Abstract
Group II introns act as both large catalytic RNAs and mobile retroelements. They are found in organelle and bacterial genomes and are spliced via a lariat intermediate, in a mechanism similar to that of spliceosomal introns. However, their distribution and insertion patterns, particularly for bacterial group II introns, suggest that they function and behave more like retroelements than organelle introns. RmInt1 is an efficient mobile intron found within the ISRm2011-2 insertion sequence in the symbiotic bacterium Sinorhizobium meliloti. This group II intron is excised, in vivo and in vitro, as intron lariats. However, the complete splicing reaction in vivo remains to be elucidated. A lacZ reporter gene system, northern blotting and real-time reverse transcription were carried out to investigate RmInt1 splicing activity. Splicing efficiency of 0.07 ± 0.02% was recorded. These findings suggest that bacterial group II introns function more like retroelements than spliceosomal introns. Their location is consistent with a role for these introns in preventing the spread of other potentially harmful mobile elements in bacteria.
INTRODUCTION
Group II introns are large catalytic RNAs with a conserved secondary structure, consisting of six domains (DI to DVI), folded into a conserved three-dimensional structure (reviewed in 1). Unlike organellar introns, most bacterial group II introns have an internally encoded (ORF within dIV) reverse transcriptase/maturase. This intron-encoded-protein (IEP) is required for the folding of the intron RNA into a catalytically active structure in vivo (2–5). Some of these group II introns act as mobile retroelements, through a target DNA-primed mechanism involving a stable ribonucleoprotein (RNP) complex containing both the intron RNA and the IEP (6–7).
Group II introns are typically spliced through two sequential transesterification reactions similar to those of nuclear spliceosomal introns (2). In the first step, the 2′-OH group of a branch point nucleotide residue, usually a bulged adenosine in dVI, attacks the 5′-splice junction, resulting in cleavage of the 5′-exon and the formation of an intron-3′-exon-branched lariat intermediate. The released 5′-exon remains associated with the intron via base pairing of the intron-binding sites (IBS1 and 2) to the exon-binding sites (EBS1 and 2) located in domain DI. In the second step, the free 3′-OH of the 5′-exon attacks the 3′-splice junction, leading to release of the intron lariat and ligation of the 5′- and 3′-exons.
The first bacterial group II intron was discovered in 1993 (8), and, by 2002, more than 200 full-length group II introns had been identified in eubacteria (9). Most of these introns were identified through sequence analysis in microbial genome sequencing projects, and some are found in Archaebacteria (10–11). Only a minority of bacterial group II introns have been unequivocally shown to be functional or to undergo splicing in vivo: Ll.ltrB from Lactococcus lactis (12–14); the Sinorhizobium meliloti RmInt1 (15); B.a.I1 and B.a.I2 from Bacillus anthracis (16); a Clostridium difficile intron (17) and the Avi.groEL intron from Azotobacter vinelandii, which inserts within the termination codon of the essential groEL gene (18,19). However, most bacterial group II introns insert between genes or directly after Rho-independent terminators (9). It is therefore possible that many of these bacterial group II introns are transcribed only weakly, if at all, in the cell. By contrast, organelle group II introns frequently insert themselves into essential genes, such as those encoding NAD dehydrogenase, ribulose-bisphosphate carboxylase and the subunits of cytochrome oxidase, which require efficient transcription and splicing (9).
RmInt1 is a mobile bacterial group II intron identified in S. meliloti, the nitrogen-fixing symbiont of alfalfa (Medicago sativa). It belongs to the IIB3 subclass (11), and has an IIB-like RNA structure with some IIA features. Moreover, unlike lactococcal and yeast introns, RmInt1 and bacterial class D introns have no C-terminal DNA-binding/DNA endonuclease region in their IEP (15,20). The mobility of the RmInt1 intron is based on a preferential pathway involving reverse splicing of the intron RNA into single-stranded DNA at a replication fork, with the nascent lagging DNA strand used as the primer for reverse transcription (21).
RmInt1 splicing has been investigated principally by analyzing excised intron products. These analyses have shown that the maturase (X domain), including the conserved C-terminal tail of the IEP, is required for splicing (22). The excised intron products have been characterized in detail and shown to consist mostly of lariat forms (23). In vitro, it has been shown that exon ligation is unusually inefficient for RmInt1 (24). This is partly because nucleotides 5 to 11 of the 3′-exon match the EBS1 5′-exon-binding site much better than the authentic IBS1 sequence in the 5′-exon (25). Furthermore exon junction products have been detected in vivo only in RT–PCR assays (15,26).
In this study, we inserted RmInt1, with its recognized exons, in frame within the coding sequence of a lacZ reporter gene. The resulting construct was expressed in S. meliloti and Escherichia coli cells. The complete splicing of RmInt1 in this new context was analyzed by investigating all the products obtained, including excised intron and ligated exons, in vivo. We found that RmInt1 splicing led to the accumulation of detectable lariat forms but that this process was inefficient, shown by a low detection of ligated exons. This inefficiency of splicing has direct consequences for the spread of bacterial group II introns, which behave more like retroelements than spliceosomal organellar introns.
MATERIALS AND METHODS
Bacterial strains and growth conditions
We used S. meliloti RMO17 (RmInt1 intron-less strain) (27) and E. coli DH5α (Bethesda Research Laboratories) for RNA extraction and β-galactosidase assays. E. coli was also used for the cloning and maintenance of plasmid constructs. S. meliloti was routinely grown at 28°C, on complete TY medium, and E. coli DH5α was grown at 37 or 28°C, on Luria-Bertani (LB) medium. When required, the medium was supplemented with antibiotics at the following concentrations: kanamycin, 200 µg/ml for S. meliloti and 50 µg/ml for E. coli; tetracycline, 10 µg/ml and ampicillin, 200 µg/ml. All plating media contained 1.5% Bacto agar.
Recombinant plasmids
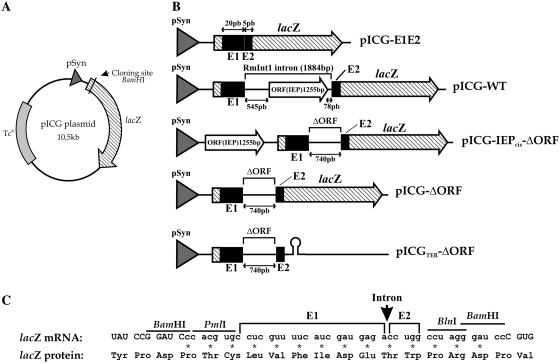
pICG plasmids are derivatives of pMP220 (28), to which the pSyn promoter (29) has been added upstream from the lacZ gene and the ribosome binding site, to ensure strong constitutive expression (29). In addition, the BlnI and polylinker BamHI sites have been removed.
pICG-E1E2 contains 25 nt corresponding to the intron flanking exons [20 bp of the 5′-exon (E1) and 5 bp of the 3′-exon (E2)] (27) from the ISRm2011-2 insertion sequence inserted into the BamHI site of the lacZ gene. pICG-WT contains the wild-type RmInt1 intron, generated in vivo by the invasion of pICG-E1E2 by the RmInt1 intron from pKG2.5 (20). pICG-DV contains a variant of the intron with a mutation in catalytic domain V, obtained from pKG2.5D5-CGA (30).
The construct used to produce the IEP protein in trans was pKGIEP (30). The series of constructs in trans generated in this study are ΔORF constructs (27), from which most of the intron ORF has been deleted. pICG-ΔORF was constructed by ligating the 0.8 kb PmlI/BlnI fragment of pKGMA4 between the corresponding sites of pICG-E1E2. pICGΔORFDV contains the ΔORF fragment with a mutation in the catalytic domain, as in pKG2.5D5-CGA (30), inserted between the PmlI and BlnI restriction sites.
The plasmids in the cis series are also based on ΔORF constructs, with the sequence encoding the IEP protein located upstream from the 5′-exon on the same plasmid. This IEP sequence was obtained by amplification from pKGIEP, with the primers 5′-GGGGAGTACTGGAAACAGGATGACTTCGGA and 5′- GGGGGCATGCTCAGGTAAACGTGTTCGTTCC, and was inserted between the ScaI and SphI sites added to the polylinker of pICG-E1E2, pICG-ΔORF and pICG-ΔORFDV, to give pICG-IEP-E1E2 (without intron), pICG-IEP-ΔORF (intron ΔORF) and pICG-IEP-ΔORFDV (CGA-mutated intron), respectively.
The plasmids of the ter series contain a synthetic Rho-independent transcription terminator (31). In these constructs, oligonucleotides 5′-GATCCCCTAGGTGAAAAAACGACAAAGCAGCACTGATTACAGTGCTGCTTTTTTTATCCCTGTG and 5′-GATCCACAGGGATAAAAAAAGCAGCACTGTAATCAGTGCTGCTTTGTCGTTTTTTCACCTAGGG were annealed and ligated downstream from the intron-flanking exons, into the BlnI/BamHI sites of pICG-E1E2, pICG-ΔORF and pICG-ΔORFDV, generating pICGTer-E1E2 (without intron), pICGTer-ΔORF (intron ΔORF) and pICGTer-ΔORFDV (CGA mutated intron), respectively.
RNA isolation and cDNA synthesis
RNA was extracted from cultures of free-living S. meliloti RMO17 and E. coli DH5α, as described earlier (23).
For reverse transcription, we annealed 10 µg of total cellular RNA with 50 ng of pd(N)6 random hexamer (Roche) in the presence of 5× first-strand reverse transcriptase buffer (Roche) and 10 mM dithiothreitol, with DEPC-treated water used to make the volume up to 8.25 µl. The mixture was first heated at 90°C for 2 min, then slowly cooled (5°C/30 s) to 25°C. cDNA synthesis was triggered by adding 0.625 mM dNTP, 15 U RNase inhibitor (Invitrogen) and 11 U AMV reverse transcriptase (Roche) and incubating for 3 h at 42°C. The reverse transcriptase was then inactivated by incubation at 85°C for 5 min. As a negative control, we replaced the enzyme with 1.75 µl of water in another reaction. After cDNA synthesis, we added 90 µl of DEPC water to the samples to give a final volume of 100 µl.
Analysis of in vivo splicing products
Primer extension analysis was performed on total RNA from RMO17 cells carrying the plasmids of the wild-type, cis and trans series, as described earlier (30). The cDNA bands corresponding to the resolved extension products were quantified with the Quantity One software package (Bio-Rad Laboratories) and excision efficiency was calculated as the the ratio of the 97 nt product from each intron construct to wild-type construct, normalized with respect to a precursor product and expressed as a percentage.
For northern blotting, total cellular RNA (5 µg) was subjected to electrophoresis in a denaturing 6% polyacrylamide gel and blotted onto a BiodyneTM membrane (Pal Corporation). The membrane was hybridized with 5′-32P-labeled oligonucleotide probes complementary to the 5′-exon (5′-AGGCCGTAATATCCAGCTGAAC), RmInt1 intron (5′-TGAAAGCCGATCCCGGAG) and the transcription terminator, representing the 3′-exon (5′-GATCCACAGGGATAAAAAAAGCAGCACTGTAATCAGTGCTGCTTTGTCGTTTTTTCACCTAGGG). The probes were radiolabeled in a final volume of 10 µl containing 75 pmol of the oligonucleotide, 20 µCi of [γ-32P]ATP (3000 Ci/mmol; 10 µCi/µl; PerkinElmer) and 10 U of T4 polynucleotide (New England Biolabs), at 37°C, for 2 h, and the labeling mixture was then passed through Sephadex G-25 columns (GE Healthcare) for probe purification. The membrane was hybridized with ∼20 000 cpm of probe/ml overnight at 42°C. It was then washed and placed against a phosphor screen, for visualization of the bands with the Personal Molecular Imager FX scanner and Quantity One software (BioRad).
β-galactosidase assays
β-galactosidase assays were performed as described by Miller (32). Briefly, overnight cultures of E. coli grown at 28 and 37°C and of S. meliloti grown at 28°C were diluted 1:50 in fresh LB and TY medium, respectively, and incubated until the cultures reached the exponential growth phase (OD600 = 0.6). Cultures were then cooled on ice for 20 min and bacterial density was recorded by measuring optical density at 600 nm. We then added aliquots of 10 µl to 300 µl of bacteria to the assay Z buffer containing 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 and 50 mM β-mercaptoethanol and adjusted the pH to 7. We added 50 µl of chloroform and 25 µl of 0.1% SDS to the sample-buffer mixture, which we then vortexed for 30 s. The samples were incubated at 28°C for 5 min and the reaction was then started by adding 0.2 ml of ONPG (4 mg/ml) (Fluka). The reaction was allowed to proceed for 10 min and was then stopped, by adding 0.5 ml of a 1 M Na2CO3 solution. Samples were centrifuged for 2 min to eliminate cell debris and absorbance was recorded at 420 nm. The results are expressed in Miller units, as described earlier (32).
SYBR green quantitative PCR
Primers were designed with Primer 3 software (http://fokker.wi.mit.edu/primer3) (33) and examined for possible secondary structures with the mfold program (http://www.bioinfo.rpi.edu/applications/mfold) (34). For lacZ1, we amplified a 149 bp fragment with 1Q_CAT (5′-CAGGAGCTAAGGAAGCTAAAATGG) and 2QR_CAT (5′-AGGCCGTAATATCCAGCTGAAC); for RmInt1 intron, a 178 bp fragment was amplified with 3Q_RIB1 (5′-GCGTAAAGCTGCGTGAATGAT) and 4QR_RIB1 (5′-TCCTCGGAGGGTTCACTTT) and, for ligated β-galactosidase gene lacZ, a 194 bp fragment was amplified with 5Q_CAT (5′-TGTACCTATAACCAGACCGTTCAG) and 6QR_LACZ (5′-GATGTGCTGCAAGGCGATT). The amplification efficiencies of all the primers were determined with serial 10-fold dilutions of the recombinant pICG-E1E2 and pICG-WT plasmids (Suplementary Figure S1). PCR efficiency was high, at 94–97.7%, with a high degree of linearity (Pearson correlation coefficient r) (Supplementary Table S1). Primer specificity was determined by melting-curve analysis and gel electrophoresis (Supplementary Figure S2).
We carried out qRT–PCR on the iCycler System (Bio-Rad). Each reaction was run in triplicate and contained 5 µl of cDNA template (equivalent to 500 ng total RNA), 0.5 mM dNTP, 5 pmol of each oligonucleotide primer, 7.5 mM MgCl2, 2.5 µl of a 1:10 000 dilution of SYBR Green, 2.3 µl of reaction buffer and 0.5 U of Platinum Taq DNA Polymerase (Invitrogen), with the final volume made up to 23 µl with DEPC water. The PCR cycling conditions were as follows: hot start, with heating at 94°C for 3 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 67°C for 30 s and extension at 72°C for 15 s. A melting curve analysis was performed in all PCR runs, to check the identity of the PCR product obtained. Controls without a template were also included. Three independent RNA preparations from each strain were tested.
Absolute mRNA levels were determined, whereas relative quantification was carried out for splicing, based on the 2−ΔΔCT method (35). Differences in relative expression were identified by carrying out Student’s t-test. Levene’s test was used to check that the variances were equal.
RESULTS
Design of plasmid constructs
We developed a splicing reporter system, by linking the splicing of RmInt1 to the constitutive expression of the lacZ gene, in pICG plasmids (see ‘Materials and Methods’ for construct descriptions and Figure 1A and B). It is not easy to insert intron RNA into reporter genes, because group II intron splicing requires base pairing between the intron sequences EBS1, EBS2 and EBS3 located in DI and the 5′- and 3′-exon sequences IBS1, IBS2 and IBS3 (EBS and IBS indicate exon-binding and intron-binding sites, respectively). We solved this problem by inserting the intron and minimal flanking exon sequences from the ISRm2011-2 insertion sequence in frame with codon 58 of the lacZ gene (Figure 1C). We used short exon sequences because they have been shown to increase RmInt1 intron excision in vitro and in vivo (24,27). In these constructs, intron splicing leads to the synthesis of a β-galactosidase protein with a short extension (14 amino acid residues) corresponding to the ligated exons, whereas, in the absence of splicing, translation is terminated due to the presence of multiple stop codons within the intron. The wild-type construct, pICG-WT, contains the full-length RmInt1 intron (1.9 kb) with the protein encoded by a sequence in DIV. pICG-IEP-ΔORF and pICG-ΔORF contain a 0.74 kb intron (27) in which the IEP is either encoded by a sequence just upstream from the 5′-exon/lacZ sequence (referred to as the cis position; Figure 1B) or constitutively produced from a compatible pKGIEP plasmid (the trans position, as described in 30). As a control for the ligated exons expected from the constructs described earlier, an intron-less construct, pICG-E1E2, consisting of the RmInt1 short flanking exons fused in frame with the lacZ gene, was used in which the IEP was either encoded by a sequence in the cis position (pICG-IEP-E1E2) or by a sequence in trans (pICG-E1E2+IEP), from compatible pKGIEP plasmids. As a negative control in these splicing assays, plasmids containing splicing-defective mutants, with a mutation in the critical conserved AGC-GUU pairing of intron RNA domain V (GUU converted to CGA, 30) were generated. These constructs were named pICGDV, pICG-IEP-ΔORFDV and pICG-ΔORFDV+IEP, for the corresponding full-length intron, and for the 0.74 kb intron with the IEP sequence expressed in cis and in trans, respectively.
Figure 1.
Constructs used in splicing analysis. (A) Schematic diagram of the pICG backbone plasmid not drawn to scale. A constitutive pSyn promoter (gray arrowhead), the E. coli lacZ gene (hatched arrow) and tetracycline resistance gene are indicated. The RmInt1 intron and flanking exon sequences are inserted into the unique BamHI site in the lacZ gene. (B) The intron-less plasmid pICG-E1E2 contains the ISRm2011-2 flanking exons (black boxes) as a control for the expected ligated exons. The pICG-WT plasmid contains the full-length RmInt1 intron with the IEP (white-filled arrow) encoded in DIV; the pICG-IEPcis-ΔORF consists of an RmInt1-ΔORF intron with the IEP sequence located upstream from the lacZ coding sequence; pICG-ΔORF consists of an RmInt1-ΔORF and pICGTer-ΔORF contains an additional Rho-independent transcription terminator 3′-to the ΔORF sequence. (C) Schematic diagram of the nucleotide sequence of the lacZ mRNA in pICG-E1E2 and the ligated exons resulting from RmInt1 splicing, presented with the corresponding amino acid sequence. This region consists of 42 nt, including 25 nt corresponding to the ISRm2011-2 flanking exons and 17 corresponding to the additional PmlI, BlnI and BamHI cloning sites linked in-frame to the lacZ gene. Upper case letters indicate the lacZ sequence, whereas lower case letter indicate the exon flanking sequences. Additional amino acids due to the addition of nucleotides to the residual lacZ remaining after intron excision are identified by asterisks. A vertical arrow indicates the site of intron insertion (wt or ΔORF) in the intron-containing constructs.
We carried out double-plasmid mobility assays to check that the pICG plasmids were fully active. The pICG-WT plasmid harboring the RmInt1 intron was transferred to the RMO17 strain (an intron-less S. meliloti strain) by conjugation, together with the acceptor construct pBB0.6+ containing the RmInt1 homing site (20). Plasmid analysis for the trans conjugants showed that almost 100% of the recipient plasmids were invaded by the RmInt1 intron (data not shown).
The in vivo excision of RmInt1
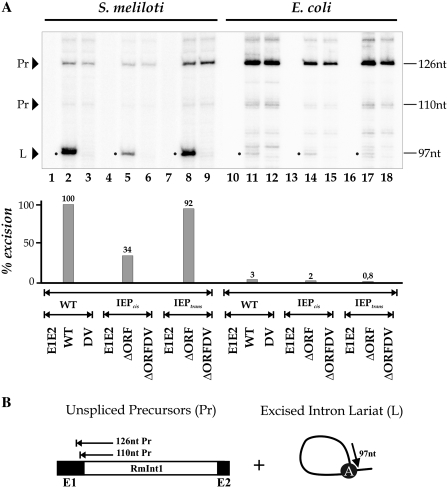
RmInt1 excision products were assessed by carrying out primer extension analysis on pICG constructs (Figure 2A), as previously described for other plasmid constructs (23,27,30). In these assays, the excised lariat intron RNA (L) is detected as an extension product of 97 nt, together with larger bands derived from unspliced precursor RNA molecules (Pr) (Figure 2B). In S. meliloti, the expected 97 nt product was obtained from constructs containing the RmInt1 intron (lanes 2, 5 and 8), whereas no band was obtained from intron-less constructs (lanes 1, 4 and 7) or from splicing-defective constructs with a mutation in DV of the intron (lanes 3, 6 and 9). RmInt1-ΔORF intron excision was three times more efficient when IEP was expressed in trans (lane 8) than when expressed in cis (lane 5), being almost as efficient as that observed with the wild-type intron construct (lane 2). This result contrasts with those for the Ll.ltrB intron, for which ΔORF intron splicing occurred at wild-type levels when the IEP sequence was expressed in cis, but at much lower levels expressed in trans from a separate plasmid (36). We analyzed the pICG constructs in E. coli, and found that the amount of excision product obtained was 95% lower than that obtained in S. meliloti, for constructs containing RmInt1 intron (lanes 11, 14 and 17). No band was observed for intron-less (lanes 10, 13 and 16) and splicing-defective (lanes 12, 15 and 18) constructs, as expected. Thus, the efficiency of RmInt1 excision depends on the bacterial host.
Figure 2.
In vivo excision of the RmInt1 intron in S. meliloti and E. coli. (A) Primer extension assays were performed on total RNA extracted from S. meliloti (lanes 1–9) and E. coli (lanes 10–18) harboring the indicated constructs. RNA was reverse-transcribed with a 32P [γ-ATP] 5′-labeled primer complementary to the first 97 nt of RmInt1. The products were analyzed by electrophoresis in a denaturing 6% polyacrylamide gel, which was dried and quantified with a PhosphorImager. The major cDNA product (97 nt, indicated with a dot) for the wild-type and ΔORF constructs (lanes 2, 5, 8, 11, 14 and 17) corresponds to the excised intron RNA. Larger products derived from unspliced precursor RNAs were also detected (110 and 126 nt). As negative controls, we used DV mutant intron constructs (lanes 3, 6, 9, 12, 15 and 18) and intron-less constructs (E1E2, lanes 1, 4, 7, 10, 13 and 16). (B) Schematic diagram of primer extension on linear unspliced precursors and excised intron lariats, showing the expected extension products. The bulged A in dVI is also indicated.
RmInt1 splicing in S. meliloti
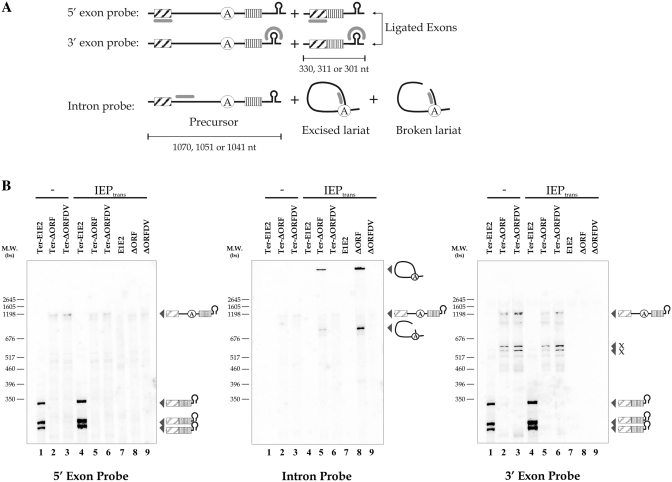
We analyzed the splicing products of the RmInt1 intron, by carrying out northern blotting (Figure 3) with S. meliloti-host RNA samples, in which in vivo excision was the most efficient (Figure 2), and RmInt1-ΔORF constructs. We also introduced a Rho-independent transcription terminator (ter series, ‘Materials and Methods’ section) into the constructs, for the identification of RNA precursors of a precise size (Figure 3B, lanes 1–6). As control for this element, we included constructs without the Rho-independent transcription terminator (lanes 7–9). A set of negative control samples, in which the IEP was absent, was used to identify the precursors and transcript forms (lanes 1–3). Precursor accumulated only for constructs containing the transcription terminator (lanes 2, 3, 5 and 6). The precursor was resolved as a triplet, because transcription is initiated by the Syn promoter at three different positions. The lower intensity of the precursor band for intron-bearing constructs (lanes 2, 3, 5 and 6) than for intron-less constructs (lanes 1 and 4, left and right panel) probably reflects a lower efficiency of the RNA polymerase, with longer templates and a greater susceptibility of the larger precursor RNA to nucleolytic degradation. Nevertheless, no traces of ligated exons were observed with the ΔORF construct complemented with the IEP (lane 5, left and right panel). Lariat intron forms were also observed for RmInt1 constructs (lanes 5 and 8, middle panel). Their electrophoretic mobility was low due to their three-dimensional configuration. The absence of ligated exons cannot be accounted for by the accumulation of intermediates of the splicing reaction steps, as no lariat-3′-exon intermediates were observed, as shown by the lack of bands on hybridization with 3′-exon probe (lanes 5 and 8, right panel). Further, a 740 nt band was shown, probably corresponding to a broken lariat form because it was detected only with the intron probe for constructs containing ΔORF complemented with the IEP in trans (lanes 5 and 8, middle panel), with no bands detected for the intron-less (lanes 1, 4 and 7, middle panel) and DV mutant (lanes 3, 6 and 9) constructs. The other bands observed on hybridization with the 3′-exon probe correspond to non-specific cleavage products (lanes 2, 3, 5 and 6, right panel, x products). Our results provide direct evidence that RmInt1 splicing in S. meliloti is inefficient, with the accumulation of the intron lariat not accompanied by the accumulation of ligated exons.
Figure 3.
RmInt1 splicing products in S. meliloti. (A) Schematic diagram showing the splicing products identified with the various probes used. The 5′- and 3′-exon probes hybridize to the precursor RNA and ligated exons, whereas the intron probe hybridizes to the precursor RNA, excised lariat intron and broken lariat intron. Exons are hatched and boxed, the intron is shown as a solid black line with a circle indicating the bulging adenosine in DVI, the transcription terminator is a stem-loop and the probes are represented by a bold gray line. The pSyn promoter includes three origins of transcription, generating three bands for the precursor or ligated exons, the size of which is indicated. (B) Northern blot hybridizations were performed with 5 µg of total RNA extracted from S. meliloti harboring the indicated pICG plasmids with (lanes 1–6; ter series) and without (lanes 7–9) the Rho-independent transcription terminator and complemented (lanes 4–9) or not (lanes 1–3) with the IEP in trans. The splicing products detected are indicated to the right of the panels. Three bands corresponding to the expected ligated exons are visible only in bacteria containing intron-less constructs of the ter series (Ter-E1E2, lanes 1 and 4). Excised and broken lariat bands are detectable only in samples containing ΔORF constructs complemented with IEP (lanes 5 and 8). Other bands corresponding to non-specific cleavage or cross-hybridization products of ∼600 nt are indicated (x).
Quantitative analysis of RmInt1 expression and splicing
No ligated exons were detected on northern blotting. We therefore used a reporter gene system to determine the extent to which exon ligation occurred during RmInt1 splicing in vivo. In this system, intron excision is linked to lacZ gene expression. Quantification was carried out in S. meliloti at its growth temperature (28°C) and in E. coli at 37 and 28°C (Supplementary Table S2). The intron-less construct of each plasmid series was used to test the system and the reaction conditions. With these constructs, β-galactosidase activity ranged from ∼4000 Miller units in the cis series in S. meliloti to 15000 Miller units in the wild-type series in E. coli grown at 37°C, demonstrating an effect of temperature on the enzymatic activity of β-galactosidase in E. coli. However, constructs harboring introns had very low levels of activity (from 5 to 20 Miller units) in both S. meliloti and E. coli and, in most cases, activity levels were similar to those obtained with the splicing-defective DV mutant. Thus, there are presumably too few ligated exons for quantification by this enzymatic method.
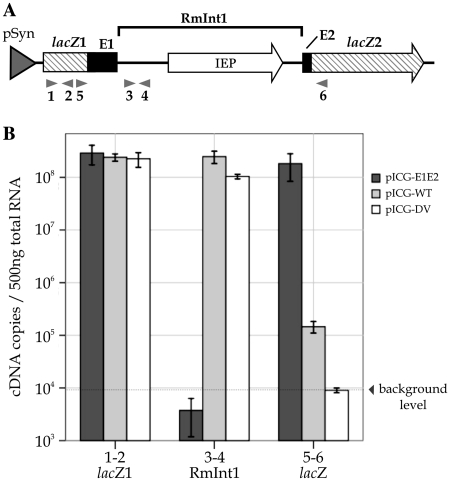
We developed a quantitative real-time reverse transcription-PCR (qRT–PCR) assay, to quantify the ligated exons generated by intron splicing and to analyze RmInt1 and lacZ transcript levels in the context of pICG plasmids, as previously carried out for Ll.ltrB in L. lactis (14). Absolute quantification was used to determine mRNA levels in the wild-type series of plasmids. Three primer pairs were used to amplify lacZ1, RmInt1 intron and ligated β-galactosidase gene lacZ produced as a consequence of the splicing of RmInt1 (Figure 4A). All transcripts displayed constitutively high levels of expression (Figure 4B). The amount of lacZ1 mRNA (primer pair 1–2), which represents total mRNA transcribed, was almost constant in all samples. The RmInt1 intron mRNA (primer pair 3–4) is produced only from constructs harboring the intron (pICG-WT and DV), both excised and being part of the precursor mRNA. RmInt1 mRNA levels were highest, due to accumulation of the more stable lariat form of the intron. The ligated β-galactosidase gene lacZ transcripts (primer pair 5–6) are produced by intron excision and ligation of the 5′- and 3′-exons containing lacZ1 and lacZ2 fragments. To avoid amplification of precursor molecules by primers 5–6, the extension step in the qRT–PCR was reduced until ligated exons were the unique product detected (‘Materials and Methods’ section). The mRNA for the intron-less construct pICG-E1E2 amplified by primers 5–6 was produced at almost the same level as that for the lacZ1, and it constituted the positive control for exon ligation. In contrast, the intron-containing construct pICG-WT generated three orders of magnitude less mRNA than pICG-E1E2 and 15 times as much mRNA as the splicing defective pICG-DV construct, which gave no distinct amplicon even if PCR was carried out over 40 cycles (not shown).
Figure 4.
Quantitative analysis of RmInt1 in S. meliloti. (A) Map of the lacZ region of pICG-WT. The RmInt1 intron with the IEP (white arrow) encoded in DIV and minimal flanking sequences from the ISRm2011-2 insertion sequence (E1 and E2) (black boxes) are located in the lacZ gene (hatched arrow). The interrupted lacZ gene is shown as lacZ1 and lacZ2 exons. Arrows under the map indicate the primers used in the real-time RT–PCR assay. Primer pair 1–2 was used to amplify the lacZ1; primers 3–4 were used to amplify the RmInt1 intron and primers 5–6 were used to amplify the ligated β-galactosidase gene lacZ. (B) Total RNA was isolated from S. meliloti containing the wild-type intron pICG-WT, the intron-less construct pICG-E1E2 and the splicing-defective mutant pICG-DV. The levels of lacZ1 (1–2), RmInt1 intron (3–4) and ligated β-galactosidase gene lacZ (5–6) mRNA were determined by an absolute quantification method. Each measurement is the mean of at least three independent RNA preparations.
We investigated the splicing efficiency of RmInt1 in S. meliloti, by calculating the ratio of the ligated lacZ gene mRNA to lacZ1 mRNA for pICG-WT and pICG-DV, normalizing the value obtained with respect to the intron-less exon-ligated control and expressing the result as a percentage. We compared the splicing efficiency of RmInt1 in S. meliloti with that in E. coli (Table 1). The splicing efficiency of RmInt1 was very low in S. meliloti, at 0.07 ± 0.02%, but was nonetheless 4.5 times higher than that in E. coli, confirming that the intron was more functional in its natural host.
Table 1.
RmInt1 splicing activity in S. meliloti and E. coli, as measured by relative quantification by qRT–PCR
| Strain and construct | Percentage of splicing ± errora |
|---|---|
| Sinorhizobium meliloti RmInt1 | |
| E1E2 | 100 |
| WT | 0.07 ± 0.02 |
| DV | 0.002 ± 0.002 |
| Escherichia coli RmInt1 | |
| E1E2 | 100 |
| WT | 0.016 ± 0.006 |
| DV | 0.004 ± 0.002 |
aThe percentage RmInt1 splicing is defined as 2−ΔΔCT where ΔΔCT = ΔCT (mRNA amplified by primer pair 5–6) − ΔCT (mRNA amplified by primer pair 1–2) expressed as a percentage with respect to each ligated-exon construct. The error is calculated formulas follows: 2−ΔΔCT · ln2 · ΣSE.
DISCUSSION
The in vivo splicing of group II introns has been studied in detail for a small number of well known group II introns, such as Ll.ltrB from L. lactis and aI5γ from S. cerevisiae (12,36–44). In most cases, these analyses were limited to the detection of the spliced form of the intron, with very few studies considering the detection (45) or quantification (14) of ligated exon forms. Group II introns often interrupt critical or, at least, functional genes of their hosts (3), affecting host viability or impairing a particular function of the cell. This characteristic has also been used to monitor splicing (38). In this study, we analyzed the splicing of the bacterial group II intron RmInt1, including intron excision and exon ligation, in S. meliloti and in E. coli, by northern blotting, β-galactosidase assays and qRT–PCR approaches. We show that the splicing of a group II intron naturally inserted into an insertion sequence (ISRm2011-2) interrupting the transposase gene is almost completely abolished, but this intron retains its invasion capacity. This suggests that bacteria may use group II introns to control the spread of other potentially harmful mobile elements.
We first developed a reporter system linking the splicing in vivo of RmInt1 intron to the expression of β-galactosidase, as previously used for other reporter genes and Ll.ltrB in E. coli (36). This system provided an intron context in which the ISRm2011-2 sequence, into which RmInt1 is inserted in its natural host, is reduced to 25 nt (20 nt for the 5′-exon and 5 nt for the 3′-exon). This intron context has been shown to improve the efficiency of RmInt1 splicing both in vivo (27) and in vitro (24–25). Splicing was also analyzed for the full-length wild-type construct and for cis constructs, in which the intron-encoded protein (IEP) sequence is located upstream from the 5′-exon, and trans constructs, in which the IEP sequence is located on a separate plasmid.
Assays of RmInt1 excision efficiency in the pICG system showed similar rates of excision for the wild-type construct and the construct in which the IEP sequence was present in trans on a separate plasmid. These findings contrast with those for LtrA in E. coli, for which lower levels of excision were reported in trans (36). As the plasmid carrying the IEP sequence in trans is present at a higher copy number than the IEP sequence in cis, the larger amount of protein produced in the trans system may rapidly become available for the binding of intron RNA, preventing degradation of the precursor RNA. RmInt1 when the IEP is in a cis position upstream from the 5′-exon was excised less efficiently, probably due to the larger distance to the promoter and the higher probability of the RNA polymerase releasing the RNA chain or dissociating from the DNA template. By contrast, RmInt1 excision in E. coli was 95% less efficient than excision of the intron from the wild-type construct in S. meliloti, suggesting that host factors may play an important role in the splicing reaction, probably during ribozyme folding, as previously reported for other group II introns (46–47).
The analysis of RmInt1 splicing products in vivo in S. meliloti confirmed that lariat form accumulates in the splicing process and that this product is free of lariat-3′-exon reaction intermediates. However, ligated exons forms, more susceptible to cell degradation and thus less prone to accumulate, were not detected. The absence of ligated exon forms contrasts with the in vivo detection of exon ligation during splicing of the lactococcal group II intron Ll.ltrB in E. coli (45), which suggests that splicing of RmInt1 may be inefficient. The lower level of accumulation of splicing products from constructs harboring a transcription terminator may be due to higher rates of RNA polymerase dissociation from short templates with a high degree of secondary structure than from templates lacking terminator sequences. We also detected a 740 nt intron band, probably corresponding to a broken lariat form. Linear group II introns have been reported to occur in vivo (48), but this form is produced predominantly during in vitro splicing reactions (49) or as a consequence of branch-site mutations in vivo (44).
The absence of ligated exons on northern blots led us to use more sensitive approaches for monitoring this product of intron RNA splicing. β-galactosidase assays in a lacZ reporter gene system have previously successfully been used for group I introns (50,51). Nevertheless, the level of exon ligation observed, in Miller units, was below background levels. Quantitative real-time qRT–PCR assays enabled us to analyze RmInt1 and lacZ mRNA levels and the intron splicing efficiency. Large amounts of lacZ1 and RmInt1 RNA were produced, but ligated lacZ gene produced three orders of magnitude less RNA, demonstrating the strong effect of RmInt1 on the genes it interrupts. We found that RmInt1 was very inefficiently spliced in vivo in S. meliloti (0.07 ± 0.02%) and in E. coli (0.016 ± 0.006%). A previous study on the splicing activity of Ll.ltrB in L. lactis (14) reported splicing efficiencies 85 to 315 times more efficient than that for RmInt1 in S. meliloti.
Our results suggest that the splicing of RmInt1 in its natural host S. meliloti, where it interrupts the transposase of ISRm2011-2, is much less efficient than that of other described bacterial group II introns (52,53). However, this inefficiency does not lead to defects in intron mobility, since a small amount of excised intron remains stable and accumulates, likely because of its association with the IEP, forming RNP particles. In fact, this intron has been shown to spread well in S. meliloti (20,26,27). In contrast, the minimal quantities of ligated exons produced during the splicing process are more likely to be susceptible to cell degradation, giving little chance of transposase translation.
Most bacterial group II introns insert into non-essential genes, but some may play a role in the cell that requires at least low levels of expression to ensure correct functioning in natural conditions. RmInt1 and, possibly, other group II introns residing in intergenic regions, within insertion sequences or after Rho-independent transcription terminators (11,54), are under no genetic pressure to maintain efficient splicing. Our findings suggest that group II introns may have an evolutionary role in circumventing splicing, thereby preventing the mobility of harmful elements in the bacterial cell.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Spanish Ministerio de Ciencia e Innovación, research grants (BIO2008-00740), CSD 2009-0006 of Programme Consolider-Ingenio 2010 and the Junta de Andalucía Proyecto de Excelencia CVI-01522 including FEDER (Fondo Europeo de Desarrollo Regional) funds, The Consejo Superior de Investigaciones Científicas, pre-doctoral fellowship (Programa I3P to I.C.). Funding for open access charge: Incentivos a Grupos de Investigación (BIO-224), Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Vicenta Millán and Ascensión Martos for technical assistance.
REFERENCES
- 1.Michel F, Costa M, Westhof E. The ribozyme core of group II introns: a structure in want of partners. Trends Biochem. Sci. 2009;34:189–199. doi: 10.1016/j.tibs.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Michel F, Ferat JL. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann K, Schmidt U. Group II introns: structural and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 4.Lambowitz AM, Zimmerly S. Mobile group II introns. Ann. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 5.Pyle AM, Lambowitz AM. Group II introns: ribozymes that splice RNA and invade DNA. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2006. pp. 469–506. [Google Scholar]
- 6.Toro N, Jiménez-Zurdo JI, García-Rodríguez FM. Bacterial group II introns: not just splicing. FEMS Microbiol. Rev. 2007;31:342–358. doi: 10.1111/j.1574-6976.2007.00068.x. [DOI] [PubMed] [Google Scholar]
- 7.Lambowitz AM, Zimmerly S. Mobile group II introns Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 8.Ferat JL, Michel F. Group II self-splicing introns in bacteria. Nature. 1993;364:358–361. doi: 10.1038/364358a0. [DOI] [PubMed] [Google Scholar]
- 9.Dai L, Toor N, Olson R, Keeping A, Zimmerly S. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–426. doi: 10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai L, Zimmerly S. ORF-less and reverse transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA. 2003;9:14–19. doi: 10.1261/rna.2126203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toro N. Bacteria and archaea group II introns: additional mobile elements in the environment. Environ. Microbiol. 2003;5:143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- 12.Mills DA, McKay LL, Dunny GM. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 1996;178:3531–3538. doi: 10.1128/jb.178.12.3531-3538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shearman C, Godon JJ, Gasson M. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis. Mol. Microbiol. 1996;21:45–53. doi: 10.1046/j.1365-2958.1996.00610.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Klein JR, McKay LL, Dunny GM. Quantitative analysis of group II intron expression and splicing in Lactococcus lactis. Appl. Environ. Microbiol. 2005;71:2576–2586. doi: 10.1128/AEM.71.5.2576-2586.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez-Abarca F, Zekri S, Toro N. Characterization and splicing in vivo of a Sinorhizobium meliloti group II intron associated with particular insertion sequences of the IS630- Tc1/IS3 retroposon superfamily. Mol. Microbiol. 1998;28:1295–1306. doi: 10.1046/j.1365-2958.1998.00894.x. [DOI] [PubMed] [Google Scholar]
- 16.Robart AR, Montgomery NK, Smith KL, Zimmerly S. Principles of 30 splice site selection and alternative splicing for an unusual group II intron from Bacillus anthracis. RNA. 2004;10:854–862. doi: 10.1261/rna.5246804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts AP, Braun V, Eichel-Streiber C, Mullany P. Demonstration that the group II intron from the clostridial conjugative transposon Tn5397 undergoes splicing in vivo. J. Bacteriol. 2001;183:1296–1299. doi: 10.1128/JB.183.4.1296-1299.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferat JL, Le Gouar M, Michel F. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol. Microbiol. 2003;49:1407–1423. doi: 10.1046/j.1365-2958.2003.03649.x. [DOI] [PubMed] [Google Scholar]
- 19.Adamidi C, Fedorova O, Pyle AM. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry. 2003;42:3409–3418. doi: 10.1021/bi027330b. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-Abarca F, García-Rodríguez FM, Toro N. Homing of a bacterial group II intron with an intron-encoded protein lacking a recognizable endonuclease domain. Mol. Microbiol. 2000;35:1405–1412. doi: 10.1046/j.1365-2958.2000.01804.x. [DOI] [PubMed] [Google Scholar]
- 21.Martínez-Abarca F, Barrientos-Durán A, Fernández-López M, Toro N. The RmInt1 group II intron has two different retrohoming pathways for mobility using predominantly the nascent lagging strand at DNA replication forks for priming. Nucleic Acids Res. 2004;32:2880–2888. doi: 10.1093/nar/gkh616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molina-Sánchez MD, Martínez-Abarca F, Toro N. Structural features in the C-terminal region of the Sinorhizobium meliloti RmInt1 group II intron-encoded protein contribute to its maturase and intron DNA-insertion function. FEMS J. 2010;277:244–254. doi: 10.1111/j.1742-4658.2009.07478.x. [DOI] [PubMed] [Google Scholar]
- 23.Molina-Sánchez MD, Martínez-Abarca F, Toro N. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J. Biol. Chem. 2006;281:28737–28744. doi: 10.1074/jbc.M602695200. [DOI] [PubMed] [Google Scholar]
- 24.Costa M, Michel F, Molina-Sanchez MD, Martinez-Abarca F, Toro N. An alternative intron-exon pairing scheme implied by unexpected in vitro activities of group II intron RmInt1 from Sinorhizobium meliloti. Biochimie. 2006;88:711–717. doi: 10.1016/j.biochi.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Costa M, Michel F, Toro N. Potential for alternative intron-exon pairings in group II intron RmInt1 from Sinorhizobium meliloti and its relatives. RNA. 2006;12:338–341. doi: 10.1261/rna.2240906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez-Abarca F, Toro N. RecA-independent ectopic transposition in vivo of a bacterial group II intron. Nucleic Acids Res. 2000;28:4397–4402. doi: 10.1093/nar/28.21.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nisa-Martínez R, Jiménez-Zurdo JI, Martínez-Abarca F, Muñoz-Adelantado E, Toro N. Dispersion of the RmInt1 group II intron in the Sinorhizobium meliloti genome upon acquisition by conjugative transfer. Nucleic Acids Res. 2007;35:214–222. doi: 10.1093/nar/gkl1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spaink HP, Okker RJH, Wijffelman CA, Pees E, Lugtenberg BJJ. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 29.Giacomini A, Ollero FJ, Squartini A, Nuti MP. Construction of multipurpose gene cartridges based on a novel synthetic promoter for high-level gene expression in Gram-negative bacteria. Gene. 1994;144:17–24. doi: 10.1016/0378-1119(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 30.Muñoz-Adelantado E, San Filippo J, Martínez-Abarca F, García-Rodríguez FM, Lambowitz AM, Toro N. Mobility of the Sinorhizobium meliloti group II intron RmInt1 occurs by reverse splicing into DNA, but requires an unknown reverse transcriptase priming mechanism. J. Mol. Biol. 2003;327:931–943. doi: 10.1016/s0022-2836(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 31.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1972. Assay of β-galactosidase; pp. 352–355. [Google Scholar]
- 33.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Cui X, Matsuura M, Wang Q, Ma H, Lambowitz AM. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J. Mol. Biol. 2004;340:211–231. doi: 10.1016/j.jmb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM. Use of targetrons to disrupt essential and non-essential genes in Staphylococcus aureus reveals temperature-sensitivity of group II intron splicing. RNA. 2006;12:1271–1281. doi: 10.1261/rna.68706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belhocine K, Mak AB, Cousineau B. Trans-splicing of the Ll.ltrB group II intron in Lactococcus lactis. Nucleic Acids Res. 2007;35:2257–2268. doi: 10.1093/nar/gkl1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19:313–319. doi: 10.1016/0092-8674(80)90505-x. [DOI] [PubMed] [Google Scholar]
- 40.Hensgens LA, Arnberg AC, Roosendaal E, van der Horst G, van der Veen R, van Ommen GJ, Grivell LA. Variation, transcription and circular RNAs of the mitochondrial gene for subunit I of cytochrome c oxidase. J. Mol. Biol. 1983;164:35–58. doi: 10.1016/0022-2836(83)90086-4. [DOI] [PubMed] [Google Scholar]
- 41.Domdey H, Apostol B, Lin RJ, Newman A, Brody E, Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984;39:611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- 42.Peebles CL, Belcher SM, Zhang M, Dietrich RC, Perlman PS. Mutation of the conserved first nucleotide of a group II intron from yeast mitochondrial DNA reduces the rate but allows accurate splicing. J. Biol. Chem. 1993;268:11929–11938. [PubMed] [Google Scholar]
- 43.Boulanger SC, Faix PH, Yang H, Zhuo J, Franzen JS, Peebles CL, Perlman PS. Length changes in the joining segment between domains 5 and 6 of a group II intron inhibit self-splicing and alter 3′ splice site selection. Mol. Cell. Biol. 1996;16:5896–5904. doi: 10.1128/mcb.16.10.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podar M, Chu VT, Pyle AM, Perlman PS. Group II intron splicing in vivo by first-step hydrolysis. Nature. 1998;391:915–918. doi: 10.1038/36142. [DOI] [PubMed] [Google Scholar]
- 45.Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G, Cavanagh S, Dunny GM, Belfort M, Lambowitz AM. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith D, Zhong J, Matsuura M, Lambowitz AM, Belfort M. Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes Dev. 2005;19:2477–2487. doi: 10.1101/gad.1345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel J, Börner T. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J. 2002;21:3794–3803. doi: 10.1093/emboj/cdf359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniels D, Michels WJ, Pyle AM. Two competing pathways for self-splicing by group II introns; a quantitative analysis of in-vitro reaction rates and products. J. Mol. Biol. 1996;256:31–49. doi: 10.1006/jmbi.1996.0066. [DOI] [PubMed] [Google Scholar]
- 50.Mercure S, Cousineau L, Montplaisir S, Belhumeur P, Lemay G. Expression of a reporter gene interrupted by the Candida albicans group I intron is inhibited by base analogs. Nucleic Acids Res. 1997;25:431–437. doi: 10.1093/nar/25.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong SH, Jeong JS, Lee YJ, Jung HI, Cho KS, Kim CM, Kwon BS, Sullenger BA, Lee SW, Kim IH. In vivo reprogramming of hTERT by trans-splicing ribozyme to target tumor cells. Mol. Ther. 2008;16:74–80. doi: 10.1038/sj.mt.6300282. [DOI] [PubMed] [Google Scholar]
- 52.Klein JR, Chen Y, Manias DA, Zhuo J, Zhou L, Peebles CL, Dunny GM. A conjugation-based system for genetic analysis of group II intron splicing in Lactococcus lactis. J. Bacteriol. 2004;186:1991–1998. doi: 10.1128/JB.186.7.1991-1998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang HR, Chao MY, Armstrong B, Wank Y, Lambowitz AM, Perlman PS. The DIVa maturase binding site in the yeast group II intron aI2 is essential for intron homing but not for in vivo splicing. Mol. Cell. Biol. 2003;23:8809–8819. doi: 10.1128/MCB.23.23.8809-8819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toor N, Robart AR, Christianson J, Zimmerly S. Self-splicing of a group IIC intron: 50 exon recognition and alternative 50 splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res. 2006;34:6561–6573. doi: 10.1093/nar/gkl820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.