Abstract
A 15-mer DNA aptamer (named TBA) adopts a G-quadruplex structure that strongly inhibits fibrin-clot formation by binding to thrombin. We have performed thermodynamic analysis, binding affinity and biological activity studies of TBA variants modified by unlocked nucleic acid (UNA) monomers. UNA-U placed in position U3, U7 or U12 increases the thermodynamic stability of TBA by 0.15–0.50 kcal/mol. In contrast, modification of any position within the two G-quartet structural elements is unfavorable for quadruplex formation. The intramolecular folding of the quadruplexes is confirmed by Tm versus ln c analysis. Moreover, circular dichroism and thermal difference spectra of the modified TBAs displaying high thermodynamic stability show bands that are characteristic for antiparallel quadruplex formation. Surface plasmon resonance studies of the binding of the UNA-modified TBAs to thrombin show that a UNA monomer is allowed in many positions of the aptamer without significantly changing the thrombin-binding properties. The biological effect of a selection of the modified aptamers was tested by a thrombin time assay and showed that most of the UNA-modified TBAs possess anticoagulant properties, and that the construct with a UNA-U monomer in position 7 is a highly potent inhibitor of fibrin-clot formation.
INTRODUCTION
G-quadruplexes constitute a unique class of highly-ordered nucleic acids structures with various folding topologies and molecularities. They are present in naturally occurring nucleic acids and play important regulatory functions in many biological processes (1–6). The occurrence of quadruplex structures in genomes of diverse organisms suggested the possibility to design potential drug components targeted towards G-quadruplexes or based on RNA or DNA G-quartets (7–15).
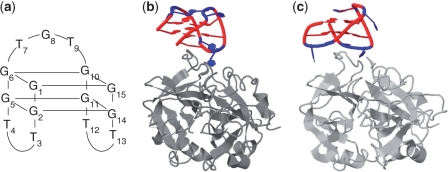
The 15-mer DNA oligonucleotide 5′-GGTTGGTGTGGTTGG is a thrombin binding aptamer (16). The aptamer, hereafter named TBA, was discovered in 1992 by in vitro selection and found to inhibit fibrin-clot formation by binding to the thrombin protein with high selectivity and affinity. According to NMR and X-ray structural studies TBA forms an intramolecular, antiparallel G-quadruplex with a chair-like conformation (17,18). The core of the quadruplex consists of two G-quartets connected by three edge-wise loops: a central TGT loop and two TT loops (Figure 1a). The aptamer interacts with two thrombin molecules, inactivating only one of them (18–20). X-ray studies indicated that inhibition of fibrinogen-clotting is a result of specific blocking of the thrombin anion exosite I by an interaction involving the central TGT loop (Figure 1b) (18–20). In the same studies it was furthermore reported that the two TT loops are involved in ionic interactions with the electropositive heparin binding site of a second thrombin molecule in the crystals to compensate the residual negative charge of the aptamer. In contrast, NMR studies indicated that the two TT loops interact with the thrombin anion exosite I (Figure 1c), while the TGT loop is in close proximity to the heparin binding site of a neighbouring thrombin molecule (18,20).
Figure 1.
Quadruplex structure of the thrombin binding aptamer (TBA) (a), and its interaction with the thrombin anion exosite I according to X-ray (b) and NMR (c) studies (20). Thrombin is marked in gray, TBA is marked in red (dG) and blue (T).
It has been suggested that the stability and rigidity of TBA is essential for interaction with the thrombin anion exosite I (21), and attempts to improve biological activity and thermal stability via chemical and structural modifications have been performed. Modifications have included 4-thio-2′-deoxyuridine (22), LNA (locked nucleic acid) (23,24), 2′-deoxy-isoguanosine (25), RNA (26,27) or 2′-O-methyl-RNA nucleotides (26,28), methylphosphonate or phosphorothioate internucleoside linkages (26,28), partial inversion of TBA polarity (5′-3′→3′-5′) with an 5′-5′ internucleoside linkage (29,30), or changes of loop size and sequence (31). Most of these modified TBAs have displayed retained or decreased thermal stability relative to the reference unmodified TBA. However variants containing e.g. 4-thio-2′-deoxyuridine or 2′-deoxy-isoguanosine nucleotides or phosphorothioate linkages have shown improved anticoagulant properties.
The chemical synthesis and thermal analysis of the UNA (unlocked nucleic acid) thymine monomer was first described by us in 1995 when a UNA monomer was shown to decrease the thermal stability (Tm value) of DNA duplexes (32). UNA is an acyclic RNA mimic (Figure 2) and in many ways the polar opposite to LNA (locked nucleic acid) within nucleic acid analogs. The missing bond between the C2′ and C3′ atoms of the ribose ring makes a UNA monomer more flexible than an unmodified RNA monomer. The synthesis of UNA-A, -C, -G and -U phosphoramidites, introduction of all UNA monomers into RNA and DNA duplexes, and thermal denaturation studies have recently been described (33,34). It was reported that UNA monomers, depending on their position in a duplex, either increase or decrease mismatch discrimination against DNA/RNA target strands, while they in a predictive and additive manner decrease the thermodynamic stability of duplexes (33–35). Moreover, a number of interesting biological applications of UNA-modified oligonucleotides, e.g. UNA-modified siRNAs, have recently been documented (36–39).
Figure 2.
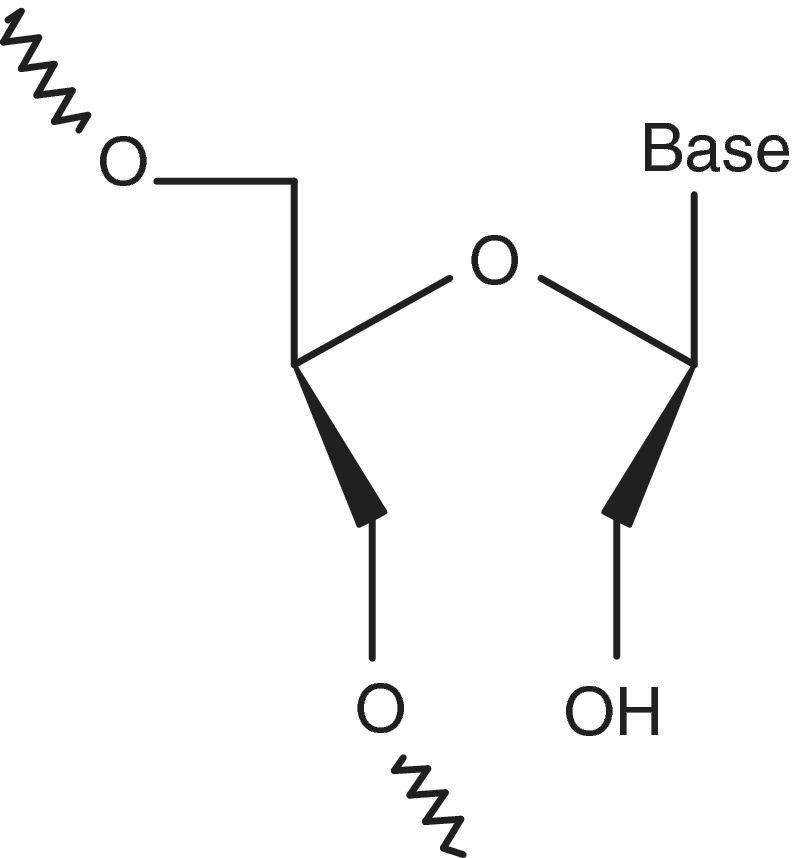
Structure of a UNA nucleotide monomer.
Herein we examine the influence of UNA on thermodynamic stability, binding affinity and biological activity of the quadruplex forming TBA. The modified variants are aptamers singly substituted with a UNA monomer in every possible position (Table 1). UV melting analysis was applied for comprehensive thermodynamic studies providing information about stoichiometry of the folding process as well as detailed thermodynamic parameters for the fifteen UNA-modified TBA variants. Thermal difference and circular dichroism (CD) spectra allowed determination of the effect of UNA substitutions on the structure of the TBA variants. Furthermore, surface plasmon resonance was used to investigate the affinity of the UNA-modified TBA analogs to thrombin. Finally, blood clotting studies with a selection of UNA-modified TBAs were performed to test their anticoagulant properties.
Table 1.
Thermodynamic parameters of quadruplex formation with UNA (X)a
| Name | Sequenceb | −ΔH° (kcal/mol) | −ΔS° (eu) | ΔG°37 (kcal/mol) | TMc (°C) | ΔΔG°37 (kcal/mol) | ΔTMc (°C) |
|---|---|---|---|---|---|---|---|
| TBA | GGTTGGTGTGGTTGG | 36.6 ± 0.4 | 116.1 ± 1.3 | –0.61 ± 0.02 | 42.2 | 0 | 0 |
| G1 | GGTTGGTGTGGTTGG | 30.9 ± 1.8 | 102.1 ± 5.7 | +0.74 ± 0.01 | 29.8 | 1.35 | −12.4 |
| G2 | GGTTGGTGTGGTTGG | n.d. | n.d. | n.d. | <20.0 | n.d. | n.d. |
| U3 | GGUTGGTGTGGTTGG | 39.4 ± 0.4 | 124.4 ± 1.1 | –0.84 ± 0.02 | 43.8 | −0.23 | 1.6 |
| U4 | GGTUGGTGTGGTTGG | 29.0 ± 2.2 | 94.2 ± 7.0 | +0.23 ± 0.02 | 34.6 | 0.84 | −7.6 |
| G5 | GGTTGGTGTGGTTGG | 41.6 ± 1.3 | 141.1 ± 4.3 | +2.15 ± 0.06 | 21.8 | 2.76 | −20.4 |
| G6 | GGTTGGTGTGGTTGG | n.d. | n.d. | n.d. | <20.0 | n.d. | n.d. |
| U7 | GGTTGGUGTGGTTGG | 38.6 ± 1.1 | 121.0 ± 3.8 | –1.11 ± 0.04 | 46.2 | −0.50 | 4.0 |
| G8 | GGTTGGTGTGGTTGG | 31.1 ± 0.9 | 101.4 ± 3.0 | +0.35 ± 0.03 | 33.6 | 0.96 | −8.6 |
| U9 | GGTTGGTGUGGTTGG | 32.7 ± 2.4 | 106.9 ± 7.7 | +0.47 ± 0.04 | 32.6 | 1.08 | −9.6 |
| G10 | GGTTGGTGTGGTTGG | n.d. | n.d. | n.d. | <20.0 | n.d. | n.d. |
| G11 | GGTTGGTGTGGTTGG | n.d. | n.d. | n.d. | <20.0 | n.d. | n.d. |
| U12 | GGTTGGTGTGGUTGG | 35.9 ± 0.62 | 113.4 ± 2.0 | −0.76 ± 0.01 | 43.7 | −0.15 | 1.5 |
| U13 | GGTTGGTGTGGTUGG | 31.6 ± 2.5 | 104.4 ± 8.2 | +0.80 ± 0.06 | 29.4 | 1.41 | −12.8 |
| G14 | GGTTGGTGTGGTTGG | n.d. | n.d. | n.d. | <20.0 | n.d. | n.d. |
| G15 | GGTTGGTGTGGTTGG | 39.3 ± 1.8 | 133.1 ± 5.8 | +2.05 ± 0.05 | 21.6 | 2.66 | −20.6 |
1 kcal = 4.184 kJ.
abuffer: 100 mM KCl, 10 mM sodium cacodylate, pH 7.0.
b5′-biotinylated oligonucleotides.
ccalculated for 10−4 M oligomer concentration.
n.d., not determined.
MATERIALS AND METHODS
Oligonucleotides
The 5′-biotinylated UNA oligonucleotides were synthesized on an automated RNA/DNA synthesizer (UNA phosphoramidite monomers and UNA-modified oligonucleotides are commercially available from www.ribotask.dk) using standard phosphoramidite chemistry. The [1-N-(4,4′-dimethoxytrityl)biotinyl-6-aminohexyl]-2-cyanoethyl(N,N-diisopropyl)phosphoramidite was applied to 5′-biotinylation of oligonucleotides. The O3′-phosphoramidites of UNA nucleotides were applied according to the previously described procedures (33) together with commercial DNA phosphoramidites. The purity of all oligonucleotides was verified by ion-exchange HPLC and determined to be 80% or greater, and the oligonucleotide compositions were confirmed by MALDI–TOF mass spectrometry.
UV melting analysis of TBA variants
Oligonucleotides were dissolved in a buffer containing 100 mM potassium chloride and 10 mM sodium cacodylate, pH 7.0. Oligonucleotide single strand concentrations were calculated from the absorbance measured above 80°C (40) and extinction coefficients which were approximated by a nearest-neighbor model (40,41) with the HyTher program. UNA-modified and unmodified DNA strands with identical sequences were assumed to have identical extinction coefficients. The samples were renatured by 5 min at 80°C and then slowly cooled to room temperature. The measurements were performed for nine different concentrations of each quadruplex in the concentration range 10−5–10−6 M using 10 mm (300 μl) quartz microcuvettes. Absorbance versus temperature curves were obtained by the UV melting method at 295 nm (42–44) in the temperature range 5–95°C on a Beckman DU 800 spectrophotometer equipped with a six-position microcell holder and a thermoprogrammer. The reversibility of transitions was ensured for all samples by measurement of heating and cooling profiles (data not shown). Three different heating rates (0.5, 0.3 and 0.2°C/min) were tested to avoid hysteresis phenomena. The rate of 0.2°C/min was selected because here the melting and annealing curves were reproducible and strictly superimposable. The lack of hysteresis phenomena implied that the quadruplex dissociation–association process was in a state of thermodynamic equilibrium (42,43). Melting curves were analyzed and the thermodynamic parameters determined by non-linear curve fitting with the program MeltWin 3.5 (45). Melting temperatures calculated for 10−4 M oligonucleotide concentration are marked by TM and melting points for any other concentration of oligonucleotide by Tm.
CD spectra
CD spectra were recorded on a Jasco J-600A spectropolarimeter using 1 ml quartz cuvettes with a 5 mm path length. The oligonucleotides were dissolved to 3.1 μM concentration in 10 mM phosphate buffered saline (138 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4), pH 7.4. All samples were renatured by 2 min at 100°C and slowly cooled to room temperature before data collection. The measurements were done at 20°C in the 200–300 nm wavelength range. The buffer spectrum was subtracted from the sample spectra. The spectra were smoothed in Microcal Origin 6.0 using a Savitzky-Golay filter.
Thermal difference spectra
The measurements were performed on a Beckman DU 800 spectrophotometer equipped with a six-position microcell holder and a thermoprogrammer using 10 mm quartz microcuvettes. The oligonucleotides were dissolved to 3.1 μM concentration in a buffer containing 100 mM potassium chloride and 10 mM sodium cacodylate, pH 7.0. Absorbance spectra were recorded at 13°C and 85°C in the 220–320 nm wavelength range. The scan speed was 1200 nm/min with a data collection of 1 pt/nm. Thermal difference spectra were obtained by subtraction of the low temperature absorbance spectra from the high temperature absorbance spectra with the Origin 6.0 program. The differential spectra were normalized by dividing the data by its maximum value (46).
Thrombin-aptamer affinity analysis by surface plasmon resonance
Real-time measurement of the interaction between thrombin and TBA and all UNA-modified TBAs was performed using a BIAcore 2000 system. Research grade SA sensor chips were from GE Healthcare. The running buffer for immobilization and sample analysis was 10 mM phosphate buffered saline (138 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4, 0.05% Tween-20), pH 7.4 at 25°C. Oligonucleotide immobilization was performed by the streptavidin-biotin coupling method. The flow rate was set to 5 μl/min, and the biotin TBA and UNA-modified TBAs were injected at a concentration of 1 µM over the streptavidin surface (SA sensor chip) for 15 min at 25°C. The unbound oligonucleotide was removed by treatment with 50 mM aqueous NaOH and the chip was primed before use. Thrombin solutions were sequentially injected over the sensor surface for 2 min at 15 µl/min and 2 min dissociation time. For each oligonucleotide, four concentrations of thrombin were injected by serially diluting samples from 200 to 25 nM along with a bovine serum albumin (BSA) sample as a non-specific target and a blank sample containing only running buffer for referencing. After each run, the surface was regenerated with 50 mM aqueous NaOH for 20 s at 15 µl/min. The raw data were processed and analyzed to determine the binding constant for each oligonucleotide. To correct for refractive index changes and instrument noise, the response data from the control surface were subtracted from the responses obtained from the reaction surface using biospecific interaction analysis evaluation 4.1. The KD values were calculated by global fitting of the four concentrations of thrombin over a constant density oligonucleotide surface. A 1:1 binding mode with mass transfer fitting was used to obtain the kinetic data.
The effects of UNA modified TBA on thrombin time
The inhibitory effect of TBA and the UNA modified variants on clotting time was measured by a thrombin time assay. The time (in seconds) for clotting at 37°C was measured on a STA-R EVOLUTION instrument (Stago) after mixing the identical volumes of citrate plasma and Trombin reagent (STA-THROMBIN 00611, Diagnostica Stago). A pool of plasma from four healthy individuals was used. The thrombin reagent was pre-incubated with TBA or the TBA variants at 0.33 µM concentration for 5 min before addition to plasma and measurement of clotting time (= thrombin time). The anti thrombin effect is the additional time for clotting in presence of the aptamers relative to a reference with water added.
RESULTS
In this study, all experiments are performed with aptamers containing 5′-biotin and a linker. This is required for the surface plasmon resonance experiments and allowed us to directly correlate data obtained from different types of studies. From preliminary (not published) experiments we know that 5′-biotin decreases thermodynamic stability and biological activity of the TBA in study herein but we assume that the effect is the same for all the 5′-biotinylated TBA variants investigated.
Thermodynamic and spectroscopic features of UNA-modified TBA variants
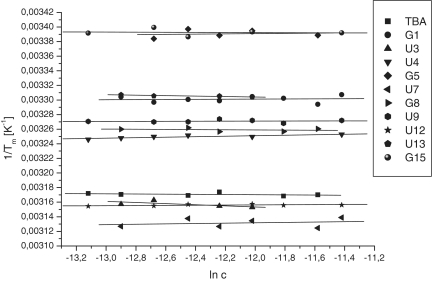
UNA monomers were introduced as single substitutions at all possible positions of TBA resulting in 15 different TBA variants. Melting curves for each variant provided the thermodynamic parameters shown in Table 1. Only three UNA-modified TBAs, namely U3, U7 and U12 (marked by underlined italic font to differentiate between name of the aptamer and UNA monomer position within an aptamer), together with unmodified TBA, showed a negative value of Gibbs’ free energy indicating formation of quadruplex structure at 37°C. Moreover, these three modified variants display increased thermodynamic stability relative to TBA (by 0.23, 0.50 and 0.15 kcal/mol, respectively). Substitution of position T7 by UNA-U was the most energetically favorable for quadruplex formation. In contrast, modification of any of the guanosine monomers forming G-quartets resulted in significant destabilization of the quadruplex structure by at least 1.35 kcal/mol. Thus, UNA monomers only stabilize the TBA quadruplex structure when placed in specific positions of the loops. Plotting 1/Tm as a function of the concentration of each of the 15 oligonucleotide variants showed no concentration dependence of Tm (Figure 3).
Figure 3.
Concentration dependence of thermal denaturation temperatures (Tm values).
CD spectra were recorded for TBA and all UNA-modified TBAs to monitor the influence of UNA on the overall structure (Figure 4). The majority of the spectra for the UNA-modified TBAs show strongly decreased intensity of the CD bands or even band loss relative to the spectrum of the unmodified TBA. However, the CD spectra of U3 and U7 show slightly more intense bands than TBA with a high-amplitude positive maximum near 293 nm. Also the spectrum of U12 shows a significant band at ∼293 nm, but its intensity is lower than in the spectrum of TBA suggesting that it originates from a less populated molecular conformation.
Figure 4.
Representative CD spectra of TBA (solid line) and UNA-modified aptamers characteristic of the three groups: U3, U7 and U12 (dashed line: U7); G1, U4, G8, U9 and U13 (dotted line: U13); G2, G5, G6, G10, G11, G14 and G15 (dash-dotted line: G14). For individual CD spectra see ‘Supplementary Data’.
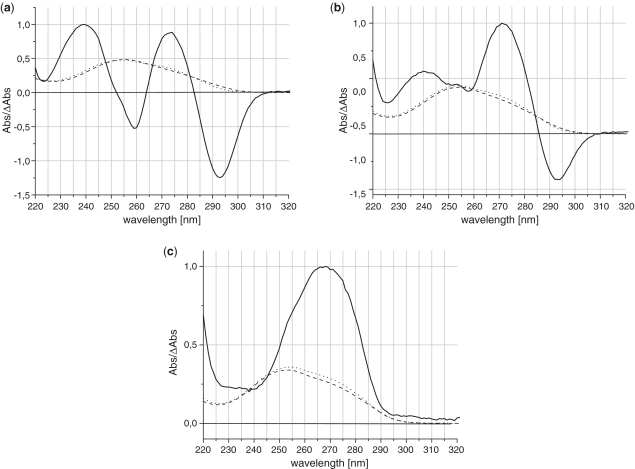
Thermal difference spectra (TDS) were recorded (Figure 5) to support the results obtained from the CD spectra. The spectra obtained divide the modified TBAs into three groups. The first group containing the U3, U7 and U12 variants presents a typical intramolecular TBA quadruplex profile. They have two positive maxima ∼240 and 273 nm, and also one moderately intense peak ∼260 nm and one highly negative peak ∼295 nm. The second group constitutes G1 and the modified TBAs possessing a UNA monomer situated in the TGT or TT loops (except the U3, U7 and U12 variants). No typical quadruplex signature was observed for this group, mainly due to disappearance of the negative peak ∼260 nm. The third group consists of TBAs modified in any of the positions entangled in G-quartets formation, except position G1. They show a complete loss of the profile characteristic for G-quadruplexes. Thus, all the data obtained from the thermal difference spectra are consistent with the conclusions from the CD spectra and the thermodynamic studies.
Figure 5.
Representative high- (dotted line) and low-temperature (dashed line) absorbance thermal difference spectra (TDS, solid line) (a) TBA, characteristic also of U3, U7 and U12; (b) G1, characteristic also of U4, G8, U9 and U13; (c) G2, characteristic also of G5, G6, G10, G11, G14 and G15. For single TDS profiles see ‘Supplementary Data’.
Affinity of UNA-modified TBAs to thrombin
To assay binding of the UNA-modified TBAs to thrombin we used surface plasmon resonance (SPR) studies. For this purpose all TBA variants contained a 5′-biotin group with a C6 spacer to facilitate binding to the surface of a streptavidin-coated sensor chip. SPR analysis was used to determine the kinetic constants at thrombin concentrations ranging from 25 to 200 nM. The kinetic profile of the unmodified TBA revealed a dissociation constant (KD) value of 102.6 nM (Table 2). U7 shows a small but significant improvement of affinity (KD = 78.2 nM), while G1 and U9 show affinities similar to the unmodified TBA. In contrast, G8 displays a significant loss of affinity (KD = 916.3 nM), while the rest of the modified TBAs showed KD values from 154.7 to 425.1 nM. No KD values were measurable for G11, U13 and G14, presumably due to lack of significant affinity towards thrombin after incorporation of UNA in these positions.
Table 2.
Comparison of binding affinity and biological effect of TBA and UNA-modified TBA variants
| Name | Sequencea | KD (nM)b | T-timec/ antithrombin effect (s) |
|---|---|---|---|
| TBA | GGTTGGTGTGGTTGG | 102.6 ± 5.1 | 28.8/9.9 |
| G1 | GGTTGGTGTGGTTGG | 95.4 ± 3.9 | 22.8/3.9 |
| G2 | GGTTGGTGTGGTTGG | 298.1 ± 5.7 | n.d. |
| U3 | GGUTGGTGTGGTTGG | 160.4 ± 4.9 | 24.0/5.1 |
| U4 | GGTUGGTGTGGTTGG | 172.3 ± 8.1 | 19.1/0.1 |
| G5 | GGTTGGTGTGGTTGG | 425.1 ± 17.7 | n.d. |
| G6 | GGTTGGTGTGGTTGG | 287.2 ± 12.3 | n.d. |
| U7 | GGTTGGUGTGGTTGG | 78.2 ± 4.9 | 32.6/13.7 |
| G8 | GGTTGGTGTGGTTGG | 913.3 ± 21.2 | 23.4/4.4 |
| U9 | GGTTGGTGUGGTTGG | 110.3 ± 4.1 | 24.4/5.4 |
| G10 | GGTTGGTGTGGTTGG | 221.8 ± 9.3 | n.d. |
| G11 | GGTTGGTGTGGTTGG | n.d. | n.d. |
| U12 | GGTTGGTGTGGUTGG | 190.3 ± 8.2 | 23.2/4.2 |
| U13 | GGTTGGTGTGGTUGG | n.d. | 18.9/0.0 |
| G14 | GGTTGGTGTGGTTGG | n.d. | n.d. |
| G15 | GGTTGGTGTGGTTGG | 154.7 ± 7.4 | 19.8/0.8 |
a5′-biotinylated oligonucleotides.
bkinetic analysis of the affinity interaction between immobilized TBA variants and thrombin determined from kon/koff kinetic analysis.
c‘T-time’ is the time required for a clot formation in the plasma from a blood sample.
‘Antithrombin effect’ is the T-time in the presence of an aptamer minus the T-time in the absence of an aptamer; no oligo added resulted in 18.9 and 19.0 s clotting times in two experimental sets and constitute reference values in the antithrombin effect calculations. Non-biotin conjugated TBA results in 34.4 s clotting time.
n.d., not determined.
The biological effect of the UNA-modified TBAs tested in a thrombin time assay
To investigate whether the UNA-modified TBAs inhibit the enzymatic activity of thrombin and to correlate the biological effect with our chemical and kinetic data, a selection of the modified TBAs was tested in a simple thrombin time assay (Table 2). The variants were chosen based mainly on their ability to form G-quadruplex structure (CD, TDS) and therefore do not include modification of G-quartets positions with the only two exceptions being G1 and G15. U7 showed an increased inhibitory effect relative to the unmodified TBA, while inhibition of coagulation by G1, U3, G8, U9 and U12 was ∼2-fold decreased, and U4, U13 and G15 showed no influence on fibrin-clot formation.
DISCUSSION
The influence of UNA monomers on thermodynamic stability of the TBA quadruplex structure
The thermodynamic studies of the TBA variants revealed significant destabilization of quadruplexes when UNA occupied any of the G-quartets forming positions (Table 1). This is expected as UNA monomers are very flexible and parallels the effect of UNA monomers on duplex thermodynamic stabilities (32–35). The magnitude of destabilization hindered the determination of comprehensive thermodynamic data for the majority of these variants. The thermodynamic parameters for G1, G5 and G15 reveal quadruplex destabilization. The relatively higher stability of the G1 and G15 variants is presumably due to the terminal positioning of the UNA-G modification with a neighboring nucleobase only at one side. In general, terminal nucleotide monomers are often characterized by slightly weaker interactions than internal monomers, and the magnitude of destabilization caused by a modification therefore should decrease when shifting towards an oligonucleotide end (35,47). Surprisingly, the UNA-G situated internally at position G5 destabilizes the quadruplex structure only to a level comparable to G15. Therefore, other differences than the terminal positioning seem to contribute to the change of the overall thermodynamic stability of the TBA variants. The G1 shows a less favorable enthalpy change which compensates the favorable increase of entropy and results in an increase of ΔG°37. In contrast, the less stable G5 and G15 display favorable enthalpy contributions accompanied by unfavorable entropic changes.
UNA modifications in positions U4, G8, U9 or U13 of the quadruplex loops also significantly decrease the thermodynamic stability of the quadruplexes which might be expected based on the reported NMR and X-ray studies which have revealed that the G8 and T9 positions are situated in proximity to G6 and G15 of the first G-quartet, while T4 and T13 are close to guanines of the first or second G-quartet (17,19,48). Furthermore, the O4 carbonyl group and the N3 imino proton of T4 and T13 are involved in hydrogen bond formation (17,19,49). The unfavorable effect of UNA placed in the G-quartets might be explained by distortion of guanine stacking and hydrogen bond interaction essential for quadruplex folding, and the destabilization effects caused by UNA observed for some specific positions in the loops might by due to disturbance of interactions between the loop and the quadruplex core or the U4:U13 hydrogen bonding interaction.
The stabilization of quadruplex structure observed for U3, U7 or U12 corresponds to ∼1.5- to 2-fold more favorable equilibrium constant for quadruplex formation relative to the TBA reference (Table 1). It has been reported that substitution of all thymidines by 2′-deoxyuridine monomers increases the thermodynamic stability of TBA by 0.16 kcal/mol per modification on average (26). UNA-U in position U12 thus stabilizes the quadruplex structure to an extent comparable to a 2′-deoxyuridine monomer, whereas the presence of a UNA-U monomer in positions U3 and U7 is more favorable than a 2′-deoxyuridine monomer for quadruplex folding. Therefore, the observed stabilization is presumably not only due to the absence of the methyl group in position 5 of the thymine nucleobases but also the increased flexibility of the UNA monomers. The enthalpies per G-quartet for TBA and U3, U7 and U12 revealed comparable values of −18.3, −19.7, −19.3 and −18.0 kcal/mol, respectively, corresponding to values reported previously for single G-quartets (50). Finally, the analysis of enthalpy-entropy contribution revealed a favorable enthalpic change which is only partially compensated by an unfavorable entropic change for U3 and U7, while the higher stability of U12 seems to have an entropic origin.
The molecularity of folding can be determined by the concentration dependence of Tm values. In general, the Tm value of an unimolecular structural motif is concentration independent, whereas an increase of sample concentration results in enhanced Tm values if structures with higher molecularity are formed (44). To verify an intramolecular folding of the TBA variants, Tm as a function of oligonucleotide concentration was studied and as no linear increase was observed an intramolecular folding mechanism for the studied structures was confirmed (Figure 3).
Structural features of the TBA variants
CD spectra can be used to distinguish between the different quadruplex folding topologies, as parallel and antiparallel structures are characterized by different CD profiles with high-amplitude band at 265 and 293 nm, respectively (27). The CD spectra for U3, U7 and U12 show a CD profile indicating antiparallel folding topology as for TBA while G2, G5, G6, G10, G11, G14 and G15 show no evidence for quadruplex formation.
It has been reported that the global shapes of TDS are specific for a given RNA/DNA structure (46). This is in accordance with our studies where three different TDS patterns were registered for the TBA variants (Figure 5). A TDS profile with two maxima at 240 and 273 nm and two minima at 260 and 295 nm that can be attributed to an intramolecular quadruplex structure (46), a TDS profile with reduced intensity of the CD bands and irregular TDS profiles with complete loss of a minimum at 260 nm that most probably indicate more unstructured states, and a TDS profile that exhibit complete loss of the typical quadruplex pattern. These results confirm those obtained from thermodynamic analysis and CD spectra.
Thrombin-aptamer kinetics and biological activity
The kinetic binding study of the UNA-modified TBA aptamers revealed that UNA modifications are allowed in about half of the positions in TBA without largely (>2-fold) changing the binding affinity (positions G1, U3, U4, U7, U9, U12 and G15). In contrast, UNA in positions G11, U13 and G14 abolish or strongly inhibit the interaction with thrombin. It has been suggested that the TBA structure is retained after binding to thrombin (48,51). We therefore can expect that changes in binding constants are connected with changes of quadruplex structure and also the thermodynamic stability of the quadruplexes assuming higher stability for ideally structured quadruplexes. According to structural studies, G1 is involved in formation of the quadruplex core and does not interact directly with the thrombin anion exosite I (18–20). It was previously reported that substitution of G1 by isoguanosine leads to an improved binding constant and retained thermodynamic stability (25). The slightly lower KD value for G1 relative to TBA is remarkable because according to the thermodynamic and structural analysis (Table 1, Figures 4 and 5), UNA-modification at position G1 significantly decreases the thermodynamic stability and disturbs the overall quadruplex folding. Apparently, increased flexibility by introduction of UNA monomer in this position is compatible with favorable interactions between the aptamer and thrombin. However, a similar behavior is observed for the terminally modified G15 with a KD value slightly higher than for the unmodified TBA.
U7 exhibits the lowest KD of all. According to X-ray data of TBA bound to thrombin, T7 is buried in a hydrophobic cluster in the fibrinogen recognition site (Figure 1b) (19,20). The increased affinity may thus be attributed to a favorable change of the orientation with the T7 UNA-modified monomer leading to a better quadruplex-protein complex fit. On the contrary, UNA-modification of position G8 in the internal loop significantly reduces affinity while variant U9 shows a KD value similar to that of TBA. According to X-ray structural data and by analogy with the observed binding properties of U7, UNA modification in positions G8 and U9 could be favorable for interaction with thrombin due to structural changes. However, modification of each of the positions destabilizes a quadruplex structure. The large difference in KD values between G8 and U9 might be explained by the observation from the crystal structure that G8 stacks with the quadruplex core whereas T9 is close to the T7 hydrophobic interaction site (19). Making G8 flexible with an UNA might reduce the stacking ability while more flexibility might ease the U9 positioning.
U3, U4 and U12 show similar, although slightly weaker affinity towards thrombin than unmodified TBA and by reference to the X-ray studies, positions U3, U4 and U12 are not directly involved in the binding to the thrombin anion exosite I. Accordingly, and confirming our thermodynamic and structural results, these UNA modifications have only minor influence on the binding constants. UNA modification of positions G2, G5, G6, G10, G11 and G14 induces significant loss of affinity towards thrombin which is likely due to the global disturbance of the quadruplex structure as documented earlier.
Our binding data fit the X-ray structure better than the NMR based model regarding the TBA–thrombin interactions. According to the NMR structure improved thrombin binding should not be induced by the U7 modification as it was suggested that the central loop is not directly involved in specific interactions with thrombin, and the inhibitory effect rather is a consequence of blocking thrombin anion exosite I by the two TT loops. On the contrary, according to the X-ray structure, modification of the two TT loops should not significantly impact the binding constant anticipating that UNA monomer induces only structural changes of local importance, while introduction of UNA into the central loop should provoke apparent changes in affinity towards thrombin, which correlate with our results.
Thrombin clotting time showed that several of the studied TBA variants possess considerable biological activity (Table 2), although most of them are less active than unmodified TBA. We observe a clear correlation between thermodynamics, kinetics and clotting time as U3, U7 and U12 are stable quadruplexes and strong thrombin binders displaying biological activity. Similarly, the thermodynamically unstable U13 possess a weak binding affinity towards thrombin and is biologically inactive. Less conclusive correlation was observed for the unstable G1, U4, G8, U9 and G15 aptamers. U4 and G15 show no biological activity but considerable binding affinity, while G8 is biologically active but displays poor thrombin binding. G1 and U9 seem to bind thrombin like TBA and show biological activity even though the thermodynamic stability is unfavorable. The less conclusive correlation between thermodynamics, kinetics and biological activity for some of our TBA variants might be due to different conditions used in the experiments as it has been found that not only potassium ions but also molecular crowding and the presence of thrombin impact quadruplex formation (52). Gratifyingly, the most thermodynamically stable TBA variant U7 with high binding affinity also showed increased anticoagulant activity relative to the unmodified TBA. In general, our results are in accordance with a previously reported study where only three TBA variants modified in positions T3, T7, or T12 by an acyclic thymidine residue [1-N-(3-hydroxy-2-hydroxymethyl-2-methylpropyl)thymidine] showed higher thermal stability relative to the unmodified aptamer (53). In the same study, variant T7 was the only of all modifications characterized as more potent in blood clotting than TBA.
CONCLUSIONS
The aptamers described in this article are the first examples of UNA-containing quadruplexes. In this study we correlate thermodynamic studies of UNA-modified TBA variants with structural aspects, thrombin binding data and biological activity. UNA introduced at positions U3, U7 or U12 make quadruplex folding more energetically favorable than observed for the parent TBA. Moreover, modification at those positions preserves the overall aptamer structure and retains the antiparallel, intramolecular quadruplex topology seen for TBA. The kinetic analyses revealed only three specific positions (G11, U13 and G14) where incorporation of UNA highly diminished or abolished thrombin binding. Moreover, based on a thrombin clotting time assay one aptamer, namely U7, was identified as more capable in blood clotting than unmodified TBA. Our findings demonstrate that UNA monomers are not only efficient modulators of quadruplex thermodynamic stability but also change the binding affinity and biological properties of TBA in a position-depending manner. The results provide a better understanding of the subtle features involved in the interaction between TBA and thrombin and facilitate the design of novel quadruplex-based drugs with higher thermodynamic stability and improved anticoagulant properties.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The Danish National Research Foundation.
Conflict of interest statement. J.W. is a co-founder of RiboTask ApS which is marketing UNA amidites and UNA oligonucleotides.
ACKNOWLEDGEMENT
We gratefully acknowledge financial support by The Danish National Research Foundation.
REFERENCES
- 1.Johnson JE, Smith JS, Kozak ML, Johnson FB. In vivo veritas: using yeast to probe the biological functions of G-quadruplexes. Biochimie. 2008;90:1250–1263. doi: 10.1016/j.biochi.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Kaminaga K, Komiyama M. G-Quadruplex formation by human telomeric repeats-containing RNA in Na+ solution. J. Am. Chem. Soc. 2008;130:11179–11184. doi: 10.1021/ja8031532. [DOI] [PubMed] [Google Scholar]
- 6.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2009;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 7.Rahman KM, Reszka AP, Gunaratnam M, Haider SM, Howard PW, Fox KR, Neidle S, Thurston DE. Biaryl polyamides as a new class of DNA quadruplex-binding ligands. Chem. Commun. 2009:4097–4099. doi: 10.1039/b902359c. [DOI] [PubMed] [Google Scholar]
- 8.de Soultrait VR, Lozach P-Y, Altmeyer R, Tarrago-Litvak L, Litvak S, Andréola ML. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002;324:195–203. doi: 10.1016/s0022-2836(02)01064-1. [DOI] [PubMed] [Google Scholar]
- 9.Girvan AC, Teng Y, Casson LK, Thomas SD, Jüliger S, Ball MW, Klein JB, Pierce WM, Barve SS, Bates PJ. AGRO100 inhibits activation of nuclear factor-κB (NF-κB) by forming a complex with NF-κB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006;5:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 10.Bates PJ, Kahlon JB, Thomas SD, Trent JO, Miller DM. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 1999;274:26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 11.Mashima T, Matsugami A, Nishikawa F, Nishikawa S, Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37:6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huppert JL. Four-stranded DNA: cancer, gene regulation and drug development. Philos. Trans. Roy. Soc. Ser. A. 2007;365:2969–2984. doi: 10.1098/rsta.2007.0011. [DOI] [PubMed] [Google Scholar]
- 13.Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian-miao O, Yu-jing L, Jia-heng T, Zhi-shu H, Kwok-Yin W, Lian-quan G. G-Quadruplexes: targets in anticancer drug design. Chem. Med. Chem. 2008;3:690–713. doi: 10.1002/cmdc.200700300. [DOI] [PubMed] [Google Scholar]
- 15.Zagotto G, Sissi C, Moro S, Dal Ben D, Parkinson GN, Fox KR, Neidle S, Palumbo M. Amide bond direction modulates G-quadruplex recognition and telomerase inhibition by 2,6 and 2,7 bis-substituted anthracenedione derivatives. Bioorg. Med. Chem. 2008;16:354–361. doi: 10.1016/j.bmc.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 16.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 17.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly JA, Feigon J, Yeates TO. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 19.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 20.Padmanabhan K, Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996 doi: 10.1107/S0907444995013977. D52, 272–282. [DOI] [PubMed] [Google Scholar]
- 21.Paborsky LR, McCurdy SN, Griffin LC, Toole JJ, Leung LL. The single-strandedDNA aptamer-binding site of human thrombin. J. Biol. Chem. 1993;268:20808–20811. [PubMed] [Google Scholar]
- 22.Raviv SM, Horvath A, Aradi J, Bagoly Z, Fazakas F, Batta Z, Muszbek L, Harsfalvi J. 4-Thio-deoxyuridylate-modified thrombin aptamer and its inhibitory effect on fibrin clot formation, platelet aggregation and thrombus growth on subendothelial matrix. J. Thromb. Homeost. 2008;6:1764–1771. doi: 10.1111/j.1538-7836.2008.03106.x. [DOI] [PubMed] [Google Scholar]
- 23.Virno A, Randazzo A, Giancola C, Bucci M, Cirino G, Mayol L. A novel thrombin binding aptamer containing a G-LNA residue. Bioorg. Med. Chem. 2007;15:5710–5718. doi: 10.1016/j.bmc.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Bonifacio L, Church F, Jarstfer M. Effect of locked-nucleic acid on a biologically active G-quadruplex. A structure-activity relationship of the thrombin aptamer. Int. J. Mol. Sci. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nallagatla SR, Heuberger B, Haque A, Switzer C. Combinatorial synthesis of thrombin-binding aptamers containing iso-guanine. J. Comb. Chem. 2009;11:364–369. doi: 10.1021/cc800178m. [DOI] [PubMed] [Google Scholar]
- 26.Sacca B, Lacroix L, Mergny J-L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang C-F, Shafer RH. Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaitseva M, Kaluzhny D, Shchyolkina A, Borisova O, Smirnov I, Pozmogova G. Conformation and thermostability of oligonucleotide d(GGTTGGTGTGGTTGG) containing thiophosphoryl internucleotide bonds at different positions. Biophys. Chem. 2010;146:1–6. doi: 10.1016/j.bpc.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Martino L, Virno A, Randazzo A, Virgilio A, Esposito V, Giancola C, Bucci M, Cirino G, Mayol L. A new modified thrombin binding aptamer containing a 5′-5′ inversion of polarity site. Nucleic Acids Res. 2006;34:6653–6662. doi: 10.1093/nar/gkl915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pagano B, Martino L, Randazzo A, Giancola C. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008;94:562–569. doi: 10.1529/biophysj.107.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smirnov I, Shafer RH. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen P, Dreiøe LH, Wengel J. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides: introduction of three novel analogues and a summary. Bioorg. Med. Chem. Lett. 1995;3:19–28. doi: 10.1016/0968-0896(94)00143-q. [DOI] [PubMed] [Google Scholar]
- 33.Langkjær N, Pasternak A, Wengel J. UNA (unlocked nucleic acid): A flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg. Med. Chem. 2009;17:5420–5425. doi: 10.1016/j.bmc.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 34.Jensen TB, Langkjær N, Wengel J. Unlocked nucleic acid (UNA) and UNA derivatives: Thermal denaturation studies. Nucleic Acids Symp. Ser. 2008;52:133–134. doi: 10.1093/nass/nrn068. [DOI] [PubMed] [Google Scholar]
- 35.Pasternak A, Wengel J. Thermodynamics of RNA duplexes modified with unlocked nucleic acid nucleotides. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq561. 18 June 2010 [Epub ahead of print]; doi:10.1093/nar/gkq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangos MM, Min K-L, Viazovkina E, Galarneau A, Elzagheid MI, Parniak MA, Damha MJ. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2′F-ANA constructs containing acyclic nucleotide inserts. J. Am. Chem. Soc. 2002;125:654–661. doi: 10.1021/ja025557o. [DOI] [PubMed] [Google Scholar]
- 37.Fluiter K, Mook ORF, Vreijling J, Langkjær N, Højland T, Wengel J, Baas F. Filling the gap in LNA antisense oligo gapmers: the effects of unlocked nucleic acid (UNA) and 4'-C-hydroxymethyl-DNA modifications on RNase H recruitment and efficacy of an LNA gapmer. Mol. BioSyst. 2009;5:838–843. doi: 10.1039/b903922h. [DOI] [PubMed] [Google Scholar]
- 38.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjær N, Babu BR, Højland T, Abramov M, Van Aerschot A, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjær N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, et al. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq341. 7 May 2010 [Epub ahead of print]; doi:10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borer PN. Optical properties of nucleic acids, absorption and circular dichroism spectra. In: Fasman GD, editor. CRC Handbook of Biochemistry and Molecular Biology: Nucleic Acids. 3rd edn. Vol. 1. Cleveland, OH: CRC Press; 1975. pp. 589–595. [Google Scholar]
- 41.Richards EG. Use of tables in calculations of absorption, optical rotatory dispersion and circular dichroism of polyribonucleotides. In: Fasman GD, editor. CRC Handbook of Biochemistry and Molecular Biology: Nucleic Acids. 3rd edn. Vol. 1. Cleveland, OH: CRC Press; 1975. pp. 596–603. [Google Scholar]
- 42.Rachwal PA, Fox KR. Quadruplex melting. Methods. 2007;43:291–301. doi: 10.1016/j.ymeth.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Mergny J-L, Lacroix L. UV melting of G-quadruplexes. In: Huntley M, editor. Current Protocols in Nucleic Acid Chemistry. Vol. 17. Chichester: John Wiley & Sons; 2009. pp. 1.1–15. [DOI] [PubMed] [Google Scholar]
- 44.Mergny J-L, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 45.McDowell JA, Turner DH. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 46.Mergny J-L, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gros J, Rosu F, Amrane S, De Cian A, Gabelica V, Lacroix L, Mergny J-L. Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang JY, McCurdy SN, Shea RG, Swaminathan S, Bolton PH. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 49.Schultze P, Macaya RF, Feigon J. Three-dimensional Solution Structure of the Thrombin-binding DNA Aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 50.Mergny J-L, Phan A-T, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 51.Wang KY, Krawczyk SH, Bischofberger N, Swaminathan S, Bolton PH. The tertiary structure of a DNA aptamer which binds to and inhibits thrombin determines activity. Biochemistry. 1993;32:11285–11292. doi: 10.1021/bi00093a004. [DOI] [PubMed] [Google Scholar]
- 52.Nagatoishi S, Tanaka Y, Tsumoto K. Circular dichroism spectra demonstrate formation of the thrombin-binding DNA aptamer G-quadruplex under stabilizing-cation-deficient conditions. Biochem. Biophys. Res. Commun. 2007;352:812–817. doi: 10.1016/j.bbrc.2006.11.088. [DOI] [PubMed] [Google Scholar]
- 53.Copolla T, Varra M, Oliviero G, Galeone A, D'Isa G, Mayol L, Morelli E, Bucci M-R, Vallecco V, Cirino G, et al. Synthesis, structural studies and biological properties of new TBA analogues containing acyclic nucleotide. Bioorg. Med. Chem. 2008;16:8244–8253. doi: 10.1016/j.bmc.2008.07.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.