Abstract
Telomerase activity, not detectable in somatic cells but frequently activated during carcinogenesis, confers immortality to tumors. Mechanisms governing expression of the catalytic subunit hTERT, the limiting factor for telomerase activity, still remain unclear. We previously proposed a model in which the binding of the transcription factor CTCF to the two first exons of hTERT results in transcriptional inhibition in normal cells. This inhibition is abrogated, however, by methylation of CTCF binding sites in 85% of tumors. Here, we showed that hTERT was unmethylated in testicular and ovarian tumors and in derivative cell lines. We demonstrated that CTCF and its paralogue, BORIS/CTCFL, were both present in the nucleus of the same cancer cells and bound to the first exon of hTERT in vivo. Moreover, exogenous BORIS expression in normal BORIS-negative cells was sufficient to activate hTERT transcription with an increasing number of cell passages. Thus, expression of BORIS was sufficient to allow hTERT transcription in normal cells and to counteract the inhibitory effect of CTCF in testicular and ovarian tumor cells. These results define an important contribution of BORIS to immortalization during tumorigenesis.
INTRODUCTION
Telomerase activity is one of the most important factors that has been linked to multiple developmental processes including cell proliferation, differentiation, aging and senescence (1,2). This complex enzyme stabilized telomeres length by adding hexameric repeats (TTAGGG) to telomeric ends of linear chromosomes, thus compensating for the continued erosion of telomeres (3). Maintenance of telomeres is required for cells to escape from replicative senescence and proliferate indefinitely. In adult humans, telomerase activity is not detectable in most somatic cells (4). In contrast, highly proliferative cells, such as germ cells and stem cells, and 85–95% of cancers express telomerase (5). Among the various components of human telomerase, only the telomerase RNA component, hTERC, and the human telomerase reverse transcriptase, hTERT, are essential for the reconstitution of telomerase activity in vitro (6–8). Moreover, it has been shown that ectopic expression of hTERT is sufficient to restore telomerase activity in telomerase-negative cells (8–10). Therefore, hTERT expression is defined as the rate-limiting factor in regulating telomerase activity (11), and many studies suggest that the regulation of hTERT occurs primarily at the transcriptional level.
Following the characterization of the hTERT genomic sequence and gene organization, a minimal promoter encompassing a 283 bp region upstream of the initiation ATG codon and numerous binding sites for transcription factors have been described (12–15).
The fact that the hTERT promoter lies within a CpG island has led to studies of transcriptional regulation through DNA methylation. Contradictory results have been published (16–21), but apparently hypermethylation of hTERT is required for its expression in the majority of telomerase-positive cells (18,22). We previously showed that the proximal exonic region of hTERT is involved in transcriptional inhibition of the gene (23) caused by binding of CTCF to the first two exons (24). In fact, hypermethylation of the hTERT exon 1 region in cancer cell lines and tumors prevents the binding and the repressive effects of CTCF. In addition, a specific 110 bp region within the core promoter was found to be hypomethylated in hTERT-expressing cells thereby allowing the transcriptional effect of the minimal promoter (25). A recent study also demonstrated that this region of the hTERT promoter upstream of the transcriptional start site is unmethylated and linked with active chromatin in cancer cells, thus explaining hTERT activity in the face of hypermethylation of the 5′ regulatory region (26). Moreover, Meeran et al. have reported that down-regulation of DNMTs in response to HDAC inhibition by treatment with potential cancer-prevention drug Sulforaphane (SFN) generates site-specific CpG demethylation primarily in the first exon of the hTERT gene, which, in turn, leads to the repressive recruitment of CTCF to the same sites we mapped earlier (23,24,27).
Although methylation of its promoter appears to be the most frequently observed mechanism of the hTERT gene regulation in tumor tissues and tumor cell lines (18,20,28), it is obvious that one or more other methylation-independent mechanisms exist for certain types of telomerase-positive cells (16,17,19,28–30) including testicular and ovarian cancers (19,28). Therefore, a mechanism that does not require hTERT methylation must be active in these telomerase-positive and hTERT umethylated tumors to prevent the repressive effects of CTCF.
Previous studies shown that BORIS (Brother of the Regulator of Imprinted Sites), also termed CTCFL (CTCF-like), is the mammalian paralogue of a highly conserved (31,32), multi-functional chromatin factor encoded by the candidate tumor suppressor gene, CTCF (33–36). BORIS was found to be present mainly in testicular and has first been found to be express in a mutually exclusive manner with CTCF during male germ cell development (37). It has recently been shown to be critical to normal spermatogenesis in mice, by regulation of the CTS (Cerebroside Sulfotranscferase) gene (38). In addition to its normal expression in testis, studies revealed that various tumors and cancer cell lines, also expressed BORIS (39–43). BORIS is also involved in epigenetic reprogramming, both in normal development and in tumorigenesis (34,37,44). In addition, it has been shown that conditional expression of BORIS induced expression of a series of Cancer Testis Antigens (CTA) genes, as MAGE-A1, NY-ESO-1 (40,42) and SPANX (45). Althought BORIS and CTCF were shown to bind to the same recognition sequences, data provided clear demonstration that BORIS is functionally different from CTCF (38). Therefore, BORIS could also bind the CTCF target site within hTERT exon1.
In this study, we examined the potential contribution of BORIS to hTERT transcriptional regulation in testicular and ovarian cancer cells. We first analyzed the methylation pattern of hTERT in primary tumors of these types, in the NCCIT and OVCAR-3 cell lines, and investigated the role of BORIS in the hTERT gene regulation. Our data indicate that expression of BORIS in tumor cells is permissive for the transcription of hTERT in spite of the presence of the CTCF repressor. We concluded that BORIS and CTCF have opposite effects on hTERT transcription. Moreover, we showed that expression of BORIS in normal cells is sufficient to allow hTERT transcription and to extend their lifespan in vitro, revealing an important role for BORIS in immortalization.
MATERIALS AND METHODS
Tissue samples and cell lines
Ten testicular and five ovarian tumors were analyzed. The use of human tissue samples for this study was according to the guidelines of the ethical committee of the Medical Faculty of Lausanne. The human tumor cell lines (HeLa, cervical adenocarcinoma; NCCIT, testis teratocarcinoma, OVCAR-3, ovary carcinoma) and normal BJ fibroblast cells were obtained from the ATCC and were grown in the medium recommended by the ATCC. HeLa, NCCIT and OVCAR-3 cell lines are telomerase-positive, whereas BJ cells are telomerase-negative. The Normal Human Bronchial Epithelial cell line (NHBE) was obtained from Lonza (Basel, Switzerland) and maintained in medium without Retinoic Acid as recommended by Lonza. The mouse normal fibroblast NIH3T3 cell line was used in transient transfection experiments and was maintained in DMEM supplemented with 10% FBS.
DNA methylation analysis
DNA was extracted from cultured cells using the DNeasy tissue kit (Qiagen, Hilden, Germany). After bisulfite modification (EpiTect Bisulfite, Qiagen, Hilden, Germany), methylation analysis of the hTERT promoter was done by amplification of a 224 bp fragment (−443 to −274 from the ATG translational start site) and analyzed by a methylation-sensitive single strand conformation assay (MS-SSCA) as previously described (46). For cell lines, methylation analysis of the promoter and first exon of hTERT (from −200 to +100) was also performed after amplification of bisulfite modified DNA with the primers 5′-CTACCCCTTCACCTTCCAA-3′ and 5′-GTTAGTTTTGGGGTTTTAGG-3′. Triplicate PCR products were cloned into the TOPO TA cloning kit (Invitrogen). Plasmid DNAs were extracted from bacterial clones with the QIAprep Spin Miniprep Kit (Qiagen). Each clone was analyzed by sequencing, with the M13 forward primer (5′-GTAAAACGACGGCCAG-3′) by the NIAID sequencing facility.
Plasmid construction and siRNA sequences
hTERT reporters
pTERT–297 contains the hTERT minimal promoter (23). The pTERT–297/ex1 vector contains the hTERT minimal promoter and 80 bp of the first exon. To generate this vector, an hTERT fragment was generated by PCR and cloned into the pGL3 basic vector (Promega). The pTERT-297/ex1mut contains a mutated version of the CTCF binding site located in exon1 (24). All constructs were used in transient transfection experiments.
BORIS and CTCF expression vectors
In the pCMV-BORIS and pCMV-CTCF vectors, BORIS cDNA and CTCF cDNA were cloned in pCMV6-XL4 by ORIGENE Technologies (Rockville, MD, USA). A pBIG-BORIS vector was created on a template containing the tetracycline-responsive, autoregulated, bidirectional expression vector pBIG2i (42). The original plasmid was used as a control. A pBORIS-puro vector contained the cDNA of BORIS under control of the CMV promoter and contains the puromycin gene as a selective marker. The empty vector was used as a negative control in transfection experiments.
siRNA BORIS and CTCF sequences
siRNAs were ordered from Sigma Life Science. An siRNA sequence targeting BORIS was: 5′-CGAGUUGAUGCCGGAAAAA[dT]-3′ and a siRNA sequence targeting CTCF was: 5′-UUGGUUCGGCAUCGUCGUU[dT]-3′.
Transient transfection and luciferase assays
Cells were seeded at a concentration of 2 × 105/3.8 cm2 for NIH3T3, HeLa, NCCIT and OVCAR-3, and 5 × 104 cells/3.8 cm2 for BJ and cultured overnight. Transient transfections of reporter plasmids (0.75 μg/well) and expression vector (1 μg/well) were carried out in triplicate using JetPEI Cationic Polymer Transfection reagent (8 μl/well) (Polyplus-transfection, Illkirch, France). All experiments were performed at least twice. The pRL-tk vector (0.25 μg/well) (Promega, Madison, WI) was co-transfected as an internal control for transfection efficiency in transient transfection experiments. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI).
NHBE cells were seeded at 3.5 × 102 cells/cm2 in 60 mm dishes. The day after, cells were transfected using FUGENE6® 3 µl for 2 µg of DNA (Roche, Rotkreuz, Switzerland) In co-transfection experiments, a reporter:expression vector ratio of 1:1 was used. Expression of pBIG-BORIS was induced by two different concentrations of doxycycline: a low concentration of 0.0625 µg/ml and a high concentration of 1µg/ml.
Co-transfection of expression plasmids and siRNA was performed in triplicate in six-well-plates for OVCAR-3 and NCCIT cell lines using the jetPRIMETM transfection reagent (Polyplus-transfection, Illkirch, France) following the manufacturer’s instructions.
Immunfluorescence
HeLa, NCCIT and OVCAR-3 cells were grown for one day on glass slides in a four-well tissue culture chamber. Cells were washed in 1× PBS and fixed 10 min in 4% paraformaldehyde/1× PBS solution, freshly prepared. After two washes with 1× PBS, cells were permeabilized for 15 min in freshly prepared 1× PBS/0.2% Triton solution. Slides were washed in 1× PBS and then cells were blocked in a 1× PBS/10% goat serum for 90 min. Rabbit anti-BORIS and mouse anti-CTCF antibodies (produced by D.L. et al.) were diluted at 1/200 and 1/400, respectively, in antibody diluent solution (DakoCytomation, Carpinteria, CA) with 0.5 M NaCl. Slides were incubated at 4°C overnight. The next day, cells were washed three times in 1× PBS/1% milk/0.5% TritonX-100. Secondary antibodies including FITC-anti-rabbit and Cy3-anti-mouse (Sigma-Aldrich, St. Louis, MO) were added at a 1/100 diloution in 1× PBS and incubated for 1 h at RT in the dark. Cells were then washed three times in 1× PBS/1% milk/0.5% TritonX-100 followed by three washes in 1× PBS. Slides were mounted in 1× PBS/20% glycerol. Images were analyzed on a confocal microscope (Nikon, Chiyoda-ku, Tokyo).
RNA extraction and RT–PCR
RNA was extracted from cells using the TRIzol Reagent (Invitrogen, Basel, Switzerland). Platinum quantitative RT–PCR Thermoscript one-step system (Invitrogen) was used to amplify hTERT mRNA as previously described (24). RT–PCR products were analyzed on 2% agarose gels.
BORIS expression was screened with the primers RT-BORIS-FW 5′-AAGCCGCGAACGGAGACGAAG-3′ and RT-BORIS-REV 5′-ACGCCTTCATCCACTTCCTCTTT-3′. CTCF expression was screened with the primers RT-CTCF-FW 5′-GTGGCGCGGAGAATGATTAC-3′ and RT-CTCF-REV 5′-TCCACAATGGCTTCGACTGC-3′. RT–PCR products were analyzed on 2% agarose gels.
For quantitative RT–PCR, RNA was converted to cDNA using random primers and superscript III reverse transcriptase (Invitrogen). Quantitative real-time RT–PCR (qPCR) analysis was performed using the TaqMan Universal PCR master Mix (Applied Biosystems, Foster city, CA) and using the Applied Biosystem 7900HT Real-Time PCR system. CTCF expression was determined using the following primers and probe: 5′-TGACACAGTCATAGCCCGAAAA-3′ (FW), 5′-TGCCTTGCTCAATATAGGAATGC-3′ (REV) and 6FAM-TGATTTGGGTGTCCACTTGCGAAAGC-MGB (probe). Human Glyceraldehyde-3-phosphate dehydrogenase (hGAPDH), human BORIS (hBORIS) and human Telomerase Reverse transcriptase (hTERT) primers/probe mixtures were purchased as pre-developed assays (Applied Biosystems, Foster city, CA).
Chromatin immunoprecipitation assay
NCCIT, OVCAR-3 and HeLa cells were used for chromatin immunoprecipitation (ChIP) assays to show the in vivo binding of CTCF and BORIS on hTERT exon1. We used a ChIP Assay kit (Upstate, Charlotteville, VA, USA) and followed the manufacturers’ recommendations. One ChIP reaction used 10 µg anti-CTCF monoclonal antibodies previously described (32) or 10 µg of anti-BORIS polyclonal antibody (Abcam, Cambridge, UK). Immunopurified DNAs were used in qPCR using SYBR green mix and the following specific primers for hTERT exon1: FW 5′-GCGGCGCGAGTTTCAG-3′ and REV 5′-GCAGCACCTCGCGGTAGT-3′. The human Nmyc CTCF binding site was used as positive control for CTCF binding using the following primers: FW 5′-GGCTCTGTGAGGAGGCAAGGTG-3′REV and 5′-GCTCTCTATTTGGAGTGGCGGG-3′. Primers used to co-amplified immunoprecipitated DNA of hTERT exon1 and H19 after ChIP were previously described (25).
RESULTS
Testicular and ovarian tumor cells exhibited the same methylation profile as normal cells
Using the MS-SSCA approach, we analyzed methylation of 20 CpGs within the hTERT 5′ regulatory region (between −443 and −274) in primary tumor samples obtained from five testis and five ovaries (Figure 1A). Analyses of the hTERT promoter showed an unmethylated pattern except for one ovarian tumor that had a hypermethylated hTERT gene (Figure 1A). Methylation analysis of the testicular (NCCIT) and the ovarian (OVCAR-3) tumor cell lines identified an unmethylated hTERT 5′ regulatory region in contrast with the hypermethylation observed in the telomerase-positive HeLa cervical tumor cell line (Figure 1A). RT–PCR analyses showed that hTERT was expressed in all of the primary tumors and cell lines regardless of the methylation profile (Figure 1B). The methylation status of the hTERT CpG island in NCCIT and OVCAR-3 cell lines was examined in greater detail around the ATG within sequences previously shown to be involved in hTERT regulation (25). Figure 1C presents data from representative clones of the HeLa and BJ cell lines as reference methylation profiles from a cancer cell line and a normal cell line, respectively (24,46). Sequencing of bisulfite-modified DNA from NCCIT and OVCAR-3 revealed hypomethylation of these regions at levels close to that observed in the normal cell line BJ, with, respectively, 17% and 0% of methylation within the region B containing the CTCF binding site. This result contrasted with data we reported previously for the HeLa cell line. In NCCIT and OVCAR-3, the mechanism preventing CTCF binding to the first exon of hTERT is thus independent of DNA methylation and had to involve another mechanism.
Figure 1.
Expression and methylation patterns of hTERT in human tumors and cell lines. (A) Schematic representation of hTERT promoter sequence: +1 represents the translational start site, the black bar represents the region of promoter analysed by MS-SSCA. Controls obtained from plasmids containing hTERT sequences that are either unmethylated (0%), or 1:1 mixture of unmethylated and fully methylated (50%), or fully methylated (100%). The cell lines are: HeLa, cervical adenocarcinoma; NCCIT, testicular teratocarcinoma and OVCAR-3, ovarian carcinoma. (B) hTERT mRNA was detected by RT–PCR, with β-actin as internal control. (C) Genomic bisulfite sequencing of hTERT promoter and proximal exonic region (–200 to +100 nt bases around the ATG transcriptional start site). After PCR amplification and cloning, 10 clones of NCCIT and OVCAR-3 were analyzed. As a comparison, five clones representing the methylation in HeLa cell line and 8 clones representing the methylation in BJ normal cell line are shown (25). Each square represents one CpG site. Methylated CpG sites are indicated in black and unmethylated in white. On top of the bar is displaying the entire sequence of the region B, in which CGs are underlined.
The repressor effect of the proximal exonic region of the hTERT gene is less efficient in testicular and ovarian tumor cell lines than in HeLa cells
The inhibitory effect of the first exon of hTERT on its minimal promoter was tested in NCCIT and OVCAR-3 and compared to HeLa and BJ fibroblasts. These four cell lines were transiently transfected with the pTERT-297 and the pTERT-297/ex1 luciferase reporter vectors containing, respectively, the hTERT minimal promoter alone or with hTERT exon 1 (Figure 2A). The transcriptional activity of the hTERT minimal promoter was different in these four cell lines, ranging from 16.5 to 88% of relative luciferase activity in BJ and HeLa cells respectively. Transfection of the pTERT-297/ex1 construct resulted in a 6-fold diminution of luciferase activity in HeLa cells. Interestingly, this reduction was only 3-fold and 2.3-fold in NCCIT and OVCAR-3, respectively (Figure 2B). It is interesting to note that in BJ, the low activity of the reporter vector containing the hTERT minimal promoter was completely inhibited by the addition of the first exon containing the CTCF binding site. Therefore, in NCCIT and OVCAR-3 cell lines, the inhibitory effect of CTCF on hTERT minimal promoter transcription was less efficient than in HeLa cells, showing that cellular representation of transcription factors were distinct and might differentially influence hTERT expression.
Figure 2.
Transcriptional activity of the hTERT minimal promoter with or without the proximal exonic region. (A) Schematic representation of the luciferase reporter plasmids pTERT-297 and pTERT-297/ex1. (B) Luciferase reporter plasmids containing the hTERT minimal promoter, without (pTERT-297) or with (pTERT-297/ex1) the proximal exonic region, are transfected into HeLa, BJ, NCCIT and OVCAR-3 cell lines. For each cell line, the fold differences between the activity of pTERT-297 and pTERT-297/ex1 were calculated and added on top of the graphic bars.
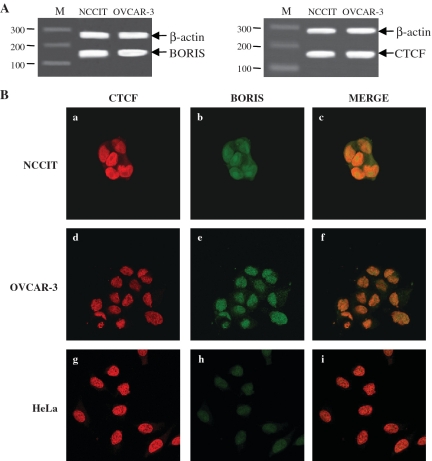
CTCF and BORIS were co-expressed in the testicular and ovarian tumor cell lines
As BORIS, a paralogue of CTCF, is expressed in male germ cells, we investigated the expression of these two factors in NCCIT and OVCAR-3. RT–PCR analyses of CTCF and BORIS transcripts showed that these two factors are expressed in both lines (Figure 3A). In normal cells, BORIS and CTCF were found to be expressed in a mutually exclusive manner (37). This prompted us to investigate the expression of these two proteins by immunofluorescence in NCCIT, OVCAR-3 and HeLa. These studies showed that BORIS and CTCF were coexpressed in the nuclei of NCCIT and OVCAR-3 (Figure 3B, a–f). In HeLa cells, CTCF was localized in the nucleus, whereas BORIS could be detected only at background levels (Figure 3B, g–i). In NCCIT and OVCAR-3, the merger of CTCF and BORIS signals showed that BORIS is likely localized in the nucleus where it could compete with CTCF.
Figure 3.
Expression and localization of CTCF and BORIS in NCCIT, OVCAR-3 and HeLa cells. (A) Transcriptional expression of CTCF and BORIS were performed by RT–PCR from extracted RNA of NCCIT and OVCAR-3. β-actin was co-amplified as internal control (B) Immunofluorescence staining of CTCF and BORIS in NCCIT (a, b, c), OVCAR-3 (d, e, f) and HeLa (g, h, i) cells. Images a, d and g: CTCF immunostaining with a mouse monoclonal CTCF antibody, followed by incubation with an anti-mouse-Cy3 antibody. Images b, e and h: BORIS immunostaining with a rabbit polyclonal BORIS antibody, followed by incubation with an anti-rabbit-FITC antibody. Images c, f and i: merge of BORIS and CTCF staining revealed a coexpression within the nucleus.
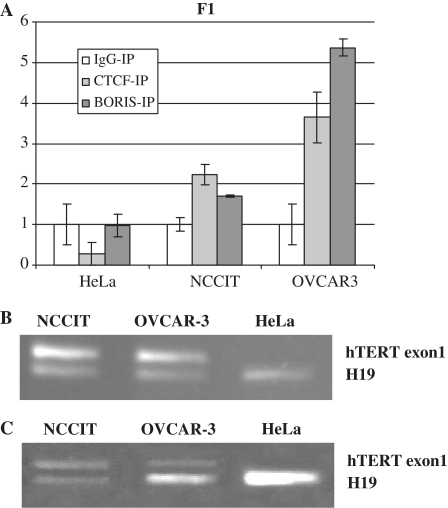
BORIS and CTCF bound in vivo to the same region of the hTERT gene
As the BORIS protein has the same 11 zinc finger domain as CTCF (34,37), we investigated the hypothesis that BORIS can replace CTCF on hTERT repressive site(s), thus allowing hTERT expression. Consequently, we studied in vivo binding of CTCF and BORIS to hTERT exon 1. Quantitative ChIP assays confirmed that CTCF did not bind the exon 1 of hTERT in HeLa cells in vivo, whereas it did bind exon 1 of hTERT in OVCAR-3 and NCCIT cells with a 3.6- and 2.2-fold change respectively (Figure 4A). Similarly, BORIS bound to the first exon of hTERT in OVCAR-3 and NCCIT cells but not in HeLa cells (Figure 4A). These results indicated that BORIS and CTCF can bind the same region of hTERT in the telomerase-positive cell lines OVCAR-3 and NCCIT. These results were confirmed by ChIP analyzed on agarose gels (Figure 4B and C) as described previously (24,25).
Figure 4.
In vivo binding of CTCF and BORIS on hTERT exon1. Chromatin ImmunoPrecipitation (ChIP) Assays were performed in HeLa, NCCIT and OVCAR-3 cell lines. After immunoprecipitation with serum or specific antibodies against CTCF or BORIS, DNAs were purified and amplified by PCR and quantitative PCR. (A) Quantitative PCR were preformed to amplify the exon1 of hTERT. Bars represent the mean value of three independent experiments. (B) PCR coamplification of the hTERT exon1 and H19 from immunoprecipitated fractions bound by CTCF antibody in NCCIT, OVCAR-3 and HeLa cells. (C) PCR coamplification of the hTERT exon1 and H19 from immunoprecipitated fractions bound by BORIS antibody in NCCIT, OVCAR-3 and HeLa cells.
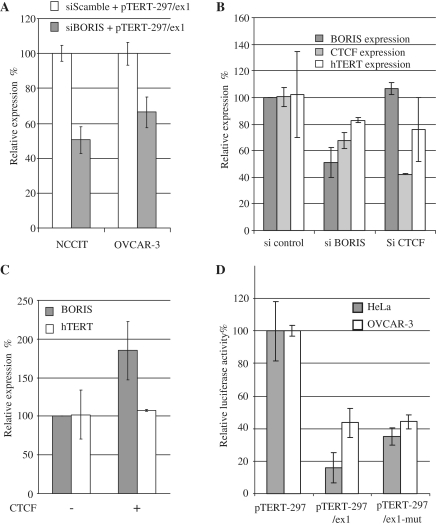
BORIS and CTCF expression levels modulated hTERT transcription
The finding that CTCF and BORIS bound hTERT exon1 in vivo, prompted us to examine the effect of BORIS downregulation on hTERT transcription. First we co-transfected NCCIT and OVCAR3 cells with reporter plasmids containing the hTERT minimal promoter and the first exon with siRNA against BORIS. The data in Figure 5A showed that a decrease in BORIS expression induced a decrease of reporter activities in both cell lines. These results demonstrated that BORIS acted as an inhibitor of the hTERT minimal promoter in transient transfection experiments. Because the use of reporter vectors did not fully reflect hTERT transcription in vivo, we performed additional transfection experiments in OVCAR-3. Then we modulated the balance between BORIS and CTCF expression levels using specific siRNAs against BORIS and CTCF. The endogenous levels of hTERT, BORIS and CTCF were then quantified by qPCR. Transcript levels in siRNA-treated cells were normalized to the levels detected after transfection with a scrambled siRNA. A 2-fold decrease in BORIS transcripts was observed in cells transfected with siRNA against BORIS and levels of CTCF transcripts were decreased by 30%. In contrast, levels of hTERT transcripts were essentially unchanged. (Figure 5B). A 2.5-fold decrease in CTCF transcripts was observed in cells transfected with siRNA against CTCF while levels of BORIS and hTERT transcripts were unaffected. These results demonstrated that control of BORIS and CTCF transcription were closely linked such that modulation of one could affect the expression of the other.
Figure 5.
Modulation of BORIS and CTCF expression levels. (A) Co-transfection of siRNA scramble or against BORIS with luciferase reporters containing the hTERT minimal promoter or the hTERT minimal promoter + exon1, in NCCIT and OVCAR-3. Relative luciferase expression is expressed in percentage with the expression of the minimal hTERT promoter referred as 100%. (B) Transfection of siRNA scramble or against CTCF or BORIS in OVCAR-3. Endogenous transcriptional levels of BORIS, CTCF and hTERT are measured 24 h after transfection by quantitative PCR. Relative expressions were calculated with the expression of each gene after transfection with the scamble siRNA referred as 100% of transcription. (C) Transcriptional level of endogenous BORIS and hTERT in OVCAR-3 after transfection of CTCF expression vector. Relative levels were calculated taking the transcriptional levels of BORIS and TERT in OVCAR-3 without over-expression of CTCF as 100%. (D) Transfection of hTERT reporters in BORIS-negative and BORIS-positive cell lines. Luciferase reporter constructs containing either the minimal promoter of hTERT, or the minimal promoter of hTERT + exon1, or the minimal promoter of hTERT + exon1 with CTCF mutated binding site, were transfected into HeLa and OVCAR-3 cell lines. Relative luciferase activities were calculated taking the level of the minimal promoter as 100%.
In a second experiment, we overexpressed CTCF in OVCAR-3 cells and measured the transcript level of endogenous hTERT and BORIS using levels in unmanipulated cells referenced as 100%. Overexpression of CTCF resulted in increased BORIS transcription, but had no effect on the transcriptional levels of hTERT (Figure 5C). This suggests that the predicted repressive effect on hTERT of increased CTCF expression was countered by the increase in BORIS expression, thereby indicating that transcription of hTERT cannot be repressed by CTCF in cells that also express BORIS. Taken together, these results demonstrated that the equilibrium between the levels of CTCF and BORIS expression have a great importance for regulating hTERT transcription.
In studies of HeLa cells that do not express BORIS, a reporter vector with a mutated CTCF binding site in exon1 exhibited increased hTERT promoter activity. Parallel studies of BORIS-positive OVCAR-3 cells revealed that the mutation had no effect on hTERT promoter activity (Figure 5D). This experiment showed that in BORIS-negative cells, mutation of the CTCF binding site diminished the inhibitory effect of CTCF on hTERT promoter, and that in BORIS-positive cells, the presence of BORIS maintained hTERT promoter activity in the presence of CTCF.
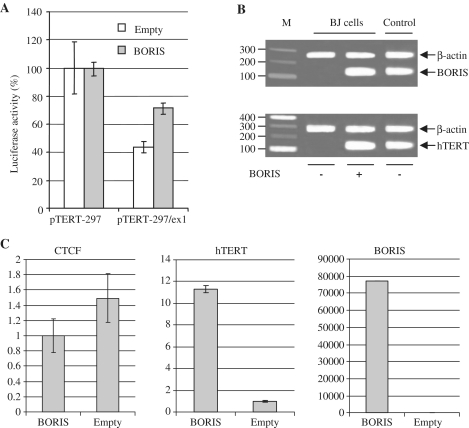
Expression of BORIS induced hTERT transcription in telomerase-negative cells
To further investigate the contributions of BORIS to hTERT transcriptional activation, NIH3T3 cells were co-transfected with a BORIS expression vector (pBIG-BORIS) and hTERT reporter constructs. Expression of BORIS was induced with doxycycline and luciferase activities were measured 24 h after transfection. The level of reporter activity of pTERT-297, which contains only the hTERT minimal promoter, was set as the reference point of 100%. The activity of the pTERT-297/ex1 vector was 2.5-fold lower in NIH3T3 cells co-transfected with the empty vector. In the co-transfection experiments with pBIG-BORIS, the reporter activity of pTERT-297/ex1 reporter activity was only 1.5-fold lower (Figure 6A), a level similar to that observed in OVCAR-3 cells transfected with pTERT-297/ex1 (Figure 2B). To confirm the stimulating effect of BORIS on hTERT transcription, the pCMV-BORIS expression vector was transfected in the BJ normal human fibroblast cells that do not express either BORIS or hTERT. After transfection with the pCMV-BORIS vector, BORIS and hTERT transcripts were easily detected by RT–PCR (Figure 6B). We also performed RT–PCR assays for the full-length and the α-spliced variants of hTERT (24). The amplified BORIS fragment obtained from BJ cells transfected with pCMV-BORIS was purified and directly sequenced, revealing the presence of only the full-length hTERT transcript (data not shown). In addition, qPCR analysis showed that high levels of BORIS transcripts were associated with substantial expression of hTERT while expression of CTCF was unchanged (Figure 6C).
Figure 6.
Transient transfection of a BORIS expression vector in normal cells. (A) Firefly Luciferase activities of pTERT-297 or pTERT-297/ex1 co-transfected with pBIG-BORIS or pBIG-empty in NIH3T3 mouse normal fibroblasts. The Renilla Luciferase was used to normalize the firefly luciferase. In each experiment, the activity was calculated by setting the activity of hTERT minimal promoter as 100%. pTERT-297: minimal hTERT promoter; pTERT-297/ex1: minimal promoter and first exon of the hTERT gene. (B) RT–PCR of BORIS and hTERT from total RNA of BJ cells 24 h after transfection with the pCMV-BORIS expression vector. (C) Real-time RT–PCR of CTCF, hTERT and BORIS from RNA of BJ cells after transfection of pCMV-BORIS or pCMV-empty. All qRT–PCR were normalized on GAPDH levels.
Expression of BORIS in NHBE cells induced expression of hTERT and promoted cell viability
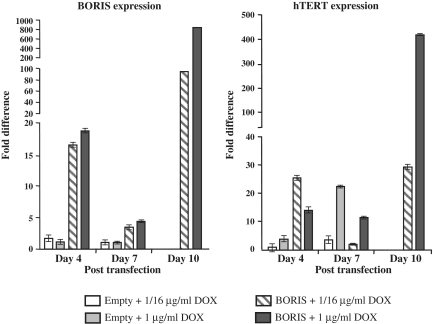
The lifespan of Normal Human Bronchial Epithelial cells (NHBE) in culture is very limited such that after 5 passages, virtually all cells detach and die. NHBE cells were transiently transfected with either a BORIS expression vector or an empty vector and BORIS expression was induced with doxycycline. After days 4, 7 and 10, the cultures were split. Half the cells were returned to culture and the other half was used for RNA extraction and qPCR to quantify BORIS and hTERT transcripts (Figure 7). Between Days 7 and 10, cells transfected with empty vector detached and died. At Day 10, cells transfected with the BORIS expression vector exhibited high levels of BORIS and hTERT transcripts. Lower levels of both BORIS and hTERT transcripts observed at Day 7 could be explained by plasmid dilution occurring with cell growth. The difference between the levels of BORIS transcripts at days 4 and 10 was ∼6-fold with 0.625 µg/ml of doxycycline and 55-fold with 1 µg/ml doxycycline. It is interesting to note that a low or a high concentration of doxycycline did not significantly affect the levels of BORIS transcripts at Day 4 while the levels of hTERT transcripts were almost 2-fold lower with the higher concentration of the drug. These observations lead us to suspect that a high level of BORIS transcripts were not necessarily associated with high levels of active BORIS protein. At Day 10, as we considered that plasmids were integrated, the levels of BORIS transcripts were 10-fold higher with a high concentration of doxycyline than with a low concentration (Figure 7). The levels of hTERT transcripts detected at low concentration of doxycycline did not differ at Days 4 or 10. With a high concentration of doxycycline, however, hTERT transcripts were 28-fold higher at Day 10 than at Day 4. Cells kept in continuous culture after day10 were viable for two more weeks, but then they stopped proliferating and finally detached. As we had found that low or high levels of BORIS transcripts affected the levels of hTERT transcripts after 10 days, the same experiments were performed using the non-inducible pBORIS-puro vector with similar results; the cells finally stopped proliferating and detached (data not shown).
Figure 7.
pBIG-BORIS inducible expression vector or the pBIG-empty vector were transfected into NHBE cell line (Normal Human Bronchial Epithelial). BORIS expression was induced with 1/16 µg/ml (0.0625 µg/ml) or 1 µg/ml of doxycycline and maintained in culture. RNAs from cells were extracted 4, 7 and 10 days after transfection. Parental cells and cells transfected with empty vector died after 7 days. The transcriptional expression of BORIS and hTERT were measured by quantitative RT–PCR and normalized on GAPDH levels.
DISCUSSION
Our studies of hTERT regulation in testicular and ovarian tumors showed that the presence of unmethylated CTCF repressor sites in the gene were not inhibitory to transcription raising the question as to how these cancers, despite a normal methylation profile, can express hTERT. Here we describe a novel mechanism that disrupts CTCF repression of hTERT—occupancy of the CTCF binding sites by BORIS. Since BORIS has the same DNA binding domain as CTCF and is known to be expressed at a high level in testis (37,38), in oocytes (47), as well as in various tumors and cancer cell lines (39–43), we investigated its potential role in the regulation of hTERT in testicular and ovarian cancers. Analysis of testicular and ovarian cancer tissues showed, at the exception of one ovarian cancer, the hTERT promoter was hypomethylated, that is in correlation with what was observed in a previous study (28). Parallel studies of the same 5′ regulatory region of hTERT in OVCAR-3 and NCCIT cell lines showed they were also hypomethylated. A more detailed bisulfite analysis of the promoter and exon 1 regions of hTERT in NCCIT and OVCAR-3 cell lines showed they were unmethylated as in normal cells, findings that contrast with the hypermethylated state of these sequences in the HeLa carcinoma cell line, that reflected the hTERT methylation profile in most cancer cells (16–18,20).
In transient transfection experiment using HeLa cells, it was shown that the activity of a reporter containing only the minimal hTERT promoter was 6-fold higher than the activity of a reporter containing the minimal promoter together with the first exon. In BJ cells, the reporter containing the minimal promoter and the exon 1 of hTERT did not exhibit any activity, most likely due to the absence of factor(s) essential for activation of the hTERT minimal promoter. In the same experiment, we showed that the activities of reporters containing the minimal promoter and the exon 1 were 2- to 3-fold higher in NCCIT and OVCAR-3 than in HeLa cells. It is noteworthy that these two tumor telomerase-positive cell lines expressed relatively high level of BORIS transcripts. Therefore, the inhibitory effect of hTERT exon 1 in reporter assays was less pronounced in BORIS-positive cells than in HeLa or BJ cells. The levels of BORIS expression is correlated with reduced inhibitory effects of the first exon of the hTERT gene. Similarly, co-transfection of a BORIS expression and hTERT reporter vectors in NIH3T3 cells revealed a reduced inhibitory effect of the first exon on its promoter activity. Indeed, when BORIS was transiently expressed in NIH3T3 cells, the reduction in luciferase activity observed with the hTERT exon1 reporter was less pronounced compared to the activity resulting in co-transfection of hTERT exon1 reporter with the empty vector. Moreover, this activity was close to that observed in the both BORIS-positive cell lines NCCIT and OVCAR-3. We also showed that transfection of telomerase-negative normal fibroblast BJ cells with a BORIS expression vector was sufficient to induce transcription of the endogenous hTERT gene. These results demonstrated that BORIS has the opposite effect of CTCF on hTERT transcription. BORIS might counteract the repressive effect of CTCF by binding the target sequences in the hTERT gene thereby permitting transcription of the hTERT promoter. Moreover, NHBE cells transfected with BORIS exhibited active transcription of hTERT and the lifespan of transfected cells was prolonged.
BORIS and CTCF were found to be coexpressed in the nuclei of NCCIT and OVCAR-3 cells and both proteins bound to the first exon of hTERT in vivo. Thus in NCCIT and OVCAR-3 cells, CTCF and BORIS might compete for the same binding sites in the hTERT gene. We demonstrated in vitro that downregulation of BORIS in NCCIT or OVCAR-3 was associated with reduced hTERT minimal promoter activity. However, experiments measuring the endogenous levels of hTERT after downregulation of BORIS did not lead to the same conclusion. Indeed, the levels of hTERT transcripts were not significantly different. This could be explained by the facts that (i) BORIS expression was reduced by only 50% and (ii) downregulation of BORIS was also associated with downregulation of CTCF. Conversely, overexpression of CTCF was associated with increased levels of BORIS transcripts. Taken together, these results demonstrated that BORIS or CTCF expression could influence levels of each other in cancer cell lines. To further examine the effects of reduced BORIS expression on hTERT transcription, we performed serial siRNA transfections and observed massive cell death. Similar observations were made in recent studies showing that downregulation of BORIS by siRNA induced apoptosis in a breast cancer cell line (48). These results suggest that disturbances of the equilibrium between CTCF and BORIS in these cells might induce cell death. In order to avoid deregulation of the balance between CTCF and BORIS, we performed transient transfection of hTERT luciferase reporters containing or lacking exon1 of hTERT with or without mutation of the CTCF binding site. In HeLa cells, where BORIS levels are low, when we expressed hTERT with a mutant CTCF target site, we observed an increase in luciferase activity, indicating that CTCF cannot bind this site, its inhibitory effect is diminished. In contrast, parallel studies of the BORIS-positive cell line OVCAR-3 revealed that luciferase activity was the same in cells bearing the normal or mutant CTCF binding site. There are multiple explanations to the observation but most feasible that BORIS isoforms that have different Zinc Finger composition can bind to minimal promoter of hTERT (E.Pugacheva et al., submitted elsewhere). The mechanism by which BORIS counteracted the inhibitory effect of CTCF on the hTERT transcription resulting in activation of hTERT transcription in testicular and ovarian cancers has to be elucidated. However, such mechanism involving CTCF and other proteins counteracting its effect on hTERT transcription has already been suggested recently. Indeed, in lymphoid cells, the PAX5 and CTCF factors bound simultaneously the hTERT exon 1. In this situation, PAX5 might antagonize the chromatin-mediated transcriptional repression by CTCF on hTERT promoter (30).
Since recent studies have shown that hTERT is an endogeneous inhibitor of the mitochondrial pathway of apoptosis (49), our data might explain how BORIS acts as inhibitor of apoptosis in cancer cells by activating hTERT transcription.
Taken together, these findings show the importance of BORIS for transcriptional activation of hTERT and, possibly, to the process of immortalization during carcinogenesis. As CTCF binds directly to SIN3A, which recruit histone deacetylase and thus prevents transcription (50), we hypothesize that BORIS might have the opposite effect and open chromatin around the transcriptional start site of hTERT. This opening would then allow other factors to activate the hTERT transcription. Moreover, recent studies showed that down-regulation of DNMT by anti-cancer drug or TSA, induced a demethylation of hTERT exon1 within the CTCF binding site, allowing binding of CTCF and inhibition of hTERT (27,51). In addition, downregulation of DNMT also induced an increase in CTCF binding on BAG-1 oncogene promoter, while an overexpression of DNMT led to increase in BORIS binding (52). This opposite behavior is further reflected by changes in H3K4Me2 and H3K9me2 ratio. Taking together these studies showed that promoters regulation by CTCF/BORIS involved epigenetic changes.
In tumors, co-expression of CTCF and BORIS could be directly responsible for epigenetic deregulation leading to cancer (34). The demonstrated utility of BORIS as a target for immunotherapy (53,54) is consistent with the importance of BORIS in carcinogenesis. The importance of BORIS for hTERT activation suggests that it might be an essential factor in the immortalization of testicular and ovarian cancer cells, possibly opening new ways to understanding of carcinogenesis and for potential anti-cancer therapies.
FUNDING
Swiss National Science Foundation (3100AO-101732 and 3100A0-113505, in part); Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases. Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Dr Herbert C. Morse III for critical reading of the article.
REFERENCES
- 1.Blackburn EH. The telomere and telomerase: nucleic acid-protein complexes acting in a telomere homeostasis system. A review. Biochemistry (Mosc) 1997;62:1196–1201. [PubMed] [Google Scholar]
- 2.Greider CW. Telomere length regulation. Annu. Rev. Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 4.Yasumoto S, Kunimura C, Kikuchi K, Tahara H, Ohji H, Yamamoto H, Ide T, Utakoji T. Telomerase activity in normal human epithelial cells. Oncogene. 1996;13:433–439. [PubMed] [Google Scholar]
- 5.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 6.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 7.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 8.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 9.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 10.Counter CM, Meyerson M, Eaton EN, Ellisen LW, Caddle SD, Haber DA, Weinberg RA. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Snow BE, Hande MP, Yeung D, Erdmann NJ, Wakeham A, Itie A, Siderovski DP, Lansdorp PM, Robinson MO, et al. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- 12.Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum. Mol. Genet. 1999;8:137–142. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- 13.Horikawa I, Cable PL, Afshari C, Barrett JC. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 1999;59:826–830. [PubMed] [Google Scholar]
- 14.Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59:551–557. [PubMed] [Google Scholar]
- 15.Wick M, Zubov D, Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT) Gene. 1999;232:97–106. doi: 10.1016/s0378-1119(99)00108-0. [DOI] [PubMed] [Google Scholar]
- 16.Dessain SK, Yu H, Reddel RR, Beijersbergen RL, Weinberg RA. Methylation of the human telomerase gene CpG island. Cancer Res. 2000;60:537–541. [PubMed] [Google Scholar]
- 17.Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087–6090. [PubMed] [Google Scholar]
- 18.Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int. J. Cancer. 2002;101:335–341. doi: 10.1002/ijc.10593. [DOI] [PubMed] [Google Scholar]
- 19.Lopatina NG, Poole JC, Saldanha SN, Hansen NJ, Key JS, Pita MA, Andrews LG, Tollefsbol TO. Control mechanisms in the regulation of telomerase reverse transcriptase expression in differentiating human teratocarcinoma cells. Biochem. Biophys. Res. Commun. 2003;306:650–659. doi: 10.1016/s0006-291x(03)01033-7. [DOI] [PubMed] [Google Scholar]
- 20.Nomoto K, Maekawa M, Sugano K, Ushiama M, Fukayama N, Fujita S, Kakizoe T. Methylation status and expression of human telomerase reverse transcriptase mRNA in relation to hypermethylation of the p16 gene in colorectal cancers as analyzed by bisulfite PCR-SSCP. Jpn. J. Clin. Oncol. 2002;32:3–8. doi: 10.1093/jjco/hyf001. [DOI] [PubMed] [Google Scholar]
- 21.Shin KH, Kang MK, Dicterow E, Park NH. Hypermethylation of the hTERT promoter inhibits the expression of telomerase activity in normal oral fibroblasts and senescent normal oral keratinocytes. Br. J. Cancer. 2003;89:1473–1478. doi: 10.1038/sj.bjc.6601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilleret I, Benhattar J. Demethylation of the human telomerase catalytic subunit (hTERT) gene promoter reduced hTERT expression and telomerase activity and shortened telomeres. Exp. Cell. Res. 2003;289:326–334. doi: 10.1016/s0014-4827(03)00281-7. [DOI] [PubMed] [Google Scholar]
- 23.Renaud S, Bosman FT, Benhattar J. Implication of the exon region in the regulation of the human telomerase reverse transcriptase gene promoter. Biochem. Biophys. Res. Commun. 2003;300:47–54. doi: 10.1016/s0006-291x(02)02775-4. [DOI] [PubMed] [Google Scholar]
- 24.Renaud S, Loukinov D, Bosman FT, Lobanenkov V, Benhattar J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33:6850–6860. doi: 10.1093/nar/gki989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renaud S, Loukinov D, Abdullaev Z, Guilleret I, Bosman FT, Lobanenkov V, Benhattar J. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007;35:1245–1256. doi: 10.1093/nar/gkl1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zinn RL, Pruitt K, Eguchi S, Baylin SB, Herman JG. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
- 27.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane Causes Epigenetic Repression of hTERT Expression in Human Breast Cancer Cell Lines. PloS one. 5:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Widschwendter A, Muller HM, Hubalek MM, Wiedemair A, Fiegl H, Goebel G, Mueller-Holzner E, Marth C, Widschwendter M. Methylation status and expression of human telomerase reverse transcriptase in ovarian and cervical cancer. Gynecol. Oncol. 2004;93:407–416. doi: 10.1016/j.ygyno.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 29.Bechter OE, Eisterer W, Dlaska M, Kuhr T, Thaler J. CpG island methylation of the hTERT promoter is associated with lower telomerase activity in B-cell lymphocytic leukemia. Exp. Hematol. 2002;30:26–33. doi: 10.1016/s0301-472x(01)00760-3. [DOI] [PubMed] [Google Scholar]
- 30.Bougel S, Renaud S, Braunschweig R, Loukinov D, Morse HC, III, Bosman FT, Lobanenkov V, Benhattar J. PAX5 activates the transcription of the human telomerase reverse transcriptase gene in B cells. J. Pathol. 220:87–96. doi: 10.1002/path.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugacheva EM, Tiwari VK, Abdullaev Z, Vostrov AA, Flanagan PT, Quitschke WW, Loukinov DI, Ohlsson R, Lobanenkov VV. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum. Mol. Genet. 2005;14:953–965. doi: 10.1093/hmg/ddi089. [DOI] [PubMed] [Google Scholar]
- 33.Dunn KL, Davie JR. The many roles of the transcriptional regulator CTCF. Biochem. Cell Biol. 2003;81:161–167. doi: 10.1139/o03-052. [DOI] [PubMed] [Google Scholar]
- 34.Klenova EM, Morse HC, III, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin. Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 35.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 36.Recillas-Targa F, De La Rosa-Velazquez IA, Soto-Reyes E, Benitez-Bribiesca L. Epigenetic boundaries of tumour suppressor gene promoters: the CTCF connection and its role in carcinogenesis. J. Cell Mol. Med. 2006;10:554–568. doi: 10.1111/j.1582-4934.2006.tb00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov AA, et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl Acad. Sci. USA. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki T, Kosaka-Suzuki N, Pack S, Shin DM, Yoon J, Abdullaev Z, Pugacheva E, Morse HC, III, Loukinov D, Lobanenkov V. Expression of a testis-specific form of Gal3st1 (CST), a gene essential for spermatogenesis, is regulated by the CTCF paralogous gene BORIS. Mol. Cell. Biol. 30:2473–2484. doi: 10.1128/MCB.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Arcy V, Pore N, Docquier F, Abdullaev ZK, Chernukhin I, Kita GX, Rai S, Smart M, Farrar D, Pack S, et al. BORIS, a paralogue of the transcription factor, CTCF, is aberrantly expressed in breast tumours. Br. J. Cancer. 2008;98:571–579. doi: 10.1038/sj.bjc.6604181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong JA, Kang Y, Abdullaev Z, Flanagan PT, Pack SD, Fischette MR, Adnani MT, Loukinov DI, Vatolin S, Risinger JI, et al. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005;65:7763–7774. doi: 10.1158/0008-5472.CAN-05-0823. [DOI] [PubMed] [Google Scholar]
- 41.Smith IM, Glazer CA, Mithani SK, Ochs MF, Sun W, Bhan S, Vostrov A, Abdullaev Z, Lobanenkov V, Gray A, et al. Coordinated activation of candidate proto-oncogenes and cancer testes antigens via promoter demethylation in head and neck cancer and lung cancer. PloS one. 2009;4:e4961. doi: 10.1371/journal.pone.0004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vatolin S, Abdullaev Z, Pack SD, Flanagan PT, Custer M, Loukinov DI, Pugacheva E, Hong JA, Morse H, III, Schrump DS, et al. Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 2005;65:7751–7762. doi: 10.1158/0008-5472.CAN-05-0858. [DOI] [PubMed] [Google Scholar]
- 43.Woloszynska-Read A, James SR, Link PA, Yu J, Odunsi K, Karpf AR. DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun. 2007;7:21. [PMC free article] [PubMed] [Google Scholar]
- 44.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kouprina N, Noskov VN, Pavlicek A, Collins NK, Schoppee Bortz PD, Ottolenghi C, Loukinov D, Goldsmith P, Risinger JI, Kim JH, et al. Evolutionary diversification of SPANX-N sperm protein gene structure and expression. PloS one. 2007;2:e359. doi: 10.1371/journal.pone.0000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guilleret I, Benhattar J. Unusual distribution of DNA methylation within the hTERT CpG island in tissues and cell lines. Biochem. Biophys. Res. Commun. 2004;325:1037–1043. doi: 10.1016/j.bbrc.2004.10.137. [DOI] [PubMed] [Google Scholar]
- 47.Monk M, Hitchins M, Hawes S. Differential expression of the embryo/cancer gene ECSA(DPPA2), the cancer/testis gene BORIS and the pluripotency structural gene OCT4, in human preimplantation development. Mol. Hum. Reprod. 2008;14:347–355. doi: 10.1093/molehr/gan025. [DOI] [PubMed] [Google Scholar]
- 48.Dougherty CJ, Ichim TE, Liu L, Reznik G, Min WP, Ghochikyan A, Agadjanyan MG, Reznik BN. Selective apoptosis of breast cancer cells by siRNA targeting of BORIS. Biochem. Biophys. Res. Commun. 2008;370:109–112. doi: 10.1016/j.bbrc.2008.03.040. [DOI] [PubMed] [Google Scholar]
- 49.Massard C, Zermati Y, Pauleau AL, Larochette N, Metivier D, Sabatier L, Kroemer G, Soria JC. hTERT: a novel endogenous inhibitor of the mitochondrial cell death pathway. Oncogene. 2006;25:4505–4514. doi: 10.1038/sj.onc.1209487. [DOI] [PubMed] [Google Scholar]
- 50.Lutz M, Burke LJ, Barreto G, Goeman F, Greb H, Arnold R, Schultheiss H, Brehm A, Kouzarides T, Lobanenkov V, et al. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28:1707–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi JH, Min NY, Park J, Kim JH, Park SH, Ko YJ, Kang Y, Moon YJ, Rhee S, Ham SW, et al. TSA-induced DNMT1 down-regulation represses hTERT expression via recruiting CTCF into demethylated core promoter region of hTERT in HCT116. Biochem. Biophys. Res. Commun. 391:449–454. doi: 10.1016/j.bbrc.2009.11.078. [DOI] [PubMed] [Google Scholar]
- 52.Sun L, Huang L, Nguyen P, Bisht KS, Bar-Sela G, Ho AS, Bradbury CM, Yu W, Cui H, Lee S, et al. DNA methyltransferase 1 and 3B activate BAG-1 expression via recruitment of CTCFL/BORIS and modulation of promoter histone methylation. Cancer Res. 2008;68:2726–2735. doi: 10.1158/0008-5472.CAN-07-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghochikyan A, Mkrtichyan M, Loukinov D, Mamikonyan G, Pack SD, Movsesyan N, Ichim TE, Cribbs DH, Lobanenkov VV, Agadjanyan MG. Elicitation of T cell responses to histologically unrelated tumors by immunization with the novel cancer-testis antigen, brother of the regulator of imprinted sites. J. Immunol. 2007;178:566–573. doi: 10.4049/jimmunol.178.1.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loukinov D, Ghochikyan A, Mkrtichyan M, Ichim TE, Lobanenkov VV, Cribbs DH, Agadjanyan MG. Antitumor efficacy of DNA vaccination to the epigenetically acting tumor promoting transcription factor BORIS and CD80 molecular adjuvant. J. Cell. Biochem. 2006;98:1037–1043. doi: 10.1002/jcb.20953. [DOI] [PubMed] [Google Scholar]