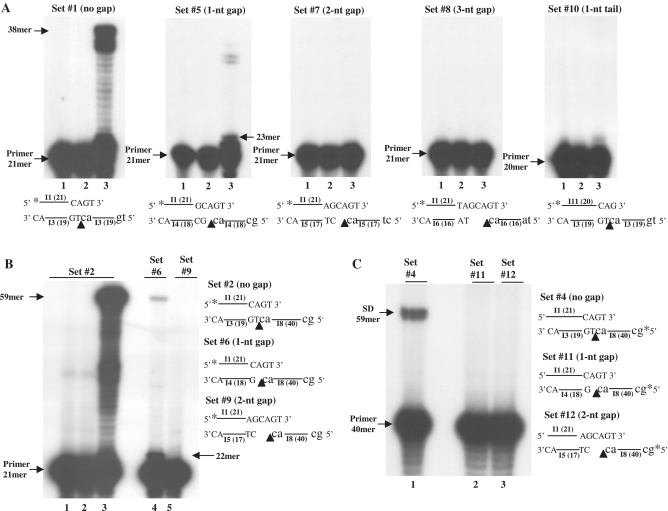
Figure 2.
Effects of various template-primers on the clamp/polymerase activity of HIV-1 RT. T/Ps designations and their schemes are indicated (see Figure 1 for details). For each tested set: 1, T/P only; 2, T/P+RT; 3, T/P+RT+dNTPs. In panel B, lanes 4 and 5 also show the outcome with T/P+RT+dNTPs . Nicks or gaps between two adjacent templates are indicated by arrow heads. (A) and (B), direct clamp/polymerase activity. (C) Strand displacement.
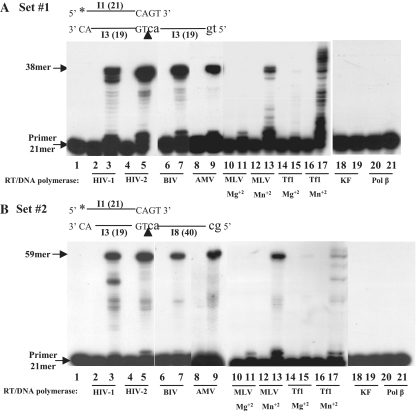
Figure 4.
The clamp/polymerase activity of different RTs and DNA polymerases. All enzymes (with equal DDDP activities, tested by a DNA–primer extension assay, see Supplementary Figure S1) were incubated with the indicated substrate as described in ‘Materials and Methods’ section. MLV RT was assayed with 5 mM MgCl2 or 0.8 mM MnCl2 while Tf1 RT with 5 mM MgCl2 or 0.5 mM MnCl2. All other enzymes were assayed with 5 mM MgCl2. Lane 1, T/P only; lanes 2–21, reactions with the various enzymes, in the absence (even-numbered lanes) or the presence of dNTPs (odd-numbered lanes). The different RTs/DNA polymerases and the presence of Mn+2 or Mg+2 (for MLV and Tf1 RTs) are indicated. (A) Assayed with set #1. (B) Assayed with set #2.
To further support the HIV-1 RT-associated clamp activity, we have also used a matching approach and tested strand displacement (SD) activity with set #4 that is similar to set #2 (Figure 1D and Supplementary Table S2). However, here, the visual monitoring of primer and templates was reversed and oligo I8 was 5′-end labeled rather than I1. Hence in this specific case, I8 is the functional primer and I1 is the functional template (Figure 1A and D). Typical to RTs, SD occurs when a DNA strand (in the present case, I3), annealed downstream to the template (I1), is dislocated by synthesis originating from an upstream primer (I8) (3,14,15). Indeed, Figure 1D shows that SD did occur, generating 59-nt products that are identical in length to the forward clamp reaction products (with set #2), as the 40-nt primer I8 was extended by 19-nt (Figure 1D, lane 3). As expected, the clamp-dependent SD synthesis is less efficient than the forward reaction, since RT-associated SD is substantially slower than regular DNA synthesis. As could be also anticipated, similar to the forward reaction with set #3 (Figure 1C, lane 8), there was no synthesis when I8 was end labeled and I3 was absent (data not shown).
The HIV-1 RT-associated clamp/polymerase activity depends on a tandem alignment of the template strands
The clamp model suggests that the two bottom template strands should be juxtaposed, so that the 3′-end of the functional template is positioned immediately next to the 5′-end of the second strand that is complementary to most of the primer. The results of the experiments shown in Figure 2A and B imply that even a single nucleotide gap between the two aligned strands in the two tested substrate sets (in set #5, relative to set#1, and in set #6 relative to set #2) strongly reduces the clamp-dependent synthesis. Quantitative analyses of the presented data reveal that the reduction in the clamp activity is by ∼98% for set #6 and by ∼90% for set #5. Interestingly, in the case of set #5, the sharp drop in activity also affects the processivity of HIV-1 RT, as the generated products are shorter than the maximal length products (32–33 nt in length relative to the 38-nt full length products). As expected, longer gaps are even more deleterious (sets #7 and #8 in Figure 2A and #9 in Figure 2B). The negative effect of template gaps was also demonstrated for SD, as in the parallel assay conducted for this activity, gaps of even a single nucleotide (as in set #11) eliminated the clamp activity (Figure 2C).
The clamp activity requires a minimal 2-nt complementarity between the termini of the DNA strands
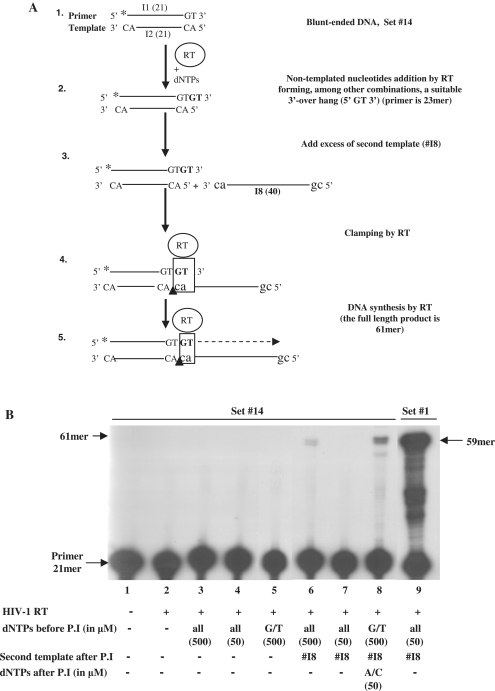
We have further studied the requirements of HIV-1 RT clamp/polymerase activity. A clamp-dependent synthesis did not occur with an overhang shorter than 2-nt, as seen with set #10 with a single-nucleotide overhang (Figure 2A). Similarly, under similar assay conditions (with 50 μM dNTPs) no extension was observed with a single-stranded primer (Supplementary Figure S3, set #13) or a blunt-ended double-stranded DNA (that is similar to set #1 but with a bottom strand longer at its 5′-end by 2-nt) (see below, set #14 in Figure 7, lane 4). Finally, we have shown that the clamp function is a general one and requires only a short complementarity between the 3′-ends of the primer and the template strand, as suggested by the substantial clamp/polymerase activity, obtained with set #15 (which differs from set #1, except for its 3′-GT tail) (Supplementary Figure S3).
Figure 7.
The reaction products of the combined non-templated addition and clamp/polymerase activity of HIV-1 RT. (A) A schematic description of the assay (see Figure 1 for details). (B) HIV-1 RT underwent pre-incubation (P.I) with the indicated T/Ps and dNTPs for 30 min at 37°C, followed by further incubations with (lanes 6, 7, 8 and 9) or without (lanes 3, 4 and 5) the acceptor oligo I8, for additional 30 min at 37°C, as described under each of the gel lanes.
The sequence of the DNA synthesized by the clamp/polymerase activity
To provide further support to the clamp/DNA synthesis, we have partially sequenced the clamp-dependent extended DNA (with set #2), by using HIV-1 RT’s DNA polymerase activity. This was done in the presence of four dNTPs combinations, each devoid a single dNTP that was replaced by the matched di-deoxy NTP (ddNTP) as chain terminators. The results obtained are consistent with the proposed clamp/DNA synthesis model (data not shown), although, HIV-1 RT is not the best enzyme for DNA sequencing, due to its relatively-low fidelity of DNA synthesis (16–18).
The effect of terminal sequences on the extent of HIV-1 RT clamp activity
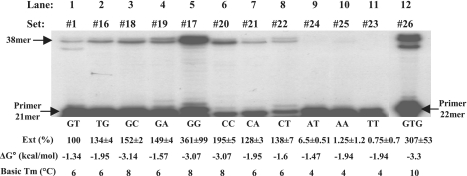
As all clamp-positive reactions, described thus far, were performed with the 5′-GT/AC-3′ primer/template terminal tails, it was of interest to test how altering these dinucleotides affects the clamp activity. First, when the order of the 3′-terminal nucleotide was reversed (TG/AC ends, set #16, Supplementary Table S2), the extent of the clamp activity was only slightly affected (Figure 3, lanes 1 and 2). Then, the effects of the terminal sequences were tested with sets of template/primers, similar set #1, except for their 3′-end dinucleotides (Supplementary Tables S1 and S2; Figure 3). The strongest activity was obtained with set #17, with a GG primer overhang (∼3.6-fold higher than set #1), while activities obtained with terminal dinucleotides containing a single G (sets #18 and #19), were lower, albeit higher than those with sets #1 and #16 (∼1.5-fold). A relatively high activity was obtained with set #20 with a CC tail (by ∼2-fold). Sets containing a single terminal C but no G (#21 and #22) yielded a lower activity relative to set #20. Substrates with primer tails with only T and/or A (sets #23, #24 and #25) were appreciably less fit for the clamp activity. It is evident that there is a better correlation between the clamp function and the apparent Tm values of the duplexes, rather than with their ΔG° values. Interestingly, the presence of two G′s in the primer’s 2-nt overhang promotes a better clamp activity than two C′s.
Figure 3.
Effects of the terminal 2-nt sequences on the efficiency of the clamp/polymerase activity of HIV-1 RT. T/Ps designations are indicated at the top of each lane. The autoradiogram was exposed for a relatively short time, in order to allow a reliable quantitative evaluation of the extent of the clamp activity with the various substrates. The primer bases forming the 3′-end tails are specified at the bottom of each lane, in addition to the calculated ΔG° values (in kcal/mol) and the Tm values (expressed in °C) for each terminal duplex. Clamp activity was determined as a percentage of the total 5′-end-labeled DNA substrate, converted to 38mer DNA products. The level of primer extension (‘Ext’) is the average of two independent experiments conducted for each substrate set. The results were normalized to the 100% value of the extension level obtained with set #1.
To check whether extending the terminal primer/template duplex to 3-nt improves the HIV-1 RT clamp activity, we have used the trinucleotides GTG/CAC sequence (set #26, Figure 3, lane 12). Although the Tm of this substrate is slightly higher than that of set #17 (with terminal GG/CC), the clamp activity was roughly similar, suggesting that substrates with 2-nt overhangs can be as efficient as slightly longer ones, depending on the terminal sequences.
The clamp/polymerase activity is typical to RTs and not to other DNA-polymerases
Another important issue is how common is the clamp activity to RTs other than HIV-1 RT and to DNA-polymerases in general. Therefore, we have also tested RTs of other lentiviruses (of HIV-2 and of BIV), RTs of the gammaretrovirus, MLV, of the betaretrovirus, AMV and of Tf1 (3,19). In addition, the KF of E. coli Pol I and Pol-β were tested, representing cellular prokaryotic and eukaryotic DNA-polymerases, respectively. As KF has an inherent 3′→5′ exonuclease activity that can distort the results (since this activity is likely to remove the nucleotides added while extending the primer during DNA synthesis), a KF mutant lacking this exonuclease activity was tested. First, we have calibrated the DDDP activity of all tested enzymes by a DNA primer-extension assay (Supplementary Figure S1); so, under the clamp assay conditions all enzymes have similar activity levels. Most RTs and DNA polymerases prefer Mg+2 over Mn+2, hence the RTs of HIV-1, HIV-2, BIV and AMV as well as Pol-β and KF were tested with MgCl2. Since the RTs of Tf1 and MLV vary in their divalent cation preferences with various substrates (8,9), they were tested with Mg+2 and with Mn+2.
The results of the assays with sets #1 and #2 (Figure 4A and B, respectively) indicate that all tested RTs have a clamp activity, whereas the two tested cellular polymerases, (both devoid of 3′→5′ exonucleolytic activity) lack any detectable level of this activity. In the case of MLV and Tf1 RTs, a substantial clamp activity is apparent with Mn+2 (with hardly any detectable activity with Mg+2). Interestingly, in case of Tf1 RT with set #1 and in the presence of Mn+2 (Figure 4A, lane 17) the generated 38-nt product is further extended to DNA strands up to 45-nt in length. This probably results from the strong template-independent synthesis, typical to this RT (9). However, such an extension is likely to take place only after completing the template-dependent synthesis, which results from the clamp activity (generating a 38 mer products), because in a control experiment, performed with Tf1 RT, under similar conditions with equimolar template-primer (I3:I1) ratios (as shown above in Figure 1), there was hardly any primer extension (data not show).
The clamp activity initiates also an RNA-dependent DNA synthesis
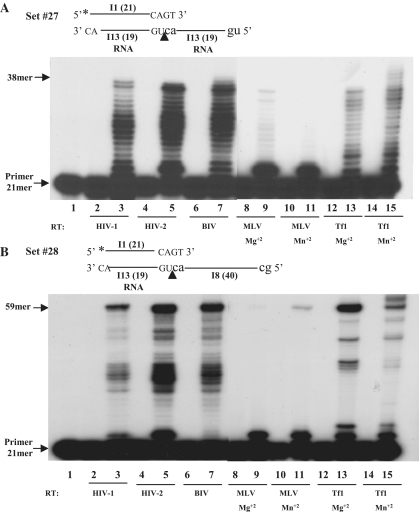
All experiments, described so far, tested the combined RT-associated clamp/DDDP activity. Since RTs possess also an RDDP activity, we further tested the clamp activity with an RNA template (designated I13, Supplementary Table S1) that is similar in sequence to the oligo-deoxynucleotide I3 template in sets #1 and #2. The results with this DNA/RNA substrate (set #27, Supplementary Table S2) show that, similar to the clamp/DDDP, the RTs of HIV-1, HIV-2, BIV and Tf1 show an efficient clamp/RDDP activity (Figure 5A). Interestingly, under the assay conditions, MLV RT shows a substantially weaker clamp/extension and full length products were achieved with only Mg+2, though a single-nucleotide elongation was substantial.
Figure 5.
The clamp/RDDP or DDDP activity of various RTs. The studied RTs (calibrated and tested under the clamp assay conditions as in Figure 4) were incubated with set #27 in (A) and with set #28 in (B). In both panels, lane 1, T/P only; lanes 2–15, reactions with the indicated RTs, in the absence (all even-numbered lanes) or the presence of dNTPs (all odd-numbered lanes).
Another related question is whether RTs can perform the clamp/DDDP activity while bridging over nicks between RNA and DNA templates. We used set #28 (Supplementary Table S2) that is similar to set #2, where the oligo-deoxynucleotide I3 is replaces by the RNA #I13 and the second template, I8, is the same one used in set #2. The results indicate that all tested RTs possess an efficient clamp activity with set #28 (Figure 5B). Interestingly, as with set #27, MLV RT could hardly extend the primer (and full length products were obtained only with Mn+2); still, with both divalent cations, an extension by a single nucleotide was efficient.
A direct test for the RT-associated clamp activity
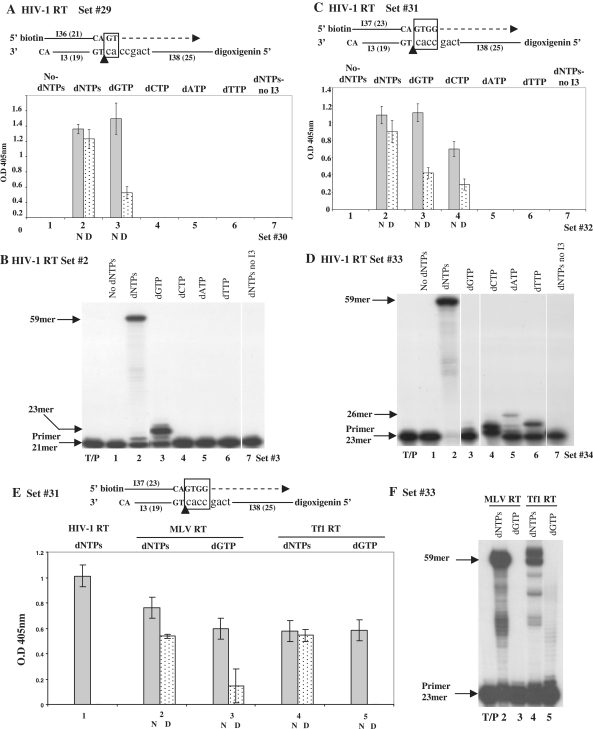
So far, the clamp activity was tested indirectly, as the resulting primer extension was examined. Hence, this analysis cannot distinguish between the direct initial RT binding to the terminal duplex (the clamping phase) and the primer extension that follows. To tell apart these two sequential reactions, we have performed an ELISA-based DNA-binding assay with HIV-1 RT, as described in ‘Materials and Methods’ section and schematically in Supplementary Figure S2, using set #29 (Supplementary Table S2). This set is similar to set #2, except that the primer is 5′-end labeled with biotin (oligo I36, Supplementary Table S1) and the second functional template is 25mer in length and is 5′-end labeled with digoxigenin (oligo I38, Supplementary Table S1). As a result of a clamp activity, the products are supposed to contain both biotin and digoxigenin tags; so, the primer moiety binds the wells of plate that were pre-coated with avidin, and the immobilized I38 oligonucleotides-containing clamp-dependent complexes are detected with anti-digoxigenin antibodies. Contrary to all analyses, shown above, here the clamp-directed signals should be potentially monitored regardless of DNA synthesis. We have also conducted the equivalent primer extension analyses of the clamp activity. In this combined clamp/polymerase analysis, the signal was detected only when dNTPs were present and, hence, polymerization took place (Figure 6A and B, columns/lanes 1 versus 2). Therefore, to directly test the formation of a clamp-dependent RT/DNA complex, each reaction had a control that was heat-denatured and re-annealed after the completion of the RT enzymatic reaction. In cases of positive signals due to DNA synthesis, the levels of reactions with or without denaturation/annealing should be fairly similar (as strand re-annealing is independent of an active RT). In contrast, signals resulting from clamping per se will be largely reduced after the denaturation/annealing process (assuming that denaturated RT cannot stabilize RT/DNA complexes). The results show that with dNTPs, HIV-1 RT induces a high signal, even after heat inactivation (Figure 6A–2N and 2D), indicating that the combined clamp activity cannot be separated from the resulting synthesis. To confirm that the two template strands are required also in this assay system, we have used set #30 (that contains only the primer and the template I38, Supplementary Table S2). Indeed, no signal was obtained (Figure 6A, column 7), similar to the data shown with set #3 (Figure 1B, lane 8 and Figure 6B, lane 7).
Figure 6.
ELISA-based DNA-binding assay with RTs of HIV-1, MLV and Tf1. The DNA-binding assay was conducted, as described schematically in Supplementary Figure S2. In (A), (C) and (E), ‘N’ marks the native and ‘D’, the heat-denatured reactions. In all schematic descriptions of the substrates used, the squares indicate the clamp regions. (A), (B), (C) and (D), tested with HIV-1 RT. The descriptions of all columns and gel lanes are indicated. Control reactions of RT+dNTPs+T/P (without I3) were conducted with set #30 (in A); set #3 (in B); set #32 (in C) and set #34 (in D), see Supplementary Table S2 for a schematic description. The bars represent the ranges calculated from at least three independent reactions. (A) DNA-binding assay with set #29. (B) Urea–PAGE analysis of the clamp/polymerase activity with set #2. (C) DNA-binding assay with set #31. (D) Urea–PAGE analysis of the clamp/polymerase activity with set #33. (E) and (F), tested with MLV RT or Tf1 RT, as indicated. (E) is a binding assay and (F) is a gel assay.
Next, we have tested if each dNTP is imperative for performing the clamp. The correct dNTP to be incorporated, dGTP, yielded a strong signal, similar to that with all four dNTPs (Figure 6A–3N). However, after denaturation, there was a sharp signal reduction, of ∼75% (Figure 6A–3D), suggesting that dGTP strongly contributes to the clamp signal by itself, although only two Gs can be incorporated (as confirmed by primer extension, Figure 6B, lane 3). All other reactions, conducted with each of the other three incorrect dNTPs, gave no detectable signal (Figure 6A and B, columns/lanes, 4–6), suggesting that only dGTP can promote the RT-directed clamp activity.
The obvious question is whether the putative dGTP-generated tetra-nucleotide duplex (GTGG/CCAC) is responsible by itself to the signal, due to its higher stability (an apparent Tm value of 14°C, versus 6°C of the initial 2-nt duplex). Therefore, we have tested set #31 (Supplementary Table S2) with a preformed 3′-end GTGG tail. Still, this duplex was not stable enough to yield a signal even with RT (with no dNTPs) (Figure 6C, column 1), suggesting that the effect of dGTP observed with set #29 (Figure 6A) is not due the higher stability of the formed duplex. However, as with set #29, adding all dNTPs generated a strong signal that was almost fully resistant to heat denaturation (Figure 6C–2N and 2D). Interestingly, dGTP (the incorrect dNTP) gave here also a high signal; similar to that obtained with all dNTPs (Figure 6C, column 3N) although there was no significant primer elongation (Figure 6D, lane 3). Moreover, the majority of the dGTP-dependent signal (∼72%) was denaturation-sensitive, suggesting that it is mostly due to a true RT-dependent clamping (Figure 6C–3D). This unexpected result suggests that dGTP is required for stabilizing the clamp-dependent RT/DNA complex, even when it is not incorporated. The correct dNTP, dCTP, that elongated the primer by 2 nucleotides (the second C was misincorporated opposite to an A, Figure 6D, lane 4) leads also to a clamp signal that is mostly denaturation-sensitive (∼60%) (Figure 6C–4N and 4D). Still, the overall dCTP-dependent signals are lower than the dGTP-dependent ones. In contrast, no clamp-dependent signals were detected with the two incorrect dNTPs, dTTP or dATP, although in the urea-PAGE analysis, dTTP was misinserted (opposite to the template G) followed by a correct insertion that was elongated by misincorporating another T (opposite to the template C), while dATP was also slightly misinserted (Figure 6D, lanes 6 and 5, respectively). Finally, as with sets #30 and #3 (Figure 6A and B, column/lane 7), no signal was obtained in the absence of oligo I3 (Figure 6C and B, column/lane 7 for sets #32 and #34, respectively, see Supplementary Table S2 for a schematic description).
The most likely conclusion from the ELISA-based assay is that the clamp activity of HIV-1 RT is largely supported by dGTP. To test whether this phenomenon is shared also by other RTs, we have performed a similar experiment with MLV and Tf1 RTs, in the presence of Mn+2, as the preferred divalent cation. Apparently, the clamp activity of these RTs is also markedly stabilized by dGTP, as denaturation caused the MLV RT-generated signal to drop by 76% and the Tf1 RT signal to totally disappear (Figure 6E, columns 3 and 5, respectively). In conclusion, the unsettled unique role of dGTP in supporting the clamp activity of RTs calls for further investigations.
HIV-1 RT can utilize the product of template-independent synthesis for its clamp activity
As shown above, 3′-end matched preformed terminal dinucleotides are required for the clamp activity. However, RTs are also capable (though to a variable extent) of adding non-templated nucleotides to the nascent DNA strand (9,18,20,21). Therefore, we have tested whether clamp activity can result also from newly-formed overhangs, generated by a prior non-templated extension of blunt-ended DNA by the RT. The experiment described schematically in Figure 7A, was conducted with set #14, a blunt-ended double strand DNA (Supplementary Table S2) in the presence of high dNTPs concentrations (500 μM each) to allow a prior template-independent DNA synthesis. RT was preincubated with the DNA and dNTPs, followed by adding the acceptor oligo I8 with the 3′-end sequence of 5′-AC-3′. Although, under the assay conditions employed, we could not detect any significant non-templated extension (Figure 7B, lane 3), adding oligo I8 resulted in weak, albeit significant, synthesis of 61-nt products (lane 6). In contrast, no significant clamp activity was observed with 50 μM dNTPs (that is too low for a non-templated extension) after adding the acceptor oligo I8 (Figure 7B, lane 4 versus lane 7).
To bias the non-templated synthesis to preferentially generate G/T-containing tails, which are suitable for the 3′-end of the acceptor strand (oligo I8), RT was preincubated with set #14 and 500 μM of each dGTP and dTTP, followed by adding oligo I8 and 50 μM of each dATP and dCTP. Here, a stronger extension of the primer to the expected products was obtained (Figure 7, lane 8), though no significant non-templated extension was observed without the acceptor strand, I8 (lane 5). This confirms the prediction that RT can utilize the end-product of a template-independent synthesis for its clamp/polymerase activity. As could be expected, the direct clamp assay is by far more sensitive than the indirect non-templated synthesis-dependent/clamp/polymerse assay, as the reaction with the preformed 3′-end overhang substrate, set #1 (with an equimolar template-primer, I3:I1, ratio) in addition to the acceptor strand, oligo I8, was much more efficient (Figure 7, lane 9 versus lanes 6 and 8).
A previous study by Golinelli and Hughes suggested that non-templated DNA synthesis by HIV-1 RT can induce non-specific strand transfers when the acceptor DNA is in an exceedingly-high excess (70-fold) over the primer (21), which is definitely above any expected in vivo physiological conditions. In contrast to this, the experimental conditions and results described in the present study are different. First, an efficient clamp/polymerase activity with preformed 2-nt overhangs could be achieved even at the acceptor (in the case of set #2, oligonucleotide I8) to primer molar ratio of 0.5:1, a ratio that is supposed to be close to the conditions that can be encountered in vivo (Figure 1C, lane 4). Moreover, even when a prior non-templated addition was required, clamp-dependent strand transfer could be accomplished in our experiments at a molar ratio of acceptor (I8) to primer as low as 4:1 (and possibly even at a lower ratio) (Figure 7B, lanes 6 and 8). Therefore, this cardinal difference between the over-saturation conditions, employed by Golinelli and Hughes, and those used by us makes it difficult to compare between those few results of theirs (which are pertinent to our study) and the results presented herein.
One explanation for the clamp/polymerase activity shown here can be the necessity for non-specific switches from one template strand to another, once the first strand was fully copied. Blunt-end duplexes can be formed during retroviral RTN due to breaks in the template RNA. Therefore, if the growing segmented DNA chains gain very short unpaired 3′-end tails due to the template-independent synthesis property of RTs, suitable acceptor DNA or RNA strands will permit extending the nascent DNA by RT, upon copying these templates. This means that these strand switches will depend on 3′-end sequence of the acceptor strand, the cellular dNTPs pool bias and the capacity of a given RT to perform non-templated additions. For example, our preliminary results suggest that MLV RT has an apparently weak template-independent DNA polymerase activity, therefore, despite its reasonable clamp activity with preformed overhangs in the presence of Mn+2 (Figure 4A and B, lanes 13) its combined non-templated addition/strand switching activity is marginal (data not shown).
Alternatively, by stabilizing the nucleic acids/RT complexes, RTs can bridge over nicks in the copied templates, even when the formed duplexes are otherwise unstable. Since the clamp-dependent DNA synthesis is accomplished in vitro by all tested RTs, it is likely to be a key factor in performing RTN. This prediction opens the door for identifying the potential specific steps during RTN that depend on this function. However, it should be also emphasized that the clamp activity characterized in this study is most likely different from the ‘classical’ (−) and (+) strand DNA transfers, known to take place during RTN, since they involve substantially longer complementary sequences between the strands. Thus, the R region, promoting (−) strand transfer, varies between 15 and 250-nt in different retroviruses, and the PBS, involved in (+) strand transfer, is usually 18-nt in length (1).
The in vitro findings described herein show that RTs can hold together template DNA strands, juxtaposed with no gaps, with the complementary primer DNA strand that has very short overlapping 3′-end tails. The formed duplexes are undoubtedly thermodynamically unstable without RT, while the same DNA complexes with RT are stable enough to allow efficient DNA synthesis. To provide structural insights into this HIV-1 RT–DNA complex, we have modeled such a 3D structure (Supplementary Figure S4). This model suggests that several RT residues may participate in forming the complex (residues 89–92, Lys154, Pro157 and Ala158). Evidently, further mutagenesis and structural studies are required to clarify the molecular basis for this activity and whether it is possible to molecularly-dissect between DNA synthesis and clamp activity of RT. Moreover, the unique contribution of dGTP to the clamp function needs to be further investigated to see whether it involves hydrolysis of dGTP (e.g. as a potential energy source). However, we could not find any apparent sequence homology between HIV-1 RT and consensus sequences of known GTPases (data not shown).
The potential involvement of dGTP in the clamp activity may be one direction in further studying the molecular processes, involved in this activity, as well as identifying specific inhibitors of these unique steps. Thus, this clamp function can be valuable as a novel and specific target for inhibiting HIV RTs and hence RTN, as this activity is probably not shared by other cellular host DNA polymerases. Finally, as RTs are widely used nowadays in bioresearch for synthesizing cDNA from RNA, the RT clamp function may be used also for the in vitro synthesis of continuous DNA strands from segmented RNA strands.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Israeli Science Foundation (grant No. 411/07).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Drs W.Beard and S.Wilson from National Institutes of Health for the gift of recombinant Pol-β. Am.H. is an incumbent of the Gregorio and Dora Shapira Chair for the Research of Malignancies at Tel Aviv University.
REFERENCES
- 1.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 2.Menendez-Arias L, Berkhout B. A special issue on retroviral reverse transcription. Virus Res. 2008;134(Issues 1-2) doi: 10.1016/j.virusres.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Herschhorn A, Hizi A. Retroviral reverse transcriptases. Cell Mol. Life Sci. 2010;67:2717–2747. doi: 10.1007/s00018-010-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz SJ, Champoux JJ. RNase H activity: structure, specificity, and function in reverse transcription. Virus Res. 2008;134:86–103. doi: 10.1016/j.virusres.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Le Grice SF. “In the beginning”: initiation of minus strand DNA synthesis in retroviruses and LTR-containing retrotransposons. Biochemistry. 2003;42:14349–14355. doi: 10.1021/bi030201q. [DOI] [PubMed] [Google Scholar]
- 7.Sevilya Z, Loya S, Hughes SH, Hizi A. The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is affected by the thumb subdomain of the small protein subunits. J. Mol. Biol. 2001;311:957–971. doi: 10.1006/jmbi.2001.4904. [DOI] [PubMed] [Google Scholar]
- 8.Avidan O, Loya S, Tonjes RR, Sevilya Z, Hizi A. Expression and characterization of a recombinant novel reverse transcriptase of a porcine endogenous retrovirus. Virology. 2003;307:341–357. doi: 10.1016/s0042-6822(02)00131-9. [DOI] [PubMed] [Google Scholar]
- 9.Kirshenboim N, Hayouka Z, Friedler A, Hizi A. Expression and characterization of a novel reverse transcriptase of the LTR retrotransposon Tf1. Virology. 2007;366:263–276. doi: 10.1016/j.virol.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Avidan O, Bochner R, Hizi A. The catalytic properties of the recombinant reverse transcriptase of bovine immunodeficiency virus. Virology. 2006;351:42–57. doi: 10.1016/j.virol.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Oz Gleenberg I, Avidan O, Goldgur Y, Herschhorn A, Hizi A. Peptides derived from the reverse transcriptase of human immunodeficiency virus type 1 as novel inhibitors of the viral integrase. J. Biol. Chem. 2005;280:21987–21996. doi: 10.1074/jbc.M414679200. [DOI] [PubMed] [Google Scholar]
- 12.Klarmann GJ, Schauber CA, Preston BD. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J. Biol. Chem. 1993;268:9793–9802. [PubMed] [Google Scholar]
- 13.DeStefano JJ, Buiser RG, Mallaber LM, Fay PJ, Bambara RA. Parameters that influence processive synthesis and site-specific termination by human immunodeficiency virus reverse transcriptase on RNA and DNA templates. Biochim. Biophys. Acta. 1992;1131:270–280. doi: 10.1016/0167-4781(92)90025-u. [DOI] [PubMed] [Google Scholar]
- 14.Fisher TS, Darden T, Prasad VR. Substitutions at Phe61 in the beta3-beta4 hairpin of HIV-1 reverse transcriptase reveal a role for the fingers subdomain in strand displacement DNA synthesis. J. Mol. Biol. 2003;325:443–459. doi: 10.1016/s0022-2836(02)01225-1. [DOI] [PubMed] [Google Scholar]
- 15.Winshell J, Paulson BA, Buelow BD, Champoux JJ. Requirements for DNA unpairing during displacement synthesis by HIV-1 reverse transcriptase. J. Biol. Chem. 2004;279:52924–52933. doi: 10.1074/jbc.M409134200. [DOI] [PubMed] [Google Scholar]
- 16.Menendez-Arias L. Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses. 2009;1:1137–1165. doi: 10.3390/v1031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 18.Patel PH, Preston BD. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc. Natl Acad. Sci. USA. 1994;91:549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hizi A, Herschhorn A. Retroviral reverse transcriptases (other than those of HIV-1 and murine leukemia virus): a comparison of their molecular and biochemical properties. Virus Res. 2008;134:203–220. doi: 10.1016/j.virusres.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–-1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 21.Golinelli MP, Hughes SH. Nontemplated base addition by HIV-1 RT can induce nonspecific strand transfer in vitro. Virology. 2002;294:122–134. doi: 10.1006/viro.2001.1322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.