Abstract
The t(14;18) translocation in follicular lymphoma is one of the most common chromosomal translocations. Most breaks on chromosome 18 are located at the 3′-UTR of the BCL2 gene and are mainly clustered in the major breakpoint region (MBR). Recently, we found that the BCL2 MBR has a non-B DNA character in genomic DNA. Here, we show that single-stranded DNA modeled from the template strand of the BCL2 MBR, forms secondary structures that migrate faster on native PAGE in the presence of potassium, due to the formation of intramolecular G-quadruplexes. Circular dichroism shows evidence for a parallel orientation for G-quadruplex structures in the template strand of the BCL2 MBR. Mutagenesis and the DMS modification assay confirm the presence of three guanine tetrads in the structure. 1H nuclear magnetic resonance studies further confirm the formation of an intramolecular G-quadruplex and a representative model has been built based on all of the experimental evidence. We also provide data consistent with the possible formation of a G-quadruplex structure at the BCL2 MBR within mammalian cells. In summary, these important features could contribute to the single-stranded character at the BCL2 MBR, thereby contributing to chromosomal fragility.
INTRODUCTION
The t(14;18) translocation is the most common chromosomal translocation in human lymphoma. It is characteristically associated with follicular lymphoma (FL), which is a sub-type of non-Hodgkin's lymphoma (1). The t(14;18) translocation results from recombination between the BCL2 gene and the J subexons of the immunoglobulin heavy chain (IgH) during V(D)J recombination in pre-B cells. This translocation juxtaposes the BCL2 locus on chromosome 18 to the enhancer element of the IgH locus on chromosome 14 (2–4). This results in the overexpression of the antiapoptotic protein, BCL2 and thereby leads to FL. Although the BCL2 gene is >200 kb in length, in the majority of FL patients, the breaks at chromosome 18 occur within a small 150-bp region located in the 3′-untranslated region (3′-UTR) of the third exon, known as the major breakpoint region (MBR) (5–7). Within the 150-bp MBR, there are three peaks of breakpoints, each ∼15–20 bp in size (6,8). The analysis of patient breakpoint junctions indicates a V(D)J recombination-mediated mechanism for generation of breaks on chromosome 14 (9,10). At the IgH locus, DH and JH subexons are typically cleaved at a pair of signal sequences (12- or 23-RSS) by the RAG complex (11). However, the mechanism of breakage at the BCL2 MBR remains a subject of active investigation. Recently, we and others showed that the RAG complex can misrecognize the BCL2 MBR but not as a cryptic RSS (12,13). We also found that the BCL2 MBR adopts a non-B DNA structure in genomic DNA, which could be recognized and cleaved by RAGs (13,14). The structure at the 150-bp BCL2 MBR contains distinctive regions of single-stranded character, as shown previously based on bisulfite reactivity (13).
The precise non-B DNA conformation at the BCL2 MBR has been a subject of active investigation (15,16). The large number of consecutive cytosines at many locations throughout the MBR, are associated with rapid opening kinetics (15). This might permit them to transiently open and assume a number of possible conformations on the non-template or template DNA strands. During transcription and DNA replication, the two DNA strands are transiently separated and this might permit either of them to assume a non-B DNA structure or result in the two DNA strands reannealing in a transiently alternative conformation [e.g. triplexes (17) or quadruplexes]. Bulk DNA structural studies often do not clearly distinguish among the various conformational possibilities. Multiple methods are required to arrive at a picture of the possible conformations. Moreover, more than one conformation might be possible involving either one or both of the DNA strands. Bisulfite reactivity can be not only due to transient or stable single-strandedness but also due to formation of B/A-intermediate duplexes (15,17), which have rapid opening kinetics, as mentioned earlier.
Both triplexes and quadruplexes have similar requirements for purine-rich DNA and Hoogsteen hydrogen bonding for stability (18). Since the region near peak I of the BCL2 MBR is abundant in guanines, we investigated the possibility that the template strand could fold into a quadruplex structure. Here, we describe a G-quadruplex structure formed by the template strand upstream of peak I of the MBR. Using various assays, we show that the intramolecular G-quadruplex formed at the MBR is parallel and dependent on potassium. Mutagenesis and DMS protection assays show the presence of three guanine quartets, which was further verified by nuclear magnetic resonance (NMR) studies. We also show that such a structure can form in the context of the whole MBR on a plasmid. Finally, we provide evidence for the ability of such a structure to decrease transcription in mammalian cells.
MATERIALS AND METHODS
Oligomeric DNA
The oligonucleotides used in the current study are listed in Supplementary Table S1. These oligomers were gel purified as described (19).
Plasmid construction
The 300-bp wild-type and mutant BCL2 MBR fragments were obtained after SalI digestion of the plasmids, pXW5 and pSCR41, respectively (14). The mutant plasmid, pSCR41, was generated using site-directed mutagenesis as described earlier (14). It contains a 3-nt mutation within the G-quadruplex forming sequence of the MBR, wherein one stretch of guanines is converted to cytosines (GGG→CCC). The 300-bp wild-type and mutant MBR fragments were then cloned into SalI digested pIRES2-EGFP (BD Biosciences, Clontech) and named as pMN11 (wild-type) and pMN12 (mutant). The orientation of the cloned insert was confirmed by DNA sequencing.
5′-end-labeling of oligomers
The 5′-end-labeling of the oligomeric DNA was done using T4 polynucleotide kinase as described earlier (20). The labeled substrates were purified using Qiagen quick nucleotide removal kit and stored at −20°C until used.
Gel mobility shift assay
The radiolabeled oligomers were incubated either in the presence or absence of 100 mM KCl in Tris–EDTA (TE) buffer (pH 8.0) at 37°C for 1 h. The different forms of the G-quadruplexes were then resolved on 15% native polyacrylamide gels in the presence or absence of 100 mM KCl, both in the gel and the buffer, at 150 V at room temperature. The gels were dried and exposed to a screen, and the signal was detected using PhosphorImager FLA9000 (Fuji, Japan).
DMS protection assay
The radiolabeled oligomers were incubated in TE in the presence of 100 mM KCl at 37°C for 1 h. Dimethyl sulfate (DMS) was added to the reaction mixture (1/200 dilution) and incubated for 15 min at room temperature. An equal volume of piperidine (10%) was added to each tube, and the reaction mixture was incubated at 90°C for 30 min. The reaction was diluted to double the volume and vacuum dried. The pellet was further washed with water thrice and dried using a speedvac concentrator. The reaction products were resolved on a 15% denaturing polyacrylamide gel, which was further dried and visualized as described earlier. For DMS on the larger fragment, the 300-bp BCL2 MBR was PCR amplified from pXW5 using primers SCR21 and SCR57 and gel purified. The fragments were heat denatured and slow annealed in the absence or presence of 100 mM KCl, and incubated at 37°C for 2 h. A 1/200 dilution of stock DMS was added to the DNA and incubated for 20 min at room temperature. The DNA was then precipitated and resuspended in 50 µl of TE buffer. An equal volume of piperidine (10%) was added to each tube and incubated at 90°C for 30 min. The DNA was again precipitated, washed with water twice and dried using speedvac concentrator. The pellet was resuspended in TE and subjected to 18 cycles of primer extension using radiolabeled MN96, with the following conditions, denaturation 94°C for 45 s, annealing 58°C for 45 s and extension 72°C for 25 s. The reaction products were mixed with formamide dye and resolved on a 10% denaturing PAGE.
Circular dichroism
The MBR wild-type, mutant and HIF-1α G-rich oligomers were incubated either in the presence or absence of 100 mM KCl, in TE at 37°C for 1 h. The circular dichroism (CD) spectra were recorded at room temperature from 220 to 300 nm and 10 cycles were accumulated for every sample, using a JASCO J-810 spectropolarimeter at a scan speed of 50 nm/min. Separate spectrum was measured for the buffer alone for 30 cycles and this was subtracted from all the experimental spectra. For the destruction of the structure, the spectra were recorded for the samples incubated with KCl at different temperatures. Furthermore, CD spectrum was recorded after the structure was reformed by incubating the same sample overnight at 37°C. The ellipticity was calculated using the software, Spectra Manager, and plotted as a function of wavelength.
NMR spectroscopy
The oligomers, MN77 (GS) and MN79 (G2 loop) were synthesized on 1 µM scale in multiples and gel purified. A total of 1 mM DNA was dissolved in TE in the presence of 100 mM KCl in a total volume of 540 μl along with 60 μl of D2O (9 : 1). The spectra were recorded on a Bruker Avance 800 MHz spectrometer with cryoprobe and TOPSPIN 2.0 software. 1D proton spectra were recorded at 5, 25, 40 or 50°C with 1024 scans each and water suppression was done using excitation sculpting. The chemical shifts were analyzed by external referencing with the help of DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) in D2O. 2D NOESY (with excitation sculpting) was performed for MN77 at 5°C with a mixing time of 250 ms on the same probe with 512 t1 increments of 96 scans each.
Model building
Intramolecularly folded parallel G-quadruplex was built using the graphics interface of Insight II (21) on an O2 SGI workstation. Loops were modeled on to the G-tetrad using the Biopolymer module within Insight II. The modeled structure was then energy minimized using Sander Module of Amber 6.0 (22). Figures were prepared using PyMOL (http://www.pymol.org).
Taq polymerase stop assay
The oligomers, MN115, MN116, MN117, MN118 or the plasmids, pXW5 and pSCR41, were heat denatured and slowly reannealed in the absence or presence of KCl or LiCl, in a buffer containing 10 mM Tris–HCl and 1.5 mM MgCl2 and radiolabeled MN96, till it reached room temperature. After addition of 200 μM dNTPs and 1 U Taq polymerase, the reactions were incubated at 47°C for 30 min. The reaction was stopped by addition of dye containing formamide and the products were resolved on 12% (for oligomeric DNA) and 8% (for plasmid DNA) denaturing PAGE and visualized as described.
Test for altered DNA structure (G-quadruplex) within cells
293T human embryonic kidney epithelial cells were transfected with pMN11 and pMN12 by calcium phosphate precipitation along with pCMV-β gal (a kind gift from Prof. K.N. Balaji, IISc, Bangalore). The number of GFP positive cells in each case was analyzed by flow cytometry and the transfection efficiency was calculated by counting X-gal stained cells (see below). The percentage GFP expression was normalized with that of the β gal expressing cells for both wild-type and mutant DNA. The data was analyzed using Graph Pad Prism software and plotted as bar diagram with standard error of mean. The experiments were repeated at least three independent times with five replicates each.
X-gal staining
The transfected cells were harvested and washed once in PBS. The cells were then incubated in PBS with 0.2% glutaraldehyde at 37°C for 15 min. The cells were washed twice with PBS containing 2 mM MgCl2 and 0.02% NP40. Finally, the cells were incubated with PBS containing 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl2 and 1 mg/ml X-gal, overnight at 37°C. The X-gal stained cells were counted and the transfection efficiency was calculated.
RESULTS
The G-rich template strand at peak I of the MBR can form a G-quadruplex structure
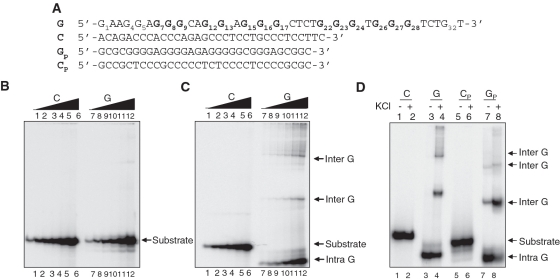
Previously, we have shown that the region upstream of peak I of the BCL2 MBR possessed bisulfite reactivity and hence a non-B DNA structure (13). Since the complementary region was rich in stretches of guanines, we wondered whether it could contribute to the formation of a G-quadruplex structure. In order to test this hypothesis, we designed an oligomeric DNA corresponding to the region of interest, representing either the sense (non-template) or antisense strand (template), referred to as C and G, respectively (Figure 1A). Increasing concentrations (5, 12.5, 25, 50, 125 and 250 nM) of [γ-32P] labeled oligomers were incubated in TE buffer (1 h, 37°C) both in the presence and absence of K+, which is known to stabilize the formation of G-quadruplexes. Products were resolved on native polyacrylamide gels both with and without KCl in the gel and buffer (Figure 1B and C). Results showed that both C and G substrates migrated according to the molecular weight, when no KCl was added in the reaction and gel (Figure 1B, lanes 1–6 and 7–12). Interestingly, when the same substrates were incubated in the presence of KCl and then electrophoresed, we observed G-rich strands migrating much faster than the C-rich strands (Figure 1C, compare lanes 7–12 to 1–6). The faster moving species could be due to formation of compact intramolecular G-quadruplexes (Figure 1C, lanes 7–12) (23–26). Apart from these, we also observe a number of additional higher molecular weight bands, which could be due to the formation of intermolecular G-quadruplexes (Figure 1C, lanes 7–12).
Figure 1.
Formation of G-quadruplex structure in the template strand of the BCL2 MBR. (A) The oligomeric sequence of the G-rich strand (indicated by ‘G’), and the complementary C-rich strand (indicated by ‘C’) are derived from the upstream region of peak I of the BCL2 MBR. GP and CP are G-rich strand and the complementary C-rich strand from the HIF-1α promoter region, which is used as a control. The stretches of guanines are in bold and have been numbered for BCL2 MBR oligomer. (B and C) Increasing concentrations (5, 12.5, 25, 50, 125 and 250 nM) of the G and C-rich strands were resolved on 15% native polyacrylamide gels in the absence (B) or presence (C) of 100 mM KCl. In case of (C), the concentration of KCl in the reactions, gel and buffer was 100 mM. (D) The G and C-rich strands from both the BCL2 MBR and HIF-1α promoter region were incubated in the absence or presence of KCl (100 mM) and resolved in the presence of KCl (100 mM). In (B–D), the substrate, intramolecular (denoted as intra G) and intermolecular (denoted as inter G) quadruplex structures are shown by arrows.
The HIF-1α promoter region has been previously shown to form G-quadruplex structures (23). We synthesized the G-rich (GP) and its complementary sequence (CP) from this region and subjected it to native gel electrophoresis in the presence of 100 mM KCl, as a positive control. Results showed that a major fraction of both the GP and G (as before) strands migrated much faster (intramolecular G-quadruplexes) compared to their respective complementary strands in presence of 100 mM KCl (Figure 1D). However, in the case of HIF-1α promoter region, interestingly the intermolecular species were observed even in the absence of KCl in the reaction, unlike the G-strand of MBR (Figure 1D, lanes 3, 4 and 7, 8).
Furthermore, shortened BCL2 MBR sequences (obtained by deletion of bases from the 5′- and 3′-ends), Gs and Cs, were designed to test their potential to fold into G-quadruplexes (Supplementary Figure S1A). Gel mobility shift assays showed the formation of intramolecular G-quadruplex on a native gel, similar to G, in the presence of KCl (Supplementary Figure S1B and C, lanes 3, 4, 7, 8). As expected, C and Cs strands did not show any shift in the mobility (Supplementary Figure S1B and C). We also performed similar reactions in the presence of 100 mM LiCl, which is known to lower the stability of G-quadruplex structures (24,27–30). As expected, we could not find the intramolecular G-quadruplex in the presence of Li+ ions in case of Gs strand (Supplementary Figure S1D and E, lanes 3, 4). Altogether, these results indicate the propensity of the BCL2 MBR template strand to form an intramolecular G-quadruplex, stabilized by K+ ions.
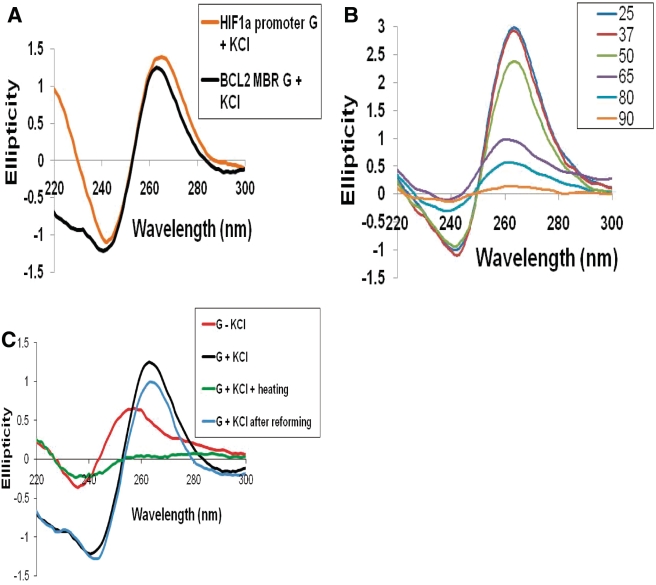
To further verify the above observations, CD was performed. The CD spectra vary depending on the specific conformation of the quadruplex. Parallel G-quadruplexes have been shown to have a peak at ∼260–265 nm and a trough at 240 nm whereas anti-parallel quadruplexes exhibit a peak at 290 nm and a dip at 260 nm (31–36). When CD spectra was acquired for the BCL2 MBR G-strand in the presence of K+, a characteristic peak at ∼263–265 nm and dip at 240 nm was observed, indicating that the G-quadruplex structure at MBR is parallel (Figure 2A). A similar pattern for the CD spectra obtained for HIF-1α (positive control) further confirms the presence of a parallel G quadruplex (Figure 2A). Furthermore, we recorded the CD spectra of the DNA in the presence of KCl, at increasing temperatures to determine whether the structure gets destroyed at higher temperature. We observed that at temperatures above 37°C, the spectrum began to slowly collapse and became more parallel to the X-axis at 90°C, indicating the destruction of the quadruplex structure (Figure 2B). More importantly, incubation of the heat denatured (90°C) sample at 37°C, overnight, restored the structure (Figure 2C). These findings further indicate that the template strand upstream of peak I in the BCL2 MBR can fold into an intramolecularly folded parallel G-quadruplex.
Figure 2.
CD studies on the MBR show formation of parallel, G-quadruplex structure on the template strand. (A) The BCL2 MBR and HIF-1α promoter G-strand (positive control) were incubated in presence of 100 mM KCl and TE at 37°C for 1 h and the CD spectra was taken using JASCO J-810 spectropolarimeter with a scan range of 220–300 nm. (B) CD spectra for the MBR template strand in TE with KCl at different temperatures. The various temperatures used are 25, 37, 50, 65, 80 and 90°C. (C) CD spectra for the MBR template strand in TE with (G + KCl) or without KCl (G − KCl), or same after heating to 90°C (G + KCl + heating) and then incubation at 37°C overnight (G + KCl after reforming), in order to reform the structure.
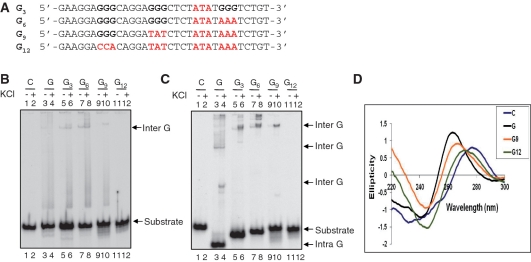
Mutation at peak I of the BCL2 MBR abolishes the intramolecular G-quadruplex
In order to test the role of the four stretches of three guanines in the G-quadruplex formation at BCL2 MBR template strand, we designed oligomers containing mutations in one (G3), two (G6), three (G9) and four (G12) stretches of guanines (Figure 3A). The [γ-32P] labeled oligomers were incubated in the presence of KCl and electrophoresed as described. In the absence of K+, all the oligomeric sequences migrated at the same position except for a very slowly migrating band at the top for G, G3 and G6 (Figure 3B, marked as Inter G). However, in the presence of KCl, we observed the formation of the faster migrating intramolecular G-quadruplex, in the case of G (Figure 3C, lanes 3 and 4), whereas none was observed for C or mutant sequences (Figure 3C, lanes 1, 2 and 5–12). As expected, the absence of even one stretch of guanines (G3) prevented the formation of intramolecular G-quadruplexes (Figure 3C, lanes 5, 6). In contrast, all of the sequences except C and G12, could form intermolecular G-quadruplex species (Figure 3C, lanes 3–10). In addition, CD studies were also carried out on all of the mutants and results showed that in case of G12, the spectra resembled that of the C strand and the characteristic peak at ∼265 nm was absent (Figure 3D). Interestingly, the pattern for G9 was similar to that of G, but the peak intensity at 265 nm was low and shifted towards the right (Figure 3D). This may be due to the fact that as the number of available guanines decreases, the structure preference changes from the intra to the intermolecular form (Figure 3D). Thus our results suggest a clear dependence of the intramolecular G-quadruplex at the template strand of the MBR on the four stretches of guanine nucleotides.
Figure 3.
Mutations in the G-quadruplex forming sequence of BCL2 MBR template strand abolishes structure formation. (A) Mutations in the G-rich strand were generated such that three (indicated as G3), six (G6), nine (G9) and 12 (G12) Gs from the 3′-end were altered to other bases (changed bases are in red color). (B and C) Different oligomeric DNA were incubated either in the presence or absence of KCl and resolved on a 15% native PAGE either in the absence (B) or presence (C) of KCl in the electrophoretic conditions. (D) CD spectra showing the effect of mutations on G-quadruplex formation. Oligomers depicted in panel A (Figure 1) were incubated with 100 mM KCl and TE at 37°C and the CD spectra were recorded as described earlier. For other details refer Figure 2 legend.
The BCL2 MBR sequence folds into a 3G plate G-quadruplex
A DMS protection assay is used for determining the guanines involved in formation of G-quadruplex structures. DMS methylates the guanine residues present in single-stranded and duplex DNA, at the N7 position. However, if the guanines are involved in Hoogsteen base pairing, which is required for G-quadruplex formation, the N7 position is not free for methylation and therefore, would not react with DMS (18). We performed the protection assay with G, G3 and G6 strands, in the presence and absence of KCl (Figure 4A). We observed that in the case of G-strand, except for G12 and G13, all of the other guanines were involved in the G-quadruplex formation, when KCl was present (Figure 4A, lane 3), while all guanines showed equal reactivity when KCl was not added (Figure 4A, lane 2). Low level DMS protection was observed for guanines at positions G7, G8, G9 and G12, G13 and G15, G16, G17 in the case of the mutants. This suggests the formation of intermolecular G-quadruplex by the mutants (Figure 4A, lanes 4–7).
Figure 4.
A DMS protection assay on BCL2 G-quadruplex structure. (A) G-strand (G), 3-nt (G3) and 6-nt (G6) mutants were treated with DMS in the presence of 100 mM KCl, cleaved by piperidine and resolved on 15% denaturing gel. Lane 1, G-strand treated with piperidine alone. Lanes 2, 4 and 6, G, G3 and G6, respectively incubated in TE without KCl, treated with DMS and piperidine. Lanes 3, 5 and 7, G, G3 and G6, respectively treated with DMS and piperidine after incubating with 100 mM KCl. (B) The substrate DNA and intramolecular G-quadruplex (in duplicates), were gel eluted, treated with DMS, cleaved by piperidine and resolved on a 15% denaturing PAGE. The positions of guanines are marked. (C) Representative 2D model of the intramolecular G quadruplex structure formed at the BCL2 MBR. The position of guanines and other nucleotides involved are marked. Arrows indicate the orientation of the strands.
Furthermore, the intramolecular G-quadruplex species and substrate DNA were gel purified and subjected to the DMS modification assay. Results showed that all the guanines were modified and hence cleaved by piperidine in the substrate (Figure 4B, lanes 2, 3). However, in case of the purified intramolecular G-quadruplexes, except for G4, G5, G12 and G13, all of the other guanines showed protection from modification by DMS, suggesting that all of these were involved in the structure formation (Figure 4B, lanes 4, 5). Therefore, the DMS modification assay reinforces that the template strand at the BCL2 MBR has the ability to form an intramolecular G-quadruplex (Figure 4C).
NMR spectroscopy confirms the formation of an intramolecular G-quadruplex at the BCL2 MBR template strand
1H NMR spectroscopy was performed on the shorter wild-type G-strand (GS, Supplementary Figure S1A). NMR spectrum of Gs showed chemical shifts between 10.8 and 12.2 ppm, which resulted from the resonance of imino hydrogen groups characteristic of G-quadruplex (Figure 5A) (37–40). Broad peaks ∼9.5–10.4 ppm were also seen, which arise from the amino protons in the G-tetrads. The peak count shows that this sequence indeed folds into an intramolecular G-quadruplex and that the structure is asymmetrical. Each peak in the region 10.8–12.2 ppm represents one imino proton and for an asymmetrical quadruplex with three G-tetrads, one should expect 12 peaks. However, the presence of more than 12 peaks suggests that more than one quadruplex conformation may be formed among the intramolecular species, where the interconversion rate is comparable to the NMR time scale, as has been reported before (41). This was further supported by the 2D NOESY spectrum, which was unable to resolve the detailed structure (Supplementary Figure S3). An interesting observation was the presence of two isolated peaks at 13 and 14.5 ppm (Figure 5A), which correspond to the G … C and A…T Watson–Crick hydrogen bonding scheme (42). These hydrogen bonds can be envisaged in the loop region of the G-quadruplex structure between A14 and T21 and G33 and C18. Temperature gradient studies on Gs strand showed the persistence of the imino peaks at the tested temperatures (Figure 5B). Interestingly, the Watson–Crick pairing was absent at 40 and 50°C (Figure 5B).
Figure 5.
1H NMR spectroscopy on the G-quadruplex at BCL2 MBR. (A) NMR spectrum of the wild-type sequence (23 mer, GS) at 5°C. The spectrum was recorded on a Bruker Avance 800 MHz spectrometer with a cryo probe and analyzed by TOPSPIN 2.0 software. (B) NMR spectra recorded at different temperatures for GS. (C) NMR spectrum of the mutant with a 2-nt loop (G2 loop) at 5°C. In all the panels A–C, the resonance peaks between 10.8 and 12.2 ppm represent those resulting from the imino hydrogen present in the G-quadruplex structure. The resonance peaks at ∼13 and 14.5 ppm result from A…T and G…C Watson–Crick base pairing.
NMR studies were also carried out on another sequence containing a 2 nt loop (G2 loop) instead of the one nucleotide loop discussed above, and it showed the formation of an intramolecular G-quadruplex similar to the wild-type on native PAGE (data not shown). However, the 1H NMR spectrum (Figure 5C) for this sequence was much more resolved than that of the wild-type sequence, perhaps indicating a more stable conformation. Here also the NMR spectrum showed the presence of Watson–Crick base pairs. Hence, NMR results point towards the formation of an intramolecularly folded G-quadruplex.
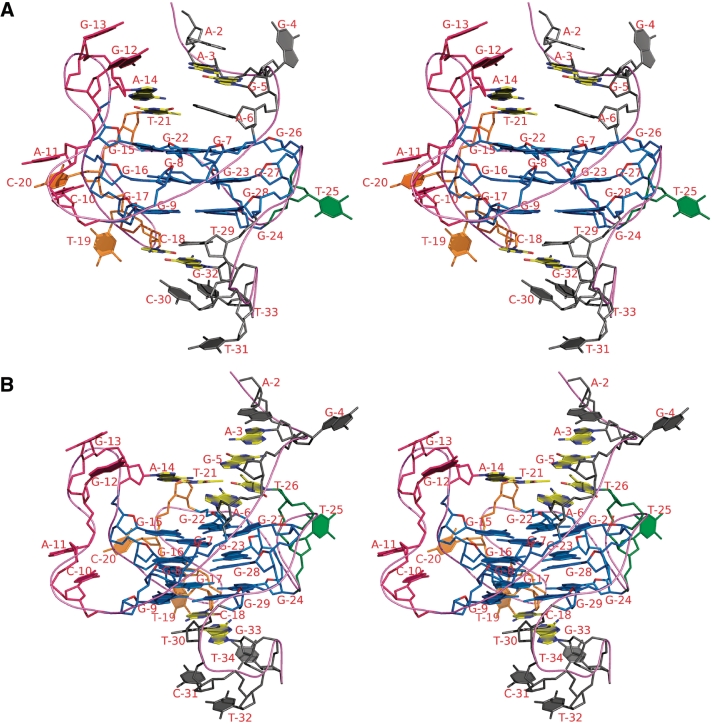
3D model of the G-quadruplex structure at the BCL2 MBR template strand
Taking into consideration, all the experimental data discussed above and the possible multiple conformations of the intramolecular G-quadruplex at the BCL2 MBR, one representative model was built for both the wild-type (Gs) and mutant (G2 loop) sequence (Figure 6A and B). The model was built using an Insight II (21) software package on an O2 SGI workstation. Energy-minimization was performed using Sander Module of Amber 6.0 (22). The proposed intramolecular G-quadruplex structure comprises three G-tetrads, G7*G15*G22*G26, G8*G16*G23*G27 and G9*G17*G24*G28 in the wild-type (Figure 6A) and G7*G15*G22*G27, G8*G16*G23*G28 and G9*G17*G24*G29 in the mutant (Figure 6B). Guanines in the G-tetrad favor anti glycosyl conformations except G15 (syn) of the second strand. The G-tetrads are interconnected by three loops. The 5-nt loop (C10, A11, G12, G13 and A14) connects strand 1 with strand 2 and the 4-nt (C18T19C20T21) loop connects second and third strand. The 1-nt (T25)/2-nt (T25, T26) loop connects third and fourth strand. Loop…loop interactions favor Watson–Crick pairing between T21 (4-nt loop) and A14 (5-nt loop) and C18 (4-nt loop) with G33 of the 3′-overhang, consistent with NMR observations. The two pairs are located above and below the G-tetrad respectively. Partial stacking involves C10 and A11 and G12 and G13 of the 5-nt loop. An extended phosphodiester conformation between A11 and G12 is necessitated to reach the 5′-end of the second strand. T19 and C20 of 4-nt loop protrude out.
Figure 6.
Proposed 3D model of a representative G-quadruplex structure adopted by the BCL2 MBR G-rich template strand. Stereo view of the proposed intramolecular G-quadruplex for wild-type (A) with 1-nt loop and mutant (B) with 2-nt loop. G-tetrad core is colored blue. Loops comprising 5, 4, and 1 or 2 nt are colored, pink, orange and green, respectively. Bases involved in WC paring are colored yellow (carbon atoms). The model was built using Insight II software package on an O2 SGI workstation (21). The model was energy-minimized using Sander module of Amber 6.0 (22).
A single nucleotide in the 1-nt loop (T25) allows the third strand to fold back to facilitate G26 to pair with G7 (strand 1) and G22 (strand 3) to form the first G-plate. In the mutant with 2 nt in the loop (2-nt), T26 forms a Watson–Crick pair with A6 of the 5′-overhang to lie above the G-plate. Interestingly, it is possible to visualize G5 … A3 (of the 5′-overhang) pairing. A2 partially stacks on this. These together with loop … loop Watson–Crick pairing may contribute to the overall stability of the structure. It may be for these reasons that the 2-nt mutant structure is more stable and displays a better resolved NMR spectrum.
Taq polymerase arrest and DMS protection at the G-quadruplex forming region at BCL2 MBR
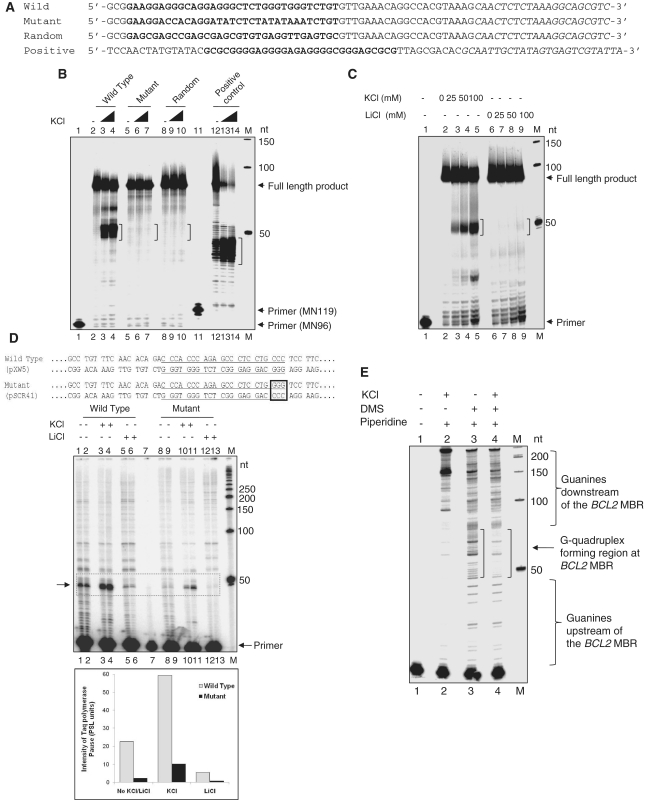
G-quadruplexes on a template strand have been shown previously to arrest progression of DNA polymerases (23,26,43). We performed a Taq DNA polymerase stop assay on an oligomeric single-stranded DNA containing the BCL2 MBR G-quadruplex forming sequence (Figure 7A). We found a major pause site in the wild-type sequence at ∼50 nt, corresponding to the G-quadruplex forming region upstream of peak I, and the pause was dependent on K+ ions (Figure 7B, lanes 2–4). Consistent with the above data, the mutant sequence did not cause any arrest in the polymerase progression (Figure 7B, lanes 5–7). More importantly, no polymerase stop was detected when a random sequence designed with an identical GC content was used for the study (Figure 7B, lanes 8–10). We have also used the G-quadruplex forming region from the HIF1α promoter as a positive control for the polymerase stop assay (Figure 7B, lanes 11–14) (23). In order to further confirm whether the observed polymerase stop was indeed due to formation of a G-quadruplex at the BCL2 MBR, the assay was also performed in the presence of increasing concentrations of KCl and LiCl. The results showed a concentration dependent increase in the intensity of the pause site in the presence of K+ ions (Figure 7C, lanes 2–5). Interestingly, when increasing concentrations of LiCl were used instead of KCl, the pause sites were absent (Figure 7C, lanes 6–9), suggesting that these were due to the formation of the G-quadruplex.
Figure 7.
G-quadruplex formation at the BCL2 MBR. (A) Oligomeric DNA containing the BCL2 MBR G-quadruplex forming sequence, MBR mutant G-strand sequence, random sequence with identical GC content and positive control sequence from HIF1α promoter region. The quadruplex forming sequence is marked in bold and the primer sequence is italicized. (B) Taq DNA polymerase stop assay on the single-stranded DNA shown in (A) in the absence of K+ ions (lanes 2, 5, 8, 12) and presence of 50 mM KCl (lanes 3, 6, 9, 13) and 100 mM KCl (lanes 4, 7, 10, 14). (C) Polymerase stop assay on the MBR wild-type G-strand in presence of increasing concentrations of KCl (lanes 2–5) and LiCl (lanes 6–9), as indicated on top of the lanes. (D) Taq DNA polymerase stop assay on plasmids containing wild-type (pXW5) and a 3-nt mutant (pSCR41) BCL2 MBR G-quadruplex forming sequence (underlined). The GGG→CCC mutation is boxed. The plasmids were heat-denatured and slowly reannealed without any salt (lanes 1, 2, 8, 9), in 50 mM KCl (lanes 3,4, 10, 11) and in 50 mM LiCl (lanes 5, 6, 12, 13). Lane 7 is the primer alone. The pause sites at the region upstream of the peak I corresponding to the G-quadruplex forming region is boxed and indicated by an arrow. Quantification of the pause sites due to the polymerase halt is shown as a bar diagram. Quantitation was done using Multi Gauge V3.0 software and the intensity is represented in PSL units. (E) A DMS protection assay on the 300-bp BCL2 MBR containing fragment. The DNA was heat denatured and reannealed in the absence (lane 3) and presence (lane 4) of 100 mM KCl and treated with DMS followed by piperidine cleavage. Lanes 1 and 2 indicate primer alone and DNA without DMS treatment, respectively. In all panels, ‘M’ is a radiolabeled 50 nt molecular weight ladder and the molecular sizes are marked.
Furthermore, G-quadruplex formation on a double-stranded plasmid DNA was studied using two plasmids, pXW5 and pSCR41, in which wild-type and mutant (3-nt mutation of one of the four tracts of three guanines) BCL2 MBR fragment were cloned, respectively. Results showed the presence of distinct polymerase pause sites in the presence of K+ ions for the wild-type DNA, whereas it was weaker by 6-fold when mutant plasmid DNA was used (Figure 7D, compare lanes 3, 4 and 10, 11). Although, bands were visible at the G-quadruplex forming region in the absence of K+ ions, the intensity was weaker (Figure 7D, lanes 1, 2), which could also be due to the K+ ions present in the Taq polymerase storage buffer. As observed above, in presence of Li+ ions, the pause sites were reduced by ∼11-fold, compared to buffer containing K+ ions (Figure 7D, lanes 5, 6). Thus the polymerase stop assay shows that the G-quadruplex formed at the template strand of the BCL2 MBR can act as a DNA polymerization block.
In order to further confirm the formation of the G-quadruplex at the region upstream of peak I, in the context of the whole MBR, we performed a DMS protection assay on a 300-bp MBR fragment derived from a plasmid, pXW5. Results showed a complete protection of the guanines when the DNA was incubated in the presence of 100 mM KCl as compared to buffer without KCl (Figure 7E, lanes 3 and 4). All of the other guanines both upstream and downstream of the MBR were not affected by the DMS modification followed by piperidine cleavage. Overall, the above studies suggest the formation of a G-quadruplex at the region upstream of peak I in the context of the entire BCL2 MBR.
Furthermore, we also tested whether RAGs could cleave the G-quadruplex forming region at peak I of the BCL2 MBR. Upon RAG cleavage on the wild-type plasmid (pXW5), we observed the presence of distinct bands at 40–50 nt, which is upstream of peak I corresponding to the G-quadruplex forming region on the template strand (Supplementary Figure S4A, lanes 1, 2). RAG cleavage was also observed in the MBR at peak II and peak III regions of the template strand, consistent with previous observations (13). Interestingly, the RAG induced cleavage at the G-quadruplex forming region produced multiple bands as opposed to single bands for peak II and peak III. This could be attributed to RAG nicking at the multiple single/double-stranded DNA transitions of possible additional non-B DNA structures at those locations. RAG nicking at a broad range of single-/double-strand transitions has been previously demonstrated (14,44). However, RAG cleavage was much weaker (∼1.5-fold) at the 3-nt mutant of peak I (GGG→CCC) (Supplementary Figure S4A, lanes 1–4 and S4B). We also tested for cleavage on the non-template strand for both the wild-type and the mutant plasmids. However, we could not find any detectable cleavage (Supplementary Figure S4C and D), which was consistent with our previous results (13). We also tested the RAG induced DSB formation in both wild-type and mutant plasmids on a native gel. However, consistent with the primer extension data on the non-template strand, no bands corresponding to DSB formation were observed (Supplementary Figure S4E). Hence, RAG cleavage on the bottom strand could be a secondary event independent of structure formation and RAGs. Replication/transcription across RAG induced nick or involvement of other enzymes like AID could result in a nick on the opposite strand within the cells to induce a DSB. However, this needs to be studied further.
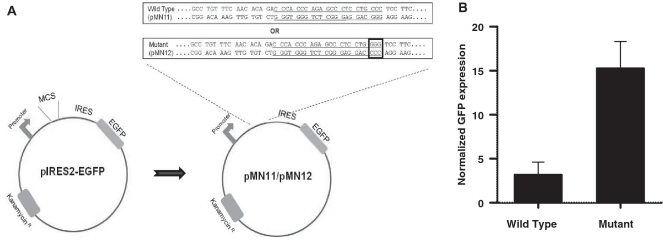
The BCL2 MBR sequence affects transcription in vivo when located upstream of a coding sequence
We examined the potential of the BCL2 MBR to fold into a non-B DNA conformation within mammalian cells by analyzing expression of a reporter gene, GFP. We cloned the wild-type and mutant (3-nt mutation of one of the four stretches of guanines) MBR containing genomic fragments upstream of the EGFP gene in the episome, pIRES2-EGFP, and used for transfecting of 293T epithelial cells (Figure 8A). pCMV-β gal was also cotransfected with the MBR constructs to determine the transfection efficiency. Upon flow cytometric analysis, we observed a significant decrease in the GFP expression in the wild-type construct compared to the mutant (Figure 8B). X-gal staining of the cells showed that there was no major difference in transfection efficiency between wild-type and mutant BCL2 MBR constructs. After normalization, the reduction in the GFP expression due to the wild-type MBR was determined to be ∼5-fold (Figure 8B). Though, this is not direct evidence for the G-quadruplex formation at the BCL2 MBR within the genome, this reporter assay implicates some forms of non-B DNA structure including a G-quadruplex at the BCL2 MBR as one plausible explanation.
Figure 8.
Non-B structure formation at the BCL2 MBR sequence within cells. (A) Plasmid constructs, pMN11 and pMN12, were prepared as described in ‘Materials and Methods’ section. The wild-type or mutant (GGG→CCC mutation in one of the G tracts, indicated in a small box) 300-bp fragments containing the MBR were placed between the EGFP gene and its promoter. If the altered structure is formed, it may interfere with transcription through the EGFP gene and thereby result in a reduced expression of the same. (B) 293T cells were cotransfected with pMN11 or pMN12 and pCMV-β gal. The percentage of GFP expressing cells was analyzed by flow cytometry and the transfection efficiency was determined by X-gal staining for the β gal expressing cells. The percentage GFP expression was normalized with the respective percentage of transfection efficiency and plotted as a bar diagram with error bars. The experiment was repeated three independent times, each containing five replicates.
DISCUSSION
The G-quadruplex structure at BCL2 MBR
In the present study, we find that the template strand of the region upstream of peak I of BCL2 MBR adopts a parallel, intramolecular G-quadruplex structure. The structure formation was dependent on potassium, disfavored by lithium and could be destroyed and recreated. One of the most interesting features of the G-quadruplex at the MBR was the presence of a single nucleotide loop. Our results showed that one nucleotide loop did not destabilize the quadruplex. Previously such G-quadruplex structures containing 1-nt loops have been described (45–50). Mutations in the guanine nucleotides adjacent to the loop completely abolished the structure, suggesting that the quadruplex formation at MBR requires the presence of at least three guanine tetrads.
Even though the gel shift assays showed the presence of a major, faster migrating species of intramolecular G-quadruplex, 1H NMR spectrum revealed that the template strand of the BCL2 MBR may adopt multiple conformations. This heterogeneity may arise due to the flexibility in the three different loops. In fact, a mutant with an extra nucleotide in the single nucleotide loop dramatically improved the resolution of the imino peaks in the 1D NMR spectra indicating that perhaps such a change favors one particular conformation. This further raises another interesting question, whether a subset of FL patients possess any mutation at the MBR. Based on our results it appears that such mutations, if present, may stabilize G-quadruplex structure formation which could make this region more susceptible to breaks induced by nucleases like the RAG complex.
One may wonder how often the BCL2 MBR could melt into a single-stranded region. Earlier studies suggested that the BCL2 MBR has a non-B DNA conformation, although the basis was not clear (13). One possible mechanism could be the random breathing of DNA, especially at G-tract rich sequences or on supercoiled DNA (51). However, it is important to point out that the conditions used in this study may not reflect an in vivo environment for DNA. During replication and transcription, DNA undergoes unwinding and thus it is possible that before reannealing some of the molecules could fold into a non-B DNA structure like a G-quadruplex. Our studies also show the ability of the BCL2 MBR to block replication in vitro as seen by the Taq polymerase stop assay. We observed that this structure can be formed within cells and can reduce the expression of a reporter gene implying that formation of a quadruplex structure can prevent the progression of RNA polymerase II and thereby lower the expression of a gene. This assay has been frequently used for demonstrating the biological relevance of such a structure formation within cells and has mostly been assessed for G-quadruplex forming sequences in the promoter regions of several genes (23,26,34,52). In our study, we have used this assay for testing the ability of the BCL2 MBR to fold into a G-quadruplex within cells. However, more detailed studies are required at the genomic level to further prove this.
Our earlier studies have also suggested formation of a triplex conformation at BCL2 MBR at peak I and its upstream sequences (17). The sequence at this region does not contain a perfect poly-purine.pyrimidine tract with mirror repeat symmetry, a feature typical of triplex formation. Because the proposed triplex was on a quasimirror repeat sequence, efforts to characterize it at the NMR level were unsuccessful. Both triplexes and G-quadruplexes share common features such as Hoogsteen hydrogen bonding and GC richness. Thus, it is possible that the BCL2 MBR may form multiple types of non-B DNA conformations.
How often are G-quadruplexes formed in the genome?
The whole genome is interspersed with sequences that have the potential to form various non-B DNA structures. Among such altered DNA structures, the estimated average incidence of quadruplex forming sequences is one per 10 000 bases (53,54). However, there is limited evidence to show that such structures exist in vivo. Telomeres are the only regions to be shown to have G-quadruplex structures in vivo. Both inter- and intramolecular structures were reported in these cases, which were shown by probing with fluorescent compounds or specific antibodies (55,56). The role of quadruplexes within the cells in regions other than telomeres is largely unclear. Recent studies have revealed that promoters of genes like BCL2, MYC, KRAS, VEGF, KIT and HIF-α may possess sequences having a potential to fold into quadruplexes. It is possible that such structures may play roles in modulating the expression of these oncogenes, both by enhancing transcription as in the case of insulin, or by repressing the expression, as for c-MYC (26,57). G-rich sequences are also present in the immunoglobulin heavy chain switch regions and ribosomal DNA (53,54,58). Many proteins have been shown to interact directly with G-quadruplexes and stabilize it, which further support the hypothesis that such structures may be formed under normal physiological conditions (59). Although the quadruplex structure observed at the MBR may require a physiological process such as replication or transcription for initial unwinding of the DNA strands in the context of genome, it is not clear whether other proteins help in stabilizing the observed structure. Further studies are required to examine this.
The roles of G-quadruplex structures in other human diseases, besides that observed in the present study, are also being investigated by various groups. CGG repeats in FMR-1 gene associated with the human neurodegenerative disease, fragile X syndrome, are shown to form a G-quadruplex structure, resulting in the expansion and silencing of the FMR-1 gene (43,60).
Connection between BCL2 MBR G-quadruplex and FL
Previously we have shown that the non-B DNA at the BCL2 MBR can be recognized by the RAG complex and result in the formation of breaks, which is a pre-requisite for the t(14;18) translocation (13,14). In the present study, we demonstrate that the template strand of this region could adopt an intramolecular G-quadruplex structure, which could be stabilized by the high concentration of K+ ions present within cells. One may wonder how often such structures are formed inside cells and whether every cell will have such structures at this location. Previously, we found that ∼25% of the human chromosomal alleles have sensitivity to bisulfite at the BCL2 MBR in B cells (14). This bisulfite sensitivity appears to reflect non-B DNA, which may be B/A intermediate DNA, transient single-strandedness or a more stable slipped DNA structure (15). G-quadraplex formation on one strand may contribute to the stability of such a slipped structure. AID action at methyl CpG sites within regions of single-stranded character may then result in T:G mismatches that are cut by the RAG complex (16). But, what makes this region so specific a target for t(14;18) translocation is still not entirely clear.
Even though the G-quadruplex formed at the template strand of BCL2 locus can be cleaved by RAGs, it may not always result in a translocation. This is because the availability of the partner chromosome could also play a role and therefore, the positioning of the chromosomes within the nucleus may be important (61).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Rapid Grant for Young Investigators scheme of Department of Biotechnology, India (102/IFD/SAN/PR-1576/2006-2007); Indian Institute of Science start up grant (to S.C.R.); CSIR, India, SRF (to M.N.); DBT, India, JRF (to B.T.M.). Funding for open access charge: Leukemia Research Foundation, USA.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Prof. N. Yathindra (IBAB, Bangalore) for his help in 3D modeling studies and comments on the article. The authors also thank M. Nishana and Sheetal Sharma for critical reading of the manuscript and members of the SCR laboratory for discussions and help.
REFERENCES
- 1.Lieber MR. Causes and consequences of chromosomal translocations. In: Kirsch I, editor. The Causes and Consequences of Chromosomal Aberrations. Boca Raton, FL: CRC Press; 1993. pp. 239–275. [Google Scholar]
- 2.Korsmeyer SJ. Chromosomal translocations in lymphoid malignancies reveal novel proto-oncogenes. Annu. Rev. Immunol. 1992;10:785–807. doi: 10.1146/annurev.iy.10.040192.004033. [DOI] [PubMed] [Google Scholar]
- 3.Bakhshi A, Jensen JP, Goldman P, Wright JJ, McBride OW, Epstein AL, Korsmeyer SJ. Cloning the chromosomal breakpoint of t(14;18) human lymphomas: clustering around JH on chromosome 14 and near a transcriptional unit on 18. Cell. 1985;41:899–906. doi: 10.1016/s0092-8674(85)80070-2. [DOI] [PubMed] [Google Scholar]
- 4.Bakhshi A, Wright JJ, Graninger W, Seto M, Owens J, Cossman J, Jensen JP, Goldman P, Korsmeyer SJ. Mechanism of the t(14;18) chromosomal translocation: structural analysis of both derivative 14 and 18 reciprocal partners. Proc. Natl Acad. Sci. USA. 1987;84:2396–2400. doi: 10.1073/pnas.84.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleary ML, Sklar J. Nucleotide sequence of a t(14;18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Proc. Natl Acad. Sci. USA. 1985;82:7439–7443. doi: 10.1073/pnas.82.21.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jager U, Bocskor S, Le T, Mitterbauer G, Bolz I, Chott A, Kneba M, Mannhalter C, Nadel B. Follicular lymphomas' BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- 7.Wyatt RT, Rudders RA, Zelenetz A, Delellis RA, Krontiris TG. BCL2 oncogene translocation is mediated by a chi-like consensus. J. Exp. Med. 1992;175:1575–1588. doi: 10.1084/jem.175.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghavan SC, Lieber MR. Chromosomal translocations and non-B DNA structures in the human genome. Cell Cycle. 2004;3:762–768. [PubMed] [Google Scholar]
- 9.Nambiar M, Kari V, Raghavan SC. Chromosomal translocations in cancer. Biochim. Biophys. Acta. 2008;1786:139–152. doi: 10.1016/j.bbcan.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Aplan PD. Causes of oncogenic chromosomal translocation. Trends Genet. 2005;22:46–55. doi: 10.1016/j.tig.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schatz DG, Oettinger MA, Schlissel MS. V(D)J recombination: molecular biology and regulation. Annu. Rev. Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 12.Marculescu R, Le T, Simon P, Jaeger U, Nadel B. V(D)J-mediated translocations in lymphoid neoplasms: a functional assessment of genomic instability by cryptic sites. J. Exp. Med. 2002;195:85–98. doi: 10.1084/jem.20011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 14.Raghavan SC, Swanson PC, Ma Y, Lieber MR. Double-strand break formation by the RAG complex at the bcl-2 major breakpoint region and at other non-B DNA structures in vitro. Mol. Cell. Biol. 2005;25:5904–5919. doi: 10.1128/MCB.25.14.5904-5919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai AG, Engelhart AE, Hatmal MM, Houston SI, Hud NV, Haworth IS, Lieber MR. Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites. J. Biol. Chem. 2009;284:7157–7164. doi: 10.1074/jbc.M806866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raghavan SC, Chastain P, Lee JS, Hegde BG, Houston S, Langen R, Hsieh CL, Haworth IS, Lieber MR. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J. Biol. Chem. 2005;280:22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- 18.Sinden RR, editor. DNA structure and function. Ist edn. San Diego, California: Academic Press; 1994. [Google Scholar]
- 19.Naik AK, Raghavan SC. P1 nuclease cleavage is dependent on length of the mismatches in DNA. DNA Repair. 2008;7:1384–1391. doi: 10.1016/j.dnarep.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Naik AK, Lieber MR, Raghavan SC. Cytosines, but not purines, determine recombination activating gene (RAG)-induced breaks on heteroduplex DNA structures: implications for genomic instability. J. Biol. Chem. 2010;285:7587–7597. doi: 10.1074/jbc.M109.089631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.InsightII2005L. 1993. Accelrys Inc, San Diego, CA, USA. [Google Scholar]
- 22.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, et al. 1999. AMBER6. University of California, San Francisco. [Google Scholar]
- 23.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 24.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 25.Oganesian L, Moon IK, Bryan TM, Jarstfer MB. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 2006;25:1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michalowski D, Chitima-Matsiga R, Held DM, Burke DH. Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res. 2008;36:7124–7135. doi: 10.1093/nar/gkn891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Töhl J, Eimer W. Interaction energies and dynamics of alkali and alkaline-earth cations in quadruplex-DNA-structures. J. Mol. Model. 1996;2:327–329. [Google Scholar]
- 29.Simonsson T. G-quadruplex DNA structures–variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 30.Sen D, Gilbert W. Guanine quartet structures. Methods Enzymol. 1992;211:191–199. doi: 10.1016/0076-6879(92)11012-8. [DOI] [PubMed] [Google Scholar]
- 31.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 32.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. Cambridge, UK: RSC Biomolecular Sciences; 2006. [Google Scholar]
- 34.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 36.Keniry MA. Quadruplex structures in nucleic acids. Biopolymers. 2000;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Feigon J, Koshlap KM, Smith FW. 1H NMR spectroscopy of DNA triplexes and quadruplexes. Methods Enzymol. 1995;261:225–255. doi: 10.1016/s0076-6879(95)61012-x. [DOI] [PubMed] [Google Scholar]
- 38.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 42.Wuthrich K. NMR of Proteins and Nucleic Acids. 1986. 1st edn. Wiley-interscience, New York, USA. [Google Scholar]
- 43.Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raghavan SC, Gu J, Swanson PC, Lieber MR. The structure-specific nicking of small heteroduplexes by the RAG complex: implications for lymphoid chromosomal translocations. DNA Repair. 2007;6:751–759. doi: 10.1016/j.dnarep.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paramasivam M, Cogoi S, Filichev VV, Bomholt N, Pedersen EB, Xodo LE. Purine twisted-intercalating nucleic acids: a new class of anti-gene molecules resistant to potassium-induced aggregation. Nucleic Acids Res. 2008;36:3494–3507. doi: 10.1093/nar/gkn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris MJ, Basu S. An unusually stable G-quadruplex within the 5'-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- 47.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Risitano A, Fox KR. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 49.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 50.Kumar N, Sahoo B, Varun KA, Maiti S. Effect of loop length variation on quadruplex-Watson Crick duplex competition. Nucleic Acids Res. 2008;36:4433–4442. doi: 10.1093/nar/gkn402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dornberger U, Leijon M, Fritzsche H. High base pair opening rates in tracts of GC base pairs. J. Biol. Chem. 1999;274:6957–6962. doi: 10.1074/jbc.274.11.6957. [DOI] [PubMed] [Google Scholar]
- 52.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36:1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang CC, Kuo IC, Ling IF, Chen CT, Chen HC, Lou PJ, Lin JJ, Chang TC. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal. Chem. 2004;76:4490–4494. doi: 10.1021/ac049510s. [DOI] [PubMed] [Google Scholar]
- 56.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 59.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 60.Sinden RR. Biological implications of the DNA structures associated with disease-causing triplet repeats. Am. J. Hum. Genet. 1999;64:346–353. doi: 10.1086/302271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat. Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.