Abstract
TIAR and HuR are mRNA-binding proteins that play important roles in the regulation of translation. They both possess three RNA recognition motifs (RRMs) and bind to AU-rich elements (AREs), with seemingly overlapping specificity. Here we show using SPR that TIAR and HuR bind to both U-rich and AU-rich RNA in the nanomolar range, with higher overall affinity for U-rich RNA. However, the higher affinity for U–rich sequences is mainly due to faster association with U-rich RNA, which we propose is a reflection of the higher probability of association. Differences between TIAR and HuR are observed in their modes of binding to RNA. TIAR is able to bind deoxy-oligonucleotides with nanomolar affinity, whereas HuR affinity is reduced to a micromolar level. Studies with U-rich DNA reveal that TIAR binding depends less on the 2′-hydroxyl group of RNA than HuR binding. Finally we show that SAXS data, recorded for the first two domains of TIAR in complex with RNA, are more consistent with a flexible, elongated shape and not the compact shape that the first two domains of Hu proteins adopt upon binding to RNA. We thus propose that these triple-RRM proteins, which compete for the same binding sites in cells, interact with their targets in fundamentally different ways.
INTRODUCTION
The regulation of mRNA stability is a major control point in gene expression, particularly under conditions of stress, immune response or proliferation (1–3). Under such conditions mRNA stability and translation are tightly controlled by the association of RNA-binding proteins (RBPs) which specifically recognize elements in the mRNA sequence (1–5). One of the best characterized regulatory elements, found predominantly in the 3′ UTR of mRNA transcripts encoding high-turnover proteins such as cytokines, lymphokines, onco-proteins and inflammatory mediators, are AU-rich elements (AREs) (6–8). AREs are specific regulatory sequences often comprising uridine- or adenine/uridine-rich stretches and have been grouped into three classes, although precise consensus sequences are yet to be clarified (7,8). Class I AREs consist of one to three copies of scattered AUUUA motifs with a nearby U-rich region. Class II AREs consist of at least two overlapping UUAUUUA(U/A)(U/A) nonamers in a U-rich region and class III AREs, which are less well characterized, have U-rich regions without the AUUUA motif. More than 4000 AREs have been mapped to the human genome, representing 5–8% of human genes (9).
Several proteins have been identified in eukaryotic cells that bind to mRNAs by targeting AREs in their 3′ UTR and play a role in regulation of mRNA stability and translational efficiency. Interestingly, their binding can result in quite different outcomes for the mRNAs. RBPs TIA-1 (T-cell restricted intracellular antigen-1) and TIAR (TIA-1 related) bind to AREs and function as translational repressors, sequestering target mRNA into stress granules (SG) following cellular stress (10–12). In contrast, AUF1 (AU-binding factor 1), TTP (tristetraprolin), and KSRP (KH-type splicing regulatory protein) binding to AREs leads to the rapid decay of the specific mRNAs (13–15). Alternatively, the HuR (Hu antigen R) protein generally has a stabilizing effect when it binds to AREs (16,17). Thus AREs appear to be the target of proteins with diverse functions leading to critically different outcomes for the mRNA.
Whether, in fact, these ARE-binding proteins compete for the same mRNA target sites is still not clearly understood. It is conceivable that the same sites are targeted, and that factors such as the relative local concentration or activation state of each of these RBPs dictate the alternative possible fates of the mRNA transcripts. Liao and colleagues have shown that competitive binding of TIAR and AUF1 determine the translation of myc (18). Alternatively, the RNA sequence preferences and/or RNA-binding modes could differ between these RBPs and a more complex interplay of protein–RNA interactions underlies their translational regulation. Indeed, co-immunoprecipitation of ARE-binding proteins and identification of their bound mRNA by microarray has revealed distinctly different populations of target mRNA in vivo (12,19–22). This is consistent with the existence of distinct binding preferences rather than simple competition for the same pool of ARE-bearing mRNA transcripts. Gorospe and colleagues have proposed different consensus sequences for each of TIAR, TIA-1, HuR and AUF1 (12,19–22). These studies suggested that HuR and TIA-1 motifs are U-rich rather than AU-rich. They also demonstrated cases where these proteins bind at overlapping as well as distinct places on the same mRNA transcript and together modulate translation (23,24). In some cases these proteins have even been shown to interact with non-ARE consensus sequences. We have demonstrated in our previous in vitro and in vivo studies that TIAR can also bind to a C-rich motif in the 3′ UTR of target mRNAs, confirming it as a novel TIAR target (19). Therefore, it is likely that ARE-binding proteins interact with their target RNA sequences with differences in their modes of binding, degree of stringency or even specificity underlying the ultimate fate of the mRNA transcript.
Two of the best-characterized ARE-binding proteins are the TIA proteins (TIA-1 and TIAR) and HuR of the Hu protein family (which includes the neuronal proteins HuB, HuC and HuD) (10,17,25). These classical RNA-recognition motif (RRM)-containing proteins are both ubiquitously expressed in mammalian cells and bind to several common mRNA targets such as TNF-α and GM-CSF (26–31). They are both nucleo-cytoplasmic shuttling ‘multi-functional’ proteins performing a variety of roles at different stages of gene expression including splicing, nucleo-cytoplasmic transport, translation and degradation of mRNA (17,25,32,33). TIA proteins are involved in the control of alternative pre-mRNA splicing, binding to U-rich RNA sequences mostly in introns and promoting the recognition of atypical 5′ splice sites (33–39). TIAR has also been reported to be able to bind strongly to a single-stranded, but not double-stranded, T-rich DNA which may position TIAR to modulate transcription and help to localize TIAR to U-rich RNA at the time of transcription (40). In the cytoplasm TIA proteins are capable of binding target sequences in the 3′-UTR of mRNA and regulating translation (12,25,32). Under conditions of stress TIA proteins play a vital role in SG formation, where untranslated mRNAs accumulate until the stress is passed (11,25,32,41,42). HuR is best known for its nuclear-cytoplasmic shuttling and its stabilizing effect on many target mRNAs (17). HuR can also increase the translation of other associated mRNAs (16), and repress the translation of other targets via miRNA recruitment (43) and by proposed interference with internal ribosome entry sites (IRESs) (44–46). The fate of the mRNA transcript is thus very different depending on whether it interacts with TIA proteins or HuR.
TIAR, which shares >80% homology with TIA-1, is a 375-amino-acid protein belonging to the RRM-containing family of RBPs. The three RRMs located at the N-terminus confer high affinity binding to U-rich RNA sequences (KD ∼ 1 nM) (19), while the C-terminal 90-amino-acid residue glutamine-rich sequence is essential for stress-granule formation (10,19,21,47–49). RRMs are ∼70–90 amino-acids long and are able to specifically bind between two and eight sequential single-stranded nucleotides (4,50). TIAR was shown to bind with highest affinity to U-rich RNA sequences, with the three RRMs contributing variously to the interaction (47). It was shown that RRM2 is both sufficient and necessary for binding to AREs and RRM3 showed binding to RNA but may have other specificities than AREs (47). RRM1 showed no binding to U-rich sequences on its own, but was subsequently shown to be able to bind T-rich DNA (40). No structural information for TIAR/RNA complexes is yet available, though structures of the individual TIAR RRMs have been elucidated using NMR (PDB ID: 2DH7; 2CQI, 1X4G). They all share canonical RRM folds of βαββαβ topology.
TIAR's glutamine-rich C-terminal region shares sequence similarity to human prion protein (51,52). When expressed alone in cells, it forms spontaneous cytoplasmic microaggregates that coaggregate other TIA proteins. It can self-oligomerize in vivo like prion proteins and is thought to be crucial for SG formation when cells are under stress (10,53). When this occurs, mRNA that is bound by TIA proteins is sequestered into the SGs. It has been proposed that the mRNA remains in this ‘holding zone’ protected from degradation until the stress is relieved and then the mRNA is either directed towards further translation or degradation (11,41,42).
The primary structures of the Hu-proteins are well conserved (RRMs share >70% amino-acid sequence identity among family members) and are arranged with two RRMs near the N-terminus, followed by a less conserved basic hinge region and a third RRM near the C-terminus (54). This arrangement of three RRM domains is strikingly similar to that seen in the TIA proteins and also confers high-affinity binding to ARE sequences (KD ∼ 1–2 nM) (55,56) although there is <30% sequence homology overall and <35% homology between any TIA-protein RRM compared to Hu-protein RRM. In the case of HuD, the relative roles of the three RRMs for ARE interactions have been interrogated. RRM1 is essential for RNA-binding, but the high-affinity interaction also requires RRM2 and RRM3 (55,57). The crystal structures of the first two RRMs of HuD protein bound to 11-nt single-stranded RNA derived from c-fos and TNF-α mRNA transcripts have been reported and provide insight into the mode of interaction of these domains with classical AREs (58). Interestingly, in the case of HuR, the RRM3 as well as the hinge region between RRM2 and RRM3 contribute significantly to ARE binding in a length-dependent manner by helping to form multimeric HuR–ARE complexes and increasing the RNA-binding affinity, respectively (56).
TIAR and HuR are thus proteins that co-exist in cells and have the capacity to bind specifically to AREs within mRNA in both the nucleus and cytoplasm. They possess analogous RRM architecture and are reported to bind AREs with similar affinity. Here we have determined whether their interactions with target RNA are truly so similar, with the control of mRNA fate potentially dependent on the local availability and activity of these proteins, or whether they have different modes of interaction with RNA. We investigated the binding of TIAR (without the glutamine-rich domain) and HuR to an AU-rich motif, a class I ARE derived from 3′UTR of TNF-α mRNA transcript, compared to a previously established class III ARE, the U-rich motif (19,47) using surface plasmon resonance (SPR). TNF-α mRNA is a known target of TIAR and HuR (28,59) and the mRNA stability and translational efficiency of this key inflammatory mediator has been shown to be regulated by TIAR, HuR and other RBPs (60). We also investigated their ability to bind to DNA compared with RNA and deoxy-U-rich oligonucleotides. TIAR has been shown to interact with DNA previously (40), but characterization of this interaction has been very limited to date. Finally, Small Angle X-ray Scattering (SAXS) was employed to obtain insight into the potential mode of interaction between these RBPs and their target RNA sequences in solution. Together, our data reveal similarities and fundamental differences in the substrate binding activities of these important mRNA binding proteins.
MATERIALS AND METHODS
Plasmid construction and protein purification
Proteins comprising TIAR and HuR RRM domains were prepared for the current studies. These include proteins with all three RRMs (referred to as TIAR123 and HuR123) and proteins comprising the two N-terminal domains only (referred to as TIAR12 and HuR12). Constructs for the expression of TIAR123 (residues 1–283) and TIAR12 (residues 1–208) were transformed into Escherichia coli strain BL21 (DE3) and the encoded proteins were expressed and purified as described previously (47). We discovered, in the course of these studies, that the TIAR12 protein was unstable and consistently degraded to a stable form representing residues 1–181 using mass spectrometry. These residues still encompass the two complete RRMs. HuR12 (residues 18–184) was cloned into pGEX-4T1, expressed in E. coli BL21 (DE3), and purified according to previously established protocols (61). A construct for the expression of HuR123 (residues 1–306) was transformed into E. coli ER2566, and purified according to previously established protocols (56). The proteins were further purified by size-exclusion and cation-exchange chromatography. The concentration of each protein was determined using the Bradford assay (BioRad) and by A280 measurements using theoretical molar extinction coefficients (ProtParam). The extinction coefficients were validated for folded protein; A280 measurements were within 10% of measurements made in 6.0 M guanidium hydrochloride. The purity of each protein was confirmed by SDS–PAGE. The active fraction of the protein was not experimentally determined, but assumed to be close to 100% upon purification.
Biosensor experiments
The dynamics of RNA/DNA–protein interactions were characterized by SPR using a BIACORE T100 instrument (Biacore Inc.). The oligonucleotides used in the analyses included U-rich and AU-rich RNA, U-rich RNA of varying length, U-rich DNA and T-rich DNA. In the early experiments the U-rich and AU-rich sequences are bounded by G-rich regions (to which the HuR and TIAR have been shown not to interact; data not shown) for the purpose of spacing the binding region from the chip surface and for providing an RNA target of comparable length to other target sequences studied previously (19). Thus the sequences included: U-rich RNA (i) 5′-GGGGGGUUUUUUUUUUUUUUUUUGGGGG-3′, (ii) 5′-UUUUUUUUUUUUUUUUU-3′, (iii) 5′-UUUUUUUUUUUUU-3′, (iv) 5′-UUUUUUUU-3′; AU-rich RNA 5′-GGGGGGUAUUUAUUAUUUAUUUAGGGGG-3′; T-rich DNA 5′-TTTTTTTTTTTTTTTTTTTT-3′ and U-rich DNA 5′-dUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdU-3′ (Table 1). The oligonucleotides were chemically synthesized carrying a 5′-biotin tag (Dharmacon Research) to allow immobilization of the RNA/DNA onto streptavidin-coated sensor chips (Series S Sensor Chip SA, Biacore Inc.). RNA were diluted to a final concentration of 1 µM in HBS buffer (10 mM HEPES, pH 7.4, 150 mM NaCl) followed by heating at 80°C for 10 min, and cooling to room temperature. The sample was then diluted 500-fold in running buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM DTT, 0.025% surfactant P20; Biacore Inc.) and injected over the sensor chip surface at 10 µl/min at 25°C to generate a ∼50 response unit (RU) RNA surface (for a low-density surface). Proteins were serially diluted in running buffer to the concentrations indicated in Figures 1–3, and injected at 25°C at a flow rate of 50 µl/min for 2–3 min. Surface regeneration to remove any protein that remained bound after 3–6 min of dissociation was achieved using a 1-min injection of 2 M NaCl at 50 µl/min. Analyses of protein concentrations were done in duplicate and any background signal from a streptavidin-only reference flow cell was subtracted from every data set. Data were analysed using a simple 1:1 Langmuir interaction model or two-state (conformational change) model using the Biacore T100 evaluation software (Biacore Inc.) to determine the kinetics (association/dissociation rate constants; ka/kd) as well as the affinities (KD) of the protein–RNA interactions.
Table 1.
Oligonucleotides used in the SPR analysis
| Oligonucleotides | Sequences (5′ Biotin - 3′) |
|---|---|
| U-rich | GGGGGGUUUUUUUUUUUUUUUUUGGGGG (28-mer) |
| UUUUUUUUUUUUUUUUU (17-mer) | |
| UUUUUUUUUUUUU (13-mer) | |
| UUUUUUUU (8-mer) | |
| AU-rich | GGGGGGUAUUUAUUAUUUAUUUAGGGGG (28-mer) |
| T-rich | TTTTTTTTTTTTTTTTTTTT (20-mer) |
| Deoxy-U-rich | dUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdUdU (20-mer) |
Both 28-mer U- and AU-rich RNAs contain poly-G linkers at both ends. The AU-rich sequence represents the HuR target site within the 3′ UTR of human TNF-α mRNA transcript (nt 464–480) (28).
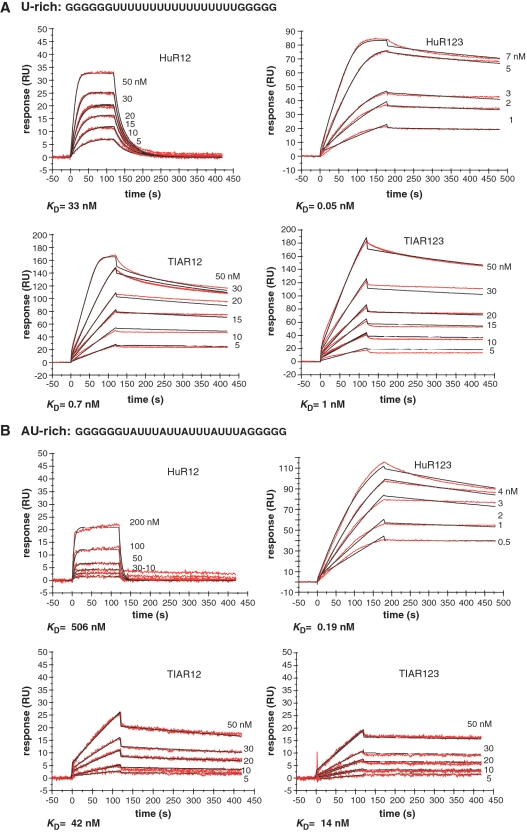
Figure 1.
Kinetic analysis of the interactions of TIAR and HuR proteins with U and AU-rich RNA using SPR. Sensorgrams of HuR12, HuR123, TIAR12 and TIAR 123 proteins to (A) a U-rich or (B) an AU-rich RNA (28-mer each) are shown. Biotinylated RNA was captured on SA-coated sensor chips and increasing concentrations of protein were injected over the surface. Injections were performed for 120 s (association phase), followed by a 300-s flow of running buffer to assess dissociation. The experiments were conducted in duplicate and showed good overlap. The red lines represent the binding responses for injections of protein analyte at specified concentrations (nanomolar) over the RNA surface. The kinetic data was fit by 1:1 Langmuir binding model that describes monovalent analyte binding to a single site on the immobilized ligand. Mass transport limitation effects were not evident. The black curves superimposed on the sensorgrams represent the model fitted curves. The rate constants ka and kd were determined simultaneously as global fitting parameters from which the KD was determined. The resulting parameter values are given in Table 2. [Note that sensorgrams for HuR12, TIAR12 and TIAR123 in panel A are reproduced from Kim et al. (19) Figure 3 with permission from the American Society for Microbiology to assist visual comparison with other sensorgrams].
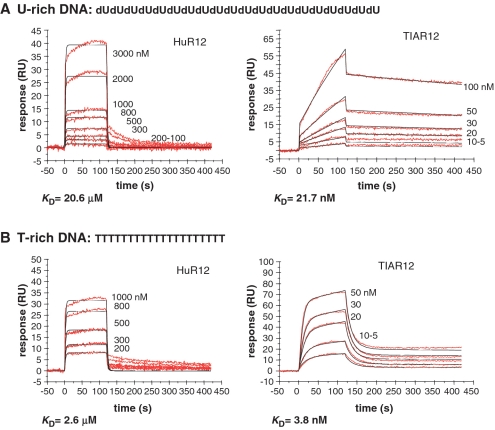
Figure 3.
Kinetic analysis of the interactions of TIAR12 and HuR12 proteins with U-rich and T-rich DNA using SPR. The binding of TIAR12 and HuR12 to (A) U- and (B) T-rich DNA is shown. Biotinylated DNA was captured on SA-coated sensor chips and increasing concentrations of protein were injected over the surface. Injections were performed for 120 s (association phase), followed by a 300-s flow of running buffer to assess dissociation. The experiments were conducted in duplicate and showed good overlap. The red lines represent the binding responses for injections of protein analyte at specified concentrations (nanomolar) over the DNA surface. The kinetic data was fit by the 1:1 Langmuir binding model (except for TIAR12 binding to T-rich DNA which was estimated by the two-state model). Mass transport limitation effects were not evident. The black curves superimposed on top of the sensorgrams represent the model fitted curves. The rate constants ka and kd were determined simultaneously as global fitting parameters from which the KD was determined.
SAXS measurements and data reduction
TIAR12 and HuR12 protein samples were subjected to SAXS analysis both in their apo forms and in combination with a 1:1 molar ratio of a 13-nt U-rich RNA (Dharmacon Research). SAXS measurements were made using the SAXS-WAXS beamline at the Australian Synchrotron, Melbourne, Australia, which is equipped with a Pilatus Detector. The scattering data was collected to provide an s range of 0.015–0.3, where s is the magnitude of the scattering vector. Samples were in 1.5-mm quartz capillaries at room temperature. Scattering was collected over a range of five concentrations between 1 and 4 mg/ml for each sample. The samples and matching buffer solutions were exposed to X-ray for 1, 5 and 1 s as the sample flowed through the capillary. The 2D scattering images were normalized for sample transmission and radially averaged. In each case the 5-s exposure provided the strongest data with no evidence of radiation damage. Scattering from the buffer and empty capillaries was subtracted after scaling scattering intensities to correspond to incident beam intensities. Data analysis was performed using the ATSAS suite of software. Scattered intensity (I) was plotted against s. Extrapolation of the I(s) profiles to zero angle [I(0)] and comparison with water as a standard indicated a molecular mass for all species consistent with no aggregation. The radius of gyration (Rg) did not vary significantly over the concentration ranges of each molecular species and all Guinear plots were linear for s.Rg < 1.3. The scattering data collected at a concentration of 4 mg/ml was used in each case. The program GNOM was used to yield the P(r) function via an indirect Fourier transform which provides the relative probabilities of the distances between the scattering centres and the maximal dimension of the scattering molecular species Dmax (62). The maximal particle dimensions were computed by constraining the function to 0 at rmax, where rmax was varied over a wide range of values in 1 Å increments. The value of rmax that yielded the highest ‘total estimate’ value as well as a plausible P(r) function was taken as the Dmax. Rg was also calculated from the second moment of the P(r) functions. The Rg values calculated from the Guinier approximation or the P(r) were favourably comparable.
Ensemble optimization method for SAXS data analysis
The SAXS data were analysed using the ensemble optimization method (EOM) (63), which is suitable for the characterization of flexible proteins in solution. This method allows for the coexistence of different conformations of the protein. A pool of 10 000 random structures of HuR12 (based on the HuD12 structure PDB ID: 1G2E) and TIAR12 (based on RRM1 and RRM2 NMR structures PDB ID: 2CQI, 2DH7) were generated using the EOM method with RRM domains defined as rigid bodies. Scattering curves generated from these structures were filtered using a genetic algorithm against the SAXS scattering curves to select an ensemble of conformers consistent with the experimental data. In order to generate a pool of protein–oligonucleotide complexes, the above process was repeated with HuR12 and TIAR12 structures with a peptide chain insertion to represent the occluded volume of RNA. After the generation of 10 000 structures the peptide was replaced with ssRNA (on the basis of PDB ID:1G2E) from which scattering curves were calculated using CRYSOL (64). These were filtered against the experimental data as described above.
Modelling of SAXS scattering data
For the HuR/RNA complex the program DAMMIF (65) was used to generate 15 ab initio dummy atoms models from its 4 mg/ml scattering curve. The models were superposed, merged and filtered using the program DAMAVER (66). This program calls a number of programs that superpose the models and also provides the normalized spatial discrepancy (NSD), a measure of how similar the models are to each other. Models are selected for inclusion for merging and filtering were required to satisfy the criterion NSD < mean NSD + 2× variation.
RESULTS
TIAR and HuR proteins both show high affinity for U-rich RNA but slow off rates from AU-rich RNA
There has been limited characterization of HuR and TIAR binding to ARE and C-rich sequences reported previously (19,47,56) but no direct comparison of the binding of these proteins to different classes of AREs, nor a focus on the difference between them. In order to directly compare the RNA-binding of HuR and TIAR to AREs, SPR was used to measure both affinity and kinetics of binding to U-rich (class III ARE) and AU-rich (class I ARE) sequences. TIAR and HuR proteins representing the three RRMs of the proteins (TIAR123 and HuR123) or the two N-terminal RRMs only (TIAR12 and HuR12) were prepared as described previously (see ‘Materials and methods’ section). These proteins were tested for their affinity for a 17-nt U-rich sequence compared with a 17-nt AU-rich sequence (corresponding to the TNF-α 3′UTR region nt 464–480) (28), using SPR (Figure 1). The eight sensorgrams (Figure 1A and B) show the binding of a range of concentrations of TIAR123, TIAR12, HuR123 and HuR12 when injected across the U- or AU-rich RNA-coated chip. The association rate constants (ka), dissociation rate constants (kd), and overall affinities (KD) for each protein, as approximated by a simple 1:1 Langmuir binding model, are shown in Table 2 and the residual plots and statistics (χ2) for the fitting is supplied in Supplementary Figure S1.
Table 2.
Kinetic and affinity constants for the interactions of HuR12, HuR123, TIAR12 and TIAR123 proteins with U- and AU-rich RNA
| Protein | RNA | ka (1/Ms) | kd (s−1) | KD (kd/ka, nM) |
|---|---|---|---|---|
| HuR12 | U-rich | (9.66 ± 0.19) × 106 | (3.20 ± 0.06) × 10−1 | 33.1 ± 1.29 |
| AU-rich | (3.07 ± 0.06) × 105 | (1.56 ± 0.01) × 10−1 | 506 ± 13.1 | |
| HuR123 | U-rich | (1.03 ± 0.01) × 107 | (5.15 ± 0.03) × 10−4 | 0.05 ± 0.001 |
| AU-rich | (4.34 ± 0.1) × 106 | (8.26 ± 0.11) × 10−4 | 0.2 ± 0.007 | |
| TIAR12 | U-rich | (4.10 ± 0.11) × 106 | (2.83 ± 0.08) × 10−3 | 0.69 ± 0.04 |
| AU-rich | (1.71 ± 0.02) × 104 | (7.26 ± 0.04) × 10−4 | 42.5 ± 0.63 | |
| TIAR123 | U-rich | (1.58 ± 0.02) × 106 | (1.56 ± 0.01) × 10−3 | 0.99 ± 0.02 |
| AU-rich | (1.15 ± 0.01) × 104 | (1.63 ± 0.02) × 10−4 | 14.1 ± 0.22 |
The association and dissociation rate constants (ka and kd) were determined as global fitting parameters for a 1:1 binding model. The equilibrium dissociation constant (KD) was determined as kd/ka. [Note that binding data for HuR12, TIAR12 and TIAR123 to U-rich RNA are reproduced from Kim et al. (19) with permission from the American Society for Microbiology to assist direct comparison].
All four proteins bound the U- and AU-rich RNA with KD in the nanomolar range but with quite different affinity and kinetics. HuR123 bound with very low nanomolar affinity to both RNA sequences as expected from previous studies (55,56), and the full-length protein (HuR123) bound with significantly higher affinity (∼1000-fold) than HuR12 comprising just the first two domains. Substrate release was the major mechanism contributing to enhanced binding of the full-length protein to both RNA substrates, evidenced by the dramatically slower dissociation rate constants of the full-length HuR123 compared to HuR12. TIAR123 also bound to both U- and AU-rich RNA with nanomolar affinity, though with 20- and 70-fold lower affinity than observed for HuR123 respectively. Here, it is interesting to note that the construct comprising only the first two domains (TIAR12) bound with an affinity similar to full-length (TIAR123) protein. In the case of TIAR, the first two domains alone appear to confer tight binding which is reflected in the TIAR12 sensorgrams showing very slow off-rates compared with HuR12.
In all cases, affinity to the U-rich sequence was higher (∼10 fold) than for the AU-rich sequence suggesting that this may be the preferred binding sequence of both HuR and TIAR between the two sequences tested. This higher affinity for U-rich sequence is consistent with a SELEX study by Dember et al. (47) and in vitro selection experiment and gel-shift assay by Park-Lee et al. (57), which determined that both TIAR and HuD proteins preferentially bind a U-rich motif. However, detailed kinetic measurements reveal that the higher affinities for U-rich sequences are largely due to the faster on-rates to the U-rich sequences. Examination of the dissociation rate constants reveals that these are similar or lower for the AU-rich sequences than the U-rich but, together with the lower association rate constants; an overall higher KD (lower affinity) is obtained for AU-rich oligonucleotide binding.
HuR12 proteins bind U-rich RNA in a length-dependent manner
Whilst the overall affinity of HuR and TIAR was higher for U-rich compared to AU-rich 17-nt RNA, this was clearly dictated by the much higher on-rates to U-rich RNA compared to AU-rich RNA. On-rates are usually determined by the diffusion of the proteins and their long range electrostatic interactions with the binding partner. These would not be expected to differ between U-rich and AU-rich RNA. The other factor influencing the association rate constant is the probability of a productive interaction occurring. This would be expected to be higher for U-rich RNA as productive binding could take place at any position along the length of the RNA that the RBP encounters. An interaction with the AU-rich sequence, however, may only be productive where the RBP encounters the RNA with adenosine positioned at its adenosine specific site.
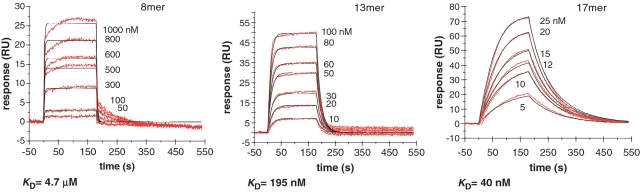
In order to verify that this effect could account for the magnitude of the enhanced association rate constant we observed for binding experiments with U-rich RNA, we conducted a series of SPR experiments (with HuR12 for proof of principle) with U-rich RNA of increasing length. The three sensorgrams (Figure 2) show the binding of a range of concentrations of HuR12 when injected across the 8, 13 and 17-mer U-rich RNA-coated chip. The association rate constants, dissociation rate constants, and overall affinities for each binding, approximated by a 1:1 Langmuir binding model, are listed in Table 3 and the residual plots and statistics (χ2) for the fitting is supplied in Supplementary Figure S1. Notably, as the length of the U-rich sequence was increased (8, 13, 17-mer) we observed increasing affinities, with association rate constants for binding occurring several orders of magnitude faster for the longest oligonucleotide, and dissociation rate constants remaining fairly constant. This is consistent with the increase in the probability of a productive interaction with the target RNA.
Figure 2.
Kinetic analysis of the interactions of HuR12 proteins with 8, 13 and 17-mer U-rich RNA using SPR. The binding of HuR12 to different lengths of U-rich RNAs (8, 13 and 17-mer) is shown. Biotinylated RNA was captured on SA-coated sensor chips and increasing concentrations of protein were injected over the surface. Injections were performed for 180 s (association phase), followed by a 360-s flow of running buffer to assess dissociation. The experiments were conducted in duplicate and showed good overlap. The red lines represent the binding responses for injections of protein analyte at specified concentrations (nanomolar) over the RNA surface. The kinetic data were fit by 1:1 Langmuir binding model which describes monovalent analyte binding to a single site on the immobilized ligand. Mass transport limitation effects were not evident. The black curves superimposed on top of the sensorgrams represent the model fitted curves. The rate constants ka and kd were determined simultaneously as global fitting parameters from which the KD was determined. The resulting parameter values are given in Table 3.
Table 3.
Kinetic and affinity constants for the interactions of HuR12 proteins with 8, 13 and 17-mer U-rich RNA
| Protein | U-rich RNA | ka (1/Ms) | kd (s−1) | KD (kd/ka, nM) |
|---|---|---|---|---|
| HuR12 | 8-mer | (8.781 ± 0.15) × 104 | (4.126 ± 0.031) × 10−1 | 4699 ± 116 |
| 13-mer | (8.657 ± 0.14) × 105 | (1.686 ± 0.027) × 10−1 | 195 ± 6.3 | |
| 17-mer | (1.588 ± 0.47) × 107 | (6.319 ± 1.9) × 10−1 | 40 ± 24 |
The association and dissociation rate constants (ka and kd) were determined as global fitting parameters for a 1:1 binding model. The equilibrium dissociation constant (KD) was determined as kd/ka.
We therefore propose that the 100-fold faster association rate constants observed for interactions with U-rich 17-nt sequences compared with AU-rich sequence, presented in the previous section, reflect the increased available binding sites for HuR and TIAR. The slower dissociation rate constants of TIAR, in particular, from AU-rich RNA may thus be a truer indication of the ‘preferred’ target sequence for these RBPs.
TIAR and HuR exhibit different binding kinetics and affinity to DNA, suggesting a different mode of interaction
Since TIAR has been reported to bind to DNA as well as to RNA, it was of interest to characterize this oligonucleotide interaction. Both HuR12 and TIAR12 were subjected to binding analysis with 20-nt U-rich DNA and T-rich DNA (to be able to differentiate between effects of removal of the 2′-hydroxyl group and the addition of the methyl group in the thymine base). The four sensorgrams (Figure 3) show the binding of a range of concentrations of HuR12 and TIAR12 when injected across the U-rich DNA and T-rich DNA-coated chip. The association and dissociation rate constants (ka and kd) and overall affinities (KD) for each binding event were estimated by a 1:1 Langmuir binding model, except for TIAR12 binding T-rich DNA which was best estimated by the two-state (conformational change) model (Table 4) and the residual plots and statistics (χ2) for the fitting is supplied in Supplementary Figure S1. Included in the table for comparison's sake are the data obtained for HuR12 and TIAR12 binding to U-rich RNA discussed earlier.
Table 4.
Kinetic and affinity constants for the interactions of TIAR12 and HuR12 proteins with U-rich RNA, U-rich DNA and T-rich DNA
| Protein | Oligo | ka (1/Ms) | kd (s−1) | KD (kd/ka, nM) |
|---|---|---|---|---|
| HuR12 | U-rich RNA | (9.66 ± 0.19) × 106 | (3.20 ± 0.06) × 10−1 | 33.1 ± 1.29 |
| U-rich DNA | (2.93 ± 0.19) × 104 | (6.03 ± 0.07) × 10−1 | 20590 ± 1573 | |
| T-rich DNA | (1.45 ± 0.03) × 105 | (3.84 ± 0.04) × 10−1 | 2644 ± 82.3 | |
| TIAR12 | U-rich RNA | (4.10 ± 0.11) × 106 | (2.83 ± 0.08) × 10−3 | 0.69 ± 0.04 |
| U-rich DNA | (2.19 ± 0.02) × 104 | (4.77 ± 0.04) × 10−4 | 21.7 ± 0.38 | |
| T-rich DNA | (7.80 ± 0.75) × 106 | (4.49 ± 0.44) × 10−1 | 3.81 ± 0.74 |
The association and dissociation rate constants (ka and kd) were determined as global fitting parameters for a 1:1 binding model or a two-state model in the case of TIAR12 binding T-rich DNA where ka and kd represents ka1 and kd1, respectively. The equilibrium dissociation constant KD was determined as kd/ka for the 1:1 binding or 1/{(ka1/kd1) × (1 + ka2/kd2)} for the two-state binding (ka1 and kd1: association and dissociation rate constants; ka2 and kd2: forward and reverse rate constants for conformational change; ka2 = 0.0026 s−1, kd2 = 1.84 × 10−4 s−1).
Interestingly, HuR12 binding to DNA was much reduced in affinity compared to RNA. Fast dissociation rate constants were apparent for all interactions by HuR12. The affinity of HuR12 for U-rich DNA was more than 500-fold lower than for U-rich RNA (KD 20.6 µM from 33 nM; Figures 1A and 3A). HuR12 binding to T-rich DNA was also reduced in affinity, but not as dramatically. Binding to T-rich DNA was ∼10-fold higher in affinity compared with U-rich DNA (KD 2.6 µM from 20.6 µM; Figure 3B). Together, these results suggest that the 2′-hydroxyl group is important for the interaction of HuR12 with target oligonucleotide, and that, in its absence; the extra methyl in thymine can contribute towards binding.
In contrast, TIAR12 showed strong nanomolar affinities to both U-rich DNA (KD 21.7 nM; Figure 3A) and T-rich DNA (KD 3.8 nM; Figure 3B) with KD values in the nanomolar range similar to those for U-rich RNA (KD 0.7 nM; Figure 1A). The kinetics of each interaction are clearly impacted by substrate selection, with the slower dissociation rate constants observed for the U-rich sequences compared with those for the T-rich DNA sequence. Almost indistinguishable results were obtained for TIAR123 (results not shown). In the case of TIAR12 interactions with oligonucleotides, the absence of the 2′-hydroxyl group results in a 30-fold loss in affinity and the presence of the methyl in thymine impacts on the kinetics of interaction, overall enhancing binding ∼6-fold. This demonstrates a fundamental difference between the modes of interaction of the first two RRMs of HuR compared with TIAR.
SAXS analysis reveals that TIAR12 bound to RNA maintains an open/flexible conformation whereas HuR12 binds RNA with a closed conformation
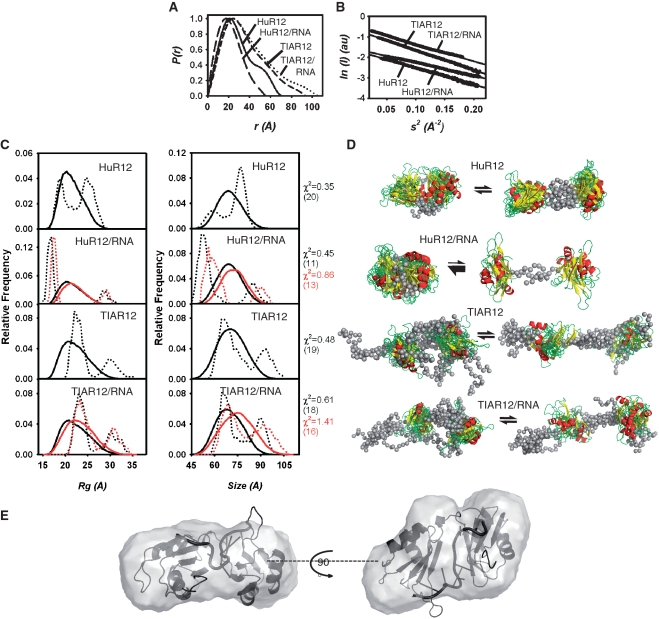
SAXS data were collected for HuR12, HuR12/RNA, TIAR12 and TIAR12/RNA in order to obtain low resolution solution structural information. A 13-nt U-rich RNA was used to permit the formation of a simple 1:1 complex. P(r) profiles calculated from the scattering data for the four samples are shown in Figure 4A. Guinier plots calculated from the scattering data for the four samples are shown in Figure 4B and show good linearity. Analysis using the EOM (63), revealed that, in all cases except for HuR12 in complex with RNA, were not consistent with a single rigid molecular conformation. This is not unexpected for molecules with separate domains that are connected by unstructured linker regions. The SAXS data were therefore analysed using EOM which allows for the coexistence of different conformations of the protein. Sets of conformers are selected using a genetic algorithm from a large number of randomly generated models that best predict the experimental data. The results of these analyses are shown in Figure 4C which show the Rg and size distribution of the ensemble of randomly generated model structures (in solid line) and the selection of structures from within the ensemble that together give rise to predicted SAXS data consistent with that experimentally obtained. Note that in the cases of protein/RNA complexes models, the analysis was done with (red lines) and without (black lines) the RNA included in the model—with very little effect on the final interpretation.
Figure 4.
SAXS analysis of (a) HuR12, (b) HuR12/RNA, (c) TIAR12 and (d) TIAR12/RNA samples. (A) P(r) profiles calculated from the scattering data for the four samples. (B) Guinier plots calculated from the scattering data for the four samples. Intensity (I) is given in arbitary units (au). (C) Ensemble optimization analysis: Rg and Rmax distributions from the best fitting ensembles calculated using EOM (63). The distribution for the pool of 10 000 conformers (solid line) and the selected best fitting ensemble (dashed line) are shown for the four samples. Black lines represent ensembles of protein models only and red lines represent ensembles of protein/RNA models. (D) superposition of the best fitting ensemble from each of the two main peaks shown as cartoon structures. The total number of structures is shown in brackets. (E) Ab initio reconstruction of HuR12/RNA complex overlayed with HuD12/RNA structure solved using X-ray crystallography (1G2E) [Rg (Guinier) = 17.1 Å, Rg (real) = 16.9 Å, Rmax = 58 Å].
The data for apo-HuR12 (Figure 4C and D) (comprised of two RRMs connected by an unstructured linker) were best fit by two populations of HuR12 structures, one more extended (Rg = 28 Å, size = 74 Å) and one more compact (Rg = 20 Å, size = 56 Å). This is consistent with a flexible protein in which RRMs are positioned at variable distances from each other over time. The SAXS data for the HuR12/RNA complex (Figure 4C and D), in contrast, were best fitted by a predominantly compact molecular shape (Rg = 16 Å, size = 51 Å). This is consistent with the HuR12/RNA complex adopting a stable uniform structure in which both RRMs are held in close proximity. In this case, where predominantly one species exists, ab initio reconstruction of the molecular shape can be performed to obtain low resolution information for the molecule or molecular complex (Figure 4E). SAXS data for HuR12/RNA complex were consistent with the structure expected based on the X-ray crystallographically derived HuD12/RNA structure (1G2E; Rg = 17.5 Å, size = 60.7 Å).
Similarly to apo-HuR12, the apo-TIAR12 data were best fit by two populations of TIAR12 structures (Figure 4C and D), one more extended (Rg = 32 Å, size = 90 Å) and one more compact (Rg = 23 Å, size = 64 Å). These greater lengths for apo-TIAR12 structures may reflect the longer linker region between the RRMs (11 in HuR12 and 14 amino acids in TIAR12) and the extra residues extending from the TIAR12 construct (five N-terminal and nine C-terminal residues). But unlike the HuR12/RNA complex, the TIAR12/RNA complex SAXS data were also best fit by two populations, similar to that seen for apo-TIAR12 (Figure 4C and D). There was no evidence that the TIAR12/RNA structure adopts a single compact structure. In order to be certain that the TIAR12 protein and 13-mer U-rich RNA used for the SAXS experiments were interacting in solution, we used size exclusion chromatography to observe complex formation (Supplementary Figure S2). The elution profiles monitored at both A260 and A280 show that TIAR12 forms a complex with the RNA of sufficient stability to shift the RNA peak, verifying the interaction. The SAXS data thus suggest that when TIAR12 interacts with U-rich 13-mer RNA, it maintains an extended flexible structure. These data indicate a fundamental difference in the interaction between the first two domains of TIAR compared with the first two domains of HuR.
DISCUSSION
Detailed kinetic analyses reveal an accurate measure of RNA-binding specificity by TIAR and HuR proteins: U- versus AU-rich RNA
In the current study, we demonstrate that TIAR and HuR proteins are able to interact with both U- and AU-rich RNA with KD values in the nanomolar range, but exhibit different affinities and kinetics of interaction. A simple inspection of KD suggests that both proteins bind to U-rich RNA sequences with higher affinity than to AU-rich sequences (Figure 1). This is consistent with findings by Park-Lee and colleagues, who reported that HuD protein, a close homologue of HuR, binds U-rich RNA with higher affinity than AU-rich RNA (57). Similarly, SELEX studies by Dember and colleagues showed that TIAR proteins preferentially bind to U-rich sequences (47). The apparently higher affinities for U-rich RNA, however, are a reflection of the much higher association rate constants for U-rich RNA. This may be partly explained by possible secondary structure formation to which the AU-rich sequence could be predisposed (67), but is better explained by the higher number of effective binding positions on the U-rich RNA which would be expected to proportionally increase the association rate constant. Indeed, our comparison of binding to U-rich sequences of increasing length show overall affinities and association rate constants increasing roughly proportionally to the increase in the number of possible binding sites.
These data help to explain some of the discrepancies between in vivo and in vitro studies of Hu and TIA protein interactions with target RNA. Whilst there may be a greater probability of these proteins forming a productive interaction at a U-rich site, the interaction with an AU-rich site, once formed, may be more stable. Hence, HuR has been shown to bind class I, II and III AREs, but is reported to enhance the stabilization of messages containing class I and class II (AU-rich) to a greater extent than class III (U-rich) AREs (17). In co-immunoprecipitation experiments, HuR targets identified by microarray were shown to contain a U-rich motif, but with a strong occurrence of adenines at several motif positions (20). Whilst this was considered to be surprisingly more U-rich than AU-rich at the time, it can be argued that the occurrence of adenosine is significant. Immunoprecipitation experiments of TIA-1 targets also revealed a common U-rich motif in which adenosine was also present (21). The equivalent study for TIAR revealed that, under stressed conditions, TIAR bound to mRNA with a consensus motif containing both uridine and adenosine [See Supplementary Data in Kim et al. (19)].
The importance of adenosine in the RNA target of Hu proteins is also apparent from the successful crystallization of HuD in complex with c-fos and TNF-α RNA, where stability of the complex plays a big role in successful crystallization. These structures show that one adenosine is preferentially accommodated at the centre of the RNA-binding site and a second may also be accommodated, though is not critical (58). Structural information for TIA proteins bound to their RNA targets is not yet available. These studies predict that, similarly to HuR, TIAR would form a more stable complex with an AU-rich sequence rather than a U-rich sequence. HuR and TIAR show similar trends in U-rich versus AU-rich preferential binding.
These studies, however, have shown differences between the HuR and TIAR when the hinge region and third RRM are removed. Our studies show that when RRM3 and the hinge region are removed from HuR, there is a dramatic loss in affinity (∼1000-fold), particularly apparent in the faster dissociation rate constant. It would thus appear that the hinge region and/or third RRM is very important to ARE binding by HuR. This is consistent with the finding of Fialcowitz-White and colleagues who showed that the hinge region contributes significantly to the HuR interaction with AU-rich RNA (56). In contrast, the affinity difference between TIAR123 and TIAR12 was minimal, suggesting that the hinge region and third RRM of TIAR are of little importance to the interaction with AREs. It must be pointed out that the domain boundaries of the HuR12 and TIAR12 constructs differ slightly. Whilst the HuR12 construct is truncated immediately following the RRM2 structural motif, the TIAR12 construct extends nine residues beyond this. It remains a possibility that this portion of TIAR's hinge region plays a role in the interaction with target RNA. This will be the subject for future investigation. The role of the RRM3 of both proteins remains to be elucidated. It has been speculated that TIAR RRMs could play a role in binding to other RNA sequences (47) and it was shown that HuR RRM3 is required for cooperative assembly of protein oligomers on RNA substrates (56).
The reported ability of TIAR to bind DNA as well as RNA prompted us to explore this interaction in comparison to that of HuR. TIAR12 and HuR12 interactions with U-rich and T-rich DNA were investigated using SPR and compared with the interactions measured for U-rich RNA. Both proteins were able to interact with DNA, but HuR12 bound with orders of magnitude lower affinity whereas TIAR12 binding remained in the nanomolar range. HuR12 preferentially bound U-rich RNA over U-rich DNA suggesting an important involvement of the 2′-hydroxyl groups. This is consistent with structural information for the HuD12/RNA complex which shows that several 2′-hydroxyl groups from target RNA form intermolecular contacts to HuD12 (58). Interestingly, HuR12 bound T-rich DNA with higher affinity than U-rich DNA. It would appear that in this case, the additional methyl group compensates, to some extent, for the loss in affinity affected by the removal of the 2′-hydroxyl group.
In contrast to HuR12, TIAR12 bound to U-rich and T-rich DNA without such a great loss in affinity compared with U-rich RNA binding. TIAR bound to U-rich DNA with 30-fold reduced affinity compared with U-rich RNA, suggesting that the 2′-hydroxyl group plays a less critical role in the TIAR/RNA interactions than it does in HuR/RNA interactions. TIAR12 also interacted with T-rich DNA with low nanomolar affinity but with different kinetics (Table 4). This suggests that the addition of methyl groups impacts the TIAR interaction with oligonucleotides—allowing both the association and dissociation to occur more readily. These results differ from affinity measurements obtained using a UV-cross-linking method reported by Suswam et al. (40) in which it was found that TIAR bound T-rich DNA with higher affinity (KDapp = 1.6 nM) than U-rich RNA (KDapp = 9.4 nM). These differences are unlikely to be due to the absence of RRM3 in our experiments, as almost identical SPR results were obtained using the TIAR123 construct (data not shown). Differences in the experimental set up may account for this reversal in apparent binding preference. In the UV-cross-linking study, the target DNA was an extended oliogonucleotide of 40 bases, whereas the target RNA was half the length. It is possible that the selected sequence and length of the oligonucleotide may have contributed to the apparently higher affinity of TIAR to the DNA sequence. The current study represents a more direct comparison of binding affinities of TIAR to DNA and RNA, and suggests that TIAR interacts with U-rich RNA with higher affinity than it interacts with DNA. Still, both interactions are in the nanomolar range and shuttling between T-rich DNA and U-rich RNA, as proposed by Suswam et al. is highly plausible (40).
TIAR and HuR bind their RNA targets in fundamentally different ways
SAXS (Small Angle X-ray Scattering) was employed to obtain further insight into the potential mode of interaction of TIAR12 and HuR12 with target RNA sequences in solution. This revealed a striking difference in the shape of the structure between TIAR12 and HuR12 upon their complex formation with the 13-mer U-rich RNA. SAXS data for TIAR12–RNA complexes are consistent with an elongated shape that is best explained if only one RRM is interacting with the RNA. It is possible that TIAR12 interacts with the RNA via only RRM2. Dember et al. (47) showed that RRM2 of TIAR is both sufficient and necessary for binding to U-rich RNA and could not detect RNA binding by RRM1 alone. They did measure slightly higher affinity by TIAR12 (KD = 40 nM) than by TIAR2 alone (KD = 50 nM) using nitrocellulose filter binding assays, suggesting that the RRM1 may contribute to binding to RNA, but the effect is negligible and may not represent a sufficiently stable interaction with the RNA to be observed by SAXS. The current data bring into question the role of RRM1 of TIAR in binding RNA, which will be addressed in future work. It is possible that, as suggested by Suswam et al. (40), the primary role of RRM1 is to interact with DNA.
The HuR12/RNA complex on the other hand, adopted a globular or more closed conformation (Figure 4B) than TIAR12/RNA, in agreement with the crystal structure of HuD12 in complex with 11-mer ARE in which the RNA is sandwiched between the RNA-binding surfaces of two RRMs (58). In the case of Hu proteins it is well documented that primary interactions with RNA occur via RRM1 and that these are augmented by RRM2 (55). Thus overall, these results strongly support the view that the RNA-binding proteins TIAR and HuR, though they share a similar triple RRM domain structure, interact with RNA targets in fundamentally different ways.
CONCLUSION
We have demonstrated that the RNA-binding regions of TIAR and HuR both readily bind AREs with nanomolar affinity, with AU-rich sequences interacting as well as or better than U-rich sequences as seen through similar or slower dissociation rate constants. However, the modes of recognition by these two proteins differ with respect to the contributions to binding made by their different RRM domains and their ability to bind to DNA versus RNA. These distinguishing features would not have been apparent from measurements of affinity alone, but are revealed upon examination of rates of interaction availed by SPR. The fundamental differences in the mode of RNA interaction by these TIAR and HuR may underlie the differences observed in their repertoire of target transcripts, as well as their distinct roles as in the nucleus and cytoplasm. Further studies involving biophysical and higher resolution structural methods will certainly help us to better understand the molecular mechanism underlying their differences, and the basis for the dynamic interplay regulating gene expression in cells.
COPYRIGHT LICENCE
In order to make a direct comparison between SPR sensorgrams showing the interaction of TIAR and HuR with U-rich RNA, we have reproduced sensorgrams from a previous publication [from Figure 3 in Kim et al., (2007) Mol. Cell. Biol. 27, 6806–6817]. Permission to re-use these sensorgrams has been granted by the copyright holder, the American Society for Microbiology, under License number 2331741184621.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Australian Research Council (DP0879279 awarded to M.C.J.W., J.A.W., M.G. and B.R.G.W.); National Health and Medical Research Council of Australia fellowship (to M.C.J.W.); National Institute on Aging-Intramural Research Program, National Institutes of Health (to M.G.); National Cancer Institute, National Institutes of Health (R01 CA102428 to G.M.W.). Funding for open access charge: Department of Biochemistry and Molecular Biology, Monash University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors also acknowledge the Australian Synchrotron where SAXS data were collected and assistance of beamline scientist Nigel Kirby.
REFERENCES
- 1.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 2.Hollams EM, Giles KM, Thomson AM, Leedman PJ. MRNA stability and the control of gene expression: implications for human disease. Neurochem Res. 2002;27:957–980. doi: 10.1023/a:1020992418511. [DOI] [PubMed] [Google Scholar]
- 3.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 6.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl Acad. Sci. USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CY, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell. Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell. Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loflin P, Chen CY, Shyu AB. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 19.Kim HS, Kuwano Y, Zhan M, Pullmann R, Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M, et al. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell. Biol. 2007;27:6806–6817. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazan-Mamczarz K, Kuwano Y, Zhan M, White EJ, Martindale JL, Lal A, Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck AR, Medley QG, O'Brien S, Anderson P, Streuli M. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res. 1996;24:3829–3835. doi: 10.1093/nar/24.19.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosset C, Boniface R, Duchez P, Solanilla A, Cosson B, Ripoche J. In vivo studies of translational repression mediated by the granulocyte-macrophage colony-stimulating factor AU-rich element. J. Biol. Chem. 2004;279:13354–13362. doi: 10.1074/jbc.M308003200. [DOI] [PubMed] [Google Scholar]
- 28.Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 29.Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J. Biol. Chem. 2004;279:12974–12979. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- 30.Masuda K, Abdelmohsen K, Gorospe M. RNA-binding proteins implicated in the hypoxic response. J. Cell Mol. Med. 2009;13:2759–2769. doi: 10.1111/j.1582-4934.2009.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson P, Kedersha N. Stressful initiations. J. Cell. Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 33.Forch P, Valcarcel J. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis. 2001;6:463–468. doi: 10.1023/a:1012441824719. [DOI] [PubMed] [Google Scholar]
- 34.Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, Parkinson J, Frey BJ, Rommens JM, Blencowe BJ. A systematic analysis of intronic sequences downstream of 5' splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–1258. doi: 10.1101/gr.073155.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izquierdo JM, Valcarcel J. Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J. Biol. Chem. 2007;282:19410–19417. doi: 10.1074/jbc.M700688200. [DOI] [PubMed] [Google Scholar]
- 36.Izquierdo JM, Valcarcel J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J. Biol. Chem. 2007;282:1539–1543. doi: 10.1074/jbc.C600198200. [DOI] [PubMed] [Google Scholar]
- 37.Le Guiner C, Lejeune F, Galiana D, Kister L, Breathnach R, Stevenin J, Del Gatto-Konczak F. TIA-1 and TIAR activate splicing of alternative exons with weak 5' splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 2001;276:40638–40646. doi: 10.1074/jbc.M105642200. [DOI] [PubMed] [Google Scholar]
- 38.Shukla S, Dirksen WP, Joyce KM, Le Guiner-Blanvillain C, Breathnach R, Fisher SA. TIA proteins are necessary but not sufficient for the tissue-specific splicing of the myosin phosphatase targeting subunit 1. J. Biol. Chem. 2004;279:13668–13676. doi: 10.1074/jbc.M314138200. [DOI] [PubMed] [Google Scholar]
- 39.Zhu H, Hasman RA, Young KM, Kedersha NL, Lou H. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 2003;23:5959–5971. doi: 10.1128/MCB.23.17.5959-5971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suswam EA, Li YY, Mahtani H, King PH. Novel DNA-binding properties of the RNA-binding protein TIAR. Nucleic Acids Res. 2005;33:4507–4518. doi: 10.1093/nar/gki763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson P, Kedersha N. On again, off again: the SRC-3 transcriptional coactivator moonlights as a translational corepressor. Mol. Cell. 2007;25:796–797. doi: 10.1016/j.molcel.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 43.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng Z, King PH, Nabors LB, Jackson NL, Chen CY, Emanuel PD, Blume SW. The ELAV RNA-stability factor HuR binds the 5'-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 2005;33:2962–2979. doi: 10.1093/nar/gki603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rivas-Aravena A, Ramdohr P, Vallejos M, Valiente-Echeverria F, Dormoy-Raclet V, Rodriguez F, Pino K, Holzmann C, Huidobro-Toro JP, Gallouzi IE, et al. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology. 2009;392:178–185. doi: 10.1016/j.virol.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 47.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 48.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian Q, Streuli M, Saito H, Schlossman SF, Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- 50.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Prusiner SB. Scrapie prions. Annu. Rev. Microbiol. 1989;43:345–374. doi: 10.1146/annurev.mi.43.100189.002021. [DOI] [PubMed] [Google Scholar]
- 52.Prusiner SB. Creutzfeldt-Jakob disease and scrapie prions. Alzheimer Dis. Assoc. Disord. 1989;3:52–78. doi: 10.1097/00002093-198903010-00007. [DOI] [PubMed] [Google Scholar]
- 53.Eisinger-Mathason TS, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shabanowitz J, Hunt DF, Macara IG, et al. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol Cell. 2008;31:722–736. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 55.Park S, Myszka DG, Yu M, Littler SJ, Laird-Offringa IA. HuD RNA recognition motifs play distinct roles in the formation of a stable complex with AU-rich RNA. Mol. Cell. Biol. 2000;20:4765–4772. doi: 10.1128/mcb.20.13.4765-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J. Biol. Chem. 2007;282:20948–20959. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park-Lee S, Kim S, Laird-Offringa IA. Characterization of the interaction between neuronal RNA-binding protein HuD and AU-rich RNA. J. Biol. Chem. 2003;278:39801–39808. doi: 10.1074/jbc.M307105200. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 59.Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson GM, Lu J, Sutphen K, Sun Y, Huynh Y, Brewer G. Regulation of A + U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 2003;278:33029–33038. doi: 10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- 61.Yeap BB, Voon DC, Vivian JP, McCulloch RK, Thomson AM, Giles KM, Czyzyk-Krzeska MF, Furneaux H, Wilce MC, Wilce JA, et al. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J. Biol. Chem. 2002;277:27183–27192. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]
- 62.Svergun D. Determination of the regularization parameter in direct-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 63.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 64.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 65.Franke D, Svergun D. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Volkov VV, Svergun D. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fialcowitz EJ, Brewer BY, Keenan BP, Wilson GM. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J. Biol. Chem. 2005;280:22406–22417. doi: 10.1074/jbc.M500618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.