Abstract
Global demethylation is required for early zygote development to establish stem cell pluripotency, yet our findings reiterate this epigenetic reprogramming event in somatic cells through ectopic introduction of mir-302 function. Here, we report that induced mir-302 expression beyond 1.3-fold of the concentration in human embryonic stem (hES) H1 and H9 cells led to reprogramming of human hair follicle cells (hHFCs) to induced pluripotent stem (iPS) cells. This reprogramming mechanism functioned through mir-302-targeted co-suppression of four epigenetic regulators, AOF2 (also known as KDM1 or LSD1), AOF1, MECP1-p66 and MECP2. Silencing AOF2 also caused DNMT1 deficiency and further enhanced global demethylation during somatic cell reprogramming (SCR) of hHFCs. Re-supplementing AOF2 in iPS cells disrupted such global demethylation and induced cell differentiation. Given that both hES and iPS cells highly express mir-302, our findings suggest a novel link between zygotic reprogramming and SCR, providing a regulatory mechanism responsible for global demethylation in both events. As the mechanism of conventional iPS cell induction methods remains largely unknown, understanding this microRNA (miRNA)-mediated SCR mechanism may shed light on the improvements of iPS cell generation.
INTRODUCTION
Somatic cell reprogramming (SCR) requires global DNA demethylation to reset cell stemness, yet the mechanism underlying this epigenetic event is unclear. Current understanding is limited by co-transfection of three or four specific transcription factors, either Oct3/4–Sox2–Klf4–cMyc or Oct3/4–Sox2–Nanog–Lin28, to promote induced pluripotent stem (iPS) cell formation (1–3). Among these reprogramming factors, POU class 5 homeobox 1 (Oct3/4) and sex determining region Y-box 2 (Sox2) are essential. Recent studies found that both Oct4 and Sox2 are also crucial for expressing mir-302 in human embryonic stem (hES) cells (4,5). Mir-302 belongs to a class of small, non-coding RNAs known as microRNAs (miRNA) that function as cytoplasmic gene silencers by suppressing translation of targeted messenger RNAs (mRNA). The majority of mir-302-targeted genes are transcripts of developmental signals and oncogenes (6); nevertheless, their interactions and overall functions remain unknown. The genomic sequence encoding mir-302 is located in the 4q25 locus of human chromosome 4, a conserved region frequently associated with longevity (7). In humans, mir-302 is predominantly expressed in hES and iPS cells, but not in differentiated cells (8,9). Loss of mir-302 has been observed prior to hES cell differentiation and proliferation during early embryonic development (8). Analogously in mice, its homologous mir-291/294/295 family presents a similar expression profile (10). Therefore, it is conceivable that embryonic stem cell-specific miRNAs such as mir-302 and mir-291/294/295 play a pivotal role in regulating cell stemness and pluripotency, whose functions may be applied to enhance the efficiency of SCR for iPS cell generation.
The initiation of SCR involves a highly coordinated DNA demethylation and histone methylation mechanism that is able to alter a genome-wide scale of chromatin structure and gene activity. To this, mir-302 may silence certain epigenetic regulators to affect the status of genomic DNA methylation. Utilizing high throughput analysis with online miRNA-target prediction programs TARGETSCAN (http://www.targetscan.org/) and PICTAR-VERT (http://pictar.mdc-berlin.de/), we found that lysine-specific histone demethylases (namely AOF, KDM or LSD) and methyl CpG-binding proteins (MECP) are two major groups of the epigenetic regulators targeted by mir-302. AOF contains two familial members AOF1 and AOF2, both of which function to repress gene transcription by demethylating histone 3 on lysine 4 (H3K4) (11–13). Inhibition of AOF2 by its antagonist tranylcypromine augments H3K4 methylation and stimulates Oct3/4 expression in embryonal carcinoma cells (11,12). In transgenic knockout mice, loss of either AOF1 or AOF2 substantially increases H3K4 methylation (13,14). AOF1-knockout mice demonstrate normal body development but fail to set up de novo DNA methylation imprints during oogenesis (13), while AOF2 deficiency causes embryonic lethality due to a progressive loss of genomic DNA methylation and lack of global cell differentiation (14). As a result, silencing of both AOF1 and AOF2 is likely to be sufficient in inducing global DNA demethylation. Our recent studies further showed that ectopic expression of the whole mir-302 familial cluster induced not only global demethylation via silencing MECP1-p66 and MECP2 but also the co-expression of Oct3/4–Sox2–Nanog genes, which led to the reprogramming of both normal and cancerous human skin cells into a hES-like pluripotent state (6,15). A similar mir-302 transfection approach was also shown to increase Oct3/4–Nanog co-expression by 2-fold in hES cells (16). Taken together, these findings suggest that mir-302 may concurrently suppress AOF1/2 and MECP1/2 to induce global demethylation and to activate the co-expression of hES-specific genes required for SCR. However, a critical link between mir-302 and AOF1/2 is still missing.
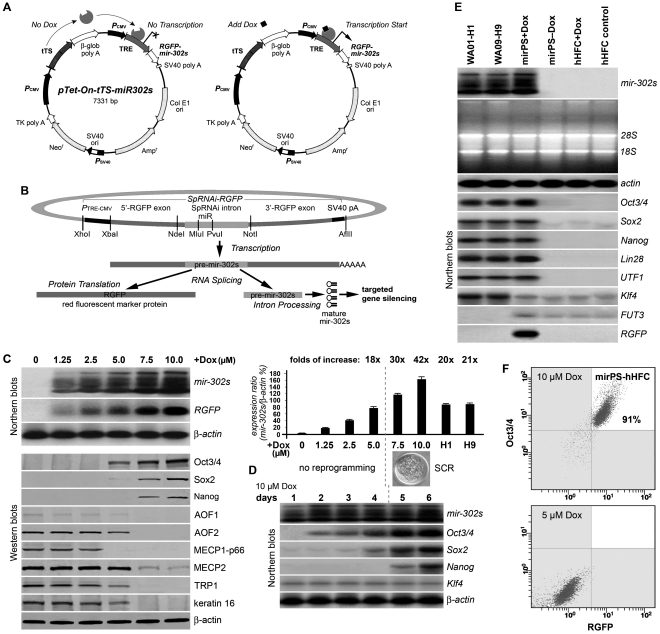
MiRNA concentration determines the efficiency of its gene targeting. To assess this dose-dependent mir-302 effect on AOF1/2 silencing and global demethylation, we adopted a novel inducible pTet-On-tTS-miR302s expression vector (Figure 1A) in conjunction with electroporation delivery to reprogram normal human hair follicle cells (hHFCs) isolated from the dermal papilla region. Skin hHFCs were chosen due to their accessibility and abundance in differentiated mesenchymal lineage cells, such as keratinocytes, melanocytes and fibroblasts. We also mimicked the natural mir-302 expression pattern by expressing all four native mir-302 familial members, mir-302a, b, c and d, in one intact intronic cluster (mir-302s; Supplementary Figure S1A) (8). Upon doxycycline (Dox) stimulation, the biogenesis of mir-302 followed the natural intronic miRNA pathway, in which mir-302 was transcribed with a gene encoded for red fluorescent protein (RGFP) and then further spliced into individual mir-302 members by spliceosomal components and cytoplasmic RNaseIII Dicers (Figure 1B) (6). MiRNA microarray analysis revealed that all mir-302 members except mir-302b* were efficiently expressed in transfected hHFCs after Dox stimulation (Supplementary Figure 1B). The procedure for generating mir-302-induced pluripotent stem (mirPS) cells is summarized in Supplementary Figure 2. Through this inducible mir-302 expression mechanism, we investigated the functional role of mir-302 in human SCR.
Figure 1.
Inducible mir-302 expression and its effect on hHFC reprogramming. (A) Construct of the Dox-inducible pTet-On-tTS-miR302s vector. (B) Mechanism of intronic mir-302 biogenesis. (C) Northern and western blot analyses of the dose-dependent mir-302 effect on the expression of core reprogramming factors and melanocytic marker genes (n = 5, P < 0.01). (D) Northern blot analysis of the time-course mir-302 effect on Oct3/4–Sox2–Nanog co-activation and SCR (n = 5, P < 0.01). (E) Northern blot analysis of hES marker gene expression induced by a high mir-302 concentration in mirPS cells compared to those in hES WA01-H1 (H1) and WA09-H9 (H9) cells (n = 5, P < 0.01). (F) FACS flow cytometry sorting of positively reprogrammed cells (mirPS-hHFC) with dual expression of mir-302 maker RGFP and hES cell marker Oct3/4.
MATERIALS AND METHODS
Cell culture and electroporation
hHFCs were isolated and dissociated from a minimum of two hair dermal papillae by 4 mg/ml collagenase I digestion for 45 min in fresh RPMI 1640 medium supplemented with 20% FBS. For culturing melanocytes, the isolated cells were cultivated in Medium 254 with the addition of human melanocytes growth supplement-2 (HMGS-2, Invitrogen, Carlsbad, CA, USA) in the absence of antibiotics at 37°C under 5% CO2. For culturing fibroblasts (hFB), the isolated cells were cultivated in Medium 106 with the addition of low serum growth supplement (LSGS, Invitrogen) in the absence of antibiotics at 37°C under 5% CO2. Cultures were passaged at 70–80% confluency by exposing cells to trypsin/EDTA solution for 1 min and rinsing once with phenol red-free DMEM medium (Invitrogen), and the detached cells were replated at 1:10 dilution in fresh Medium 254 with HMGS-2 or Medium 106 with LSGS, respectively. For electroporation, a mixture of pTet-On-tTS-mir302s (10 µg) and pTet-On-Adv-Neo(−) (50 µg) was added with the isolated hHFCs (20 000–50 000) in a hypoosmolar buffer (200 µl; Eppendorf, Westbury, NY, USA) and electroporation was performed using Eppendorf Multiporator at 300–400 V for 150 µs. The electroporated cells were first grown in phenol red-free DMEM medium (Invitrogen) supplemented with 20% knockout serum, 1% MEM non-essential amino acids, 10 ng/ml bFGF, 1 mM GlutaMax and 1 mM sodium pyruvate, for 24 h at 37°C under 5% CO2. Then, 850 µg/ml G418 and >3.75 µg/ml doxycycline (Dox) were added into the medium. The medium and antibiotics were refreshed daily for 3–5 days till the cells expressed strong red fluorescent RGFP. Next, the individual red fluorescent cell (mirPS) was monitored under a TE2000 inverted microscopic system (Nikon, Melville, NY, USA) and separately collected into a 96 well, using MO-188NE 3D micromanipulators (Nikon). In the absence of Dox, the mirPS cells were grown and passaged in knockout DMEM/F-12 medium (Invitrogen) supplemented with 20% knockout serum, 1% MEM non-essential amino acids, 100 µM ß-mercaptoethanol, 1 mM GlutaMax, 1 mM sodium pyruvate, 10 ng/ml bFGF, 100 IU/ml penicillin/ 100 µg/ml streptomycin/ 250 µg/ml G418, 0.1 µM A83-01 and 0.1 µM valproic acid (Stemgent, San Diego, CA, USA), at 37°C under 5% CO2. Alternatively, in the presence of Dox (2.5–5 µg/ml; Sigma-Aldrich, St. Louis, MO, USA), the mirPS cells were cultivated and passaged in the same feeder-free cultural condition with addition of 0.05 µM GSK inhibitor SB216763 (Stemgent). Addition of GSK inhibitor could facilitate mirPS cell proliferation but with a slight tendency to cause neural differentiation. For neural cell induction, the mirPS cells were grown in the above feeder-free cultural condition with 0.05 µM SB216763 but no Dox.
Construction of the RGFP transgene encoding mir-302s (RGFP-mir-302s)
The mir-302 familial cluster (mir-302s) was generated as reported earlier (6). The mir-302s consists of four parts, including precursor miRNAs (pre-miRNAs) of mir-302a, b, c and d. Synthetic oligonucleotides (Sigma-Genosys, St Louis, MO, USA) were listed in Supplementary Table S1. For expression, we mixed an equal amount (1:1) of the mir-302s cluster and a pre-made SpRNAi-RGFP transgene vector from our previous study (6), and then digested the mixture with MluI/Pvu1 restriction enzymes at 37°C for 4 h. The digested mixture was collected with a gel extraction filter in 30 µl of ddH2O and ligated together with T4 DNA ligase at 8°C for 16 h. This formed the RGFP-mir-302s transgene, which could be further cleaved out of the vector with XhoI/HindIII digestion.
Construction of the inducible pTet-On-tTS-mir302s vector
We first modified a Dox-inducible pSingle-tTS-shRNA vector (Clontech, Palo Alto, CA, USA) by replacing its U6 promoter with a TRE-CMV promoter isolated from a pTRE-Tight plasmid (Clontech). Then, the modified vector was digested with XhoI/HindIII restriction enzymes at 37°C for 4 h, purified by a gel extraction filter in 30 µl of ddH2O, and then mixed and ligated with the XhoI/HindIII-cleaved RGFP-mir-302s transgene (1:1) with T4 DNA ligase at 8°C for 16 h. This formed the inducible pTet-On-tTS-mir302s vector.
Immunostaining assay
Embedding, sectioning and immunostaining tissue samples were performed as reported (6). Primary antibodies included Tuj1 (Abcam Inc., Cambridge, MA, USA), ABCA2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), COL1A1 (Santa Cruz), COL2A1 (Santa Cruz), M-cadherin (Abcam), MUC2 (Abcam), MUC5B (Abcam), CD13 (Santa Cruz), CD105 (Abcam), keratin 14 (Abcam), keratin 16 (Abcam), Mitf (Abcam), TRP1 (Santa Cruz), a-SMA (Abcam), ED-A fibronectin (Santa Cruz), GAD67 (Abcam), EN4 (Abcam), SC (Abcam), BMP11 (Abcam), cTnT (Abcam), myosin HC (Abcam) and RGFP (Clontech). Fluorescent dye-labeled goat anti-rabbit or horse anti-mouse antibody was used as the secondary antibody (Invitrogen, Molecular Probes). Positive results were examined and analyzed at ×100 or ×200 magnification under a fluorescent 80i microscopic quantitation system with a Metamorph imaging program (Nikon).
Northern blot analysis
Total RNAs (10 µg) were isolated with a mirVana™ miRNA isolation kit (Ambion, Austin, TX, USA), fractionated by either 15% TBE-urea polyacrylamide gel or 3.5% low melting point agarose gel electrophoresis, and electroblotted onto a nylon membrane. Detection of mir-302 was performed with a locked nucleic acid [LNA]-DNA probe (5′-[TCACTGAAAC]ATGGAAGCACTTA-3′), while probes for other gene detection were synthesized and listed in Supplementary Table S2. All probes were purified by high-performance liquid chromatography and tail labeled with terminal transferase (20 U) for 20 min in the presence of [32P]-dATP (>3000 Ci/mM, Amersham International, Arlington Heights, IL, USA).
Anti-miRNA oligonucleotide transfection
For mir-302 knockdown assay, we encapsulated the above anti-mir-302 [LNA]-DNA probe (2–10 µg/ml) with a FuGENE HD reagent (Roche, Indianapolis, IA, USA) and applied to cell cultures at ∼30–40% confluency. Cell culture medium was refreshed 12-h post-treatment. The mir-302-knockdown rate was determined by northern blot analysis at 24-h post-treatment.
Western blot analysis
Western blotting was performed as reported (6). Major primary antibodies in this study included Oct3/4 (Santa Cruz), SSEA-1 (Santa Cruz), SSEA-3 (Santa Cruz), SSEA-4 (Santa Cruz), Sox2 (Santa Cruz), Nanog (Santa Cruz), Klf4 (Santa Cruz), Lin28 (Abcam), UTF1 (Abcam), DNMT1 (Abcam), HDAC2 (Abcam), H3K4me2/3 (Abcam), AID/AICDA (Santa Cruz), AOF1 (Abcam), AOF2 (Sigma), MECP1-p66 (Millipore, Billerica, MA), MECP2 (Abcam), TRP1 (Santa Cruz), keratin 16 (Abcam), β-actin (Chemicon, Temecula, CA, USA) and RGFP (Clontech).
Luciferase 3′-UTR reporter assay
Luciferase assays were performed using the pMir-Report miRNA Expression Reporter Vector System (Ambion), according to the manufacturer’s instruction. The mir-302 target sites (normal and/or mutant) were inserted in the 3′-UTR cloning site of the pMir-Report Luciferase Reporter vector. The two target sites were synthesized and separated by 12-CAGT-repeats. Another pMir-Report ß-gal Control vector was used as a no reporter control. We transfected 200 ng of the reporter vector into 50 000 mirPS cells in the absence or presence of Dox treatment, using a FuGene HD reagent (Roche) following the manufacturer’s suggestion. Cell lysates were harvested 48 h after transfection, and the knockdown levels of luciferase were normalized and shown by ratios of relative luciferase activity (RFA), which was calculated by the level of luciferase activity in Dox-treated (Dox-on) mirPS cells divided by that of untreated (Dox-off) mirPS cells. Mir-434-expressing cells generated by electroporating hHFCs with pTet-On-tTS-miR434-5p were served as a negative control.
Bisulfite DNA sequencing
Genomic DNAs were isolated from about two million cells using a DNA isolation kit (Roche) and 1 µg of the isolated DNAs were further treated with bisulfite (CpGenome DNA modification kit, Chemicon, Temecula, CA, USA), according to the manufacturers’ suggestions. Meanwhile, 2 µg of untreated DNAs were digested with a CCGG-cutting restriction enzyme HpaII and then analyzed by 1% agarose gel electrophoresis to determine genome-wide demethylation. The treatment with bisulfite converted all unmethylated cytosine to uracil, while methylated cytosine remained as cytosine. For bisulfite DNA sequencing analyses, we amplified the promoter regions of Oct3/4 and Nanog with PCR. Primers included 5′-GAGGCTGGAGCAGAAGGATTGCTTTGG-3′ and 5′-CCCTCCTGAC CCATCACCTCCACCACC-3′ for Oct3/4, and 5′-TGGTTAGGTTGGTTTTAAAT TTTTG-3′ and 5′-AACCCACCCTTATAAATTCTCAATTA-3′ for Nanog (1). The bisulfite-modified DNAs (50 ng) were first mixed with the primers (total 100 pmol) in 1× PCR buffer, heated to 94°C for 2 min, and immediately cooled on ice. Next, 25 cycles of PCR were performed as follows: 94°C for 1 min and 70°C for 3 min, using an Expand High Fidelity PCR kit (Roche). The amplified DNA product with a correct size was further fractionized by 3% agarose gel electrophoresis, purified with a gel extraction filter (Qiagen), and then used in DNA sequencing. A detailed profile of the DNA methylation sites was generated by comparing the unchanged cytosine in the converted DNA sequence to the unconverted one.
Microarray analysis
Microarray analyses were performed as reported earlier (6). Human genome GeneChip U133 plus 2.0 arrays (Affymetrix, Santa Clara, CA, USA) were used to detect the alterations of over 47 000 human gene expression patterns in tested cells, while miRNA microarray analyses were done by LC Sciences (San Diego, CA, USA). Total RNAs from each tested sample were isolated using a mirVana™ miRNA isolation kit (Ambion), following the manufacturer’s suggestion. The purity and quantity of isolated RNAs were assessed using 1% formaldehyde–agarose gel electrophoresis and spectrophotometer measurement (Bio-Rad). The sample signals were normalized using the total average difference between perfectly matched probes and mismatched probes. Alterations of genome-wide gene expression patterns were analyzed using Affymetrix Microarray Suite version 5.0, Expression Console™ version 1.1.1 (Affymetrix) and Genesprings (Silicon Genetics) softwares. Changes in gene expression rates >1-fold were considered as positive differential genes. For gene clustering, a plug-in program Genetrix (Epicenter Software) was used in conjunction with the Affymetrix softwares. Signals of the sample were normalized with the internal housekeeping control average in each microarray.
Implantation and teratoma formation
Approximately 5–10 mirPS or mirPS-NT cell-derived embryoid bodies (4- to 16-cell-stage) were suspended in 50 µl of a mixture of DMEM and Matrigel (2:1), followed by implantation into the uterus of a 6-week-old female pseudopregnant immunocompromised SCID-beige mouse. The pseudopregnant mice were prepared by intraperitoneal injection of 1 IU human menopausal gonadotrophin for 2 days and then human chorionic gonadotrophin (hCG) for one more day. The cells and mice were not treated with Dox before or after implantation. The mice were anesthetized with 2.5% Avertin solution, 0.4 ml per mouse during implantation. Xenografted masses were monitored 3–4 weeks after the implantation or when the sizes were grown to >100 mm3. Cysts/teratomas were dissected and the volumes were calculated using the formula (length × width2)/2. Cyst/teratoma lesions were counted, weighed and subjected to further histological analysis. Formation of teratoma-like tissue cysts was usually observed at ∼2.5-week post-implantation.
Microinjection
Microinjection was performed on the nuclei of cells plated 8–12 h before on a collagen-coated glass coverslip. Cells were microinjected using a set of Eppendorf microinjector and MO-188NE 3D hydraulic micromanipulators mounted on a Nikon TE2000 microscope system with duration and pressure settings of 0.5 s and 30 hectopascals, respectively. The microinjection buffer contained 10 µg/ml AOF2, 2 mg/ml fluorescein (FITC)-labeled dextran dye, 10 mMNaH2PO4 and 70 mM KCl, pH 7.2. After injection, cells were cultivated in the feeder-free mirPS cell cultural condition with addition of 0.05 µM SB216763.
Nuclear transfer
MirPS cell cytoplasm with different concentrations of mir-302 was prepared by varying in Dox treatments (from 1.0, 5.0, 10.0 to 20.0 µg/ml). The concentrations of cytoplasmic mir-302 were confirmed by northern blot analysis. Hybrids [mirPS-nuclear transfer (NT) cells] of mirPS cell cytoplasm and hFB nuclei were generated using a set of MO-188NE 3D hydraulic fine/coarse micromanipulators and micro-injectors/holders under a TE2000 inverted microscopic system (Nikon). The mirPS-NT cells were then grown in knockout DMEM/F-12 medium (Invitrogen) supplemented with 20% knockout serum, 1% MEM non-essential amino acids, 100 µM β-mercaptoethanol, 1 mM GlutaMax, 1 mM sodium pyruvate, 10 ng/ml bFGF, 100 IU/ml penicillin/100 µg/ml streptomycin, 0.1 µM A83-01 and 0.1 µM valproic acid, at 37°C under 5% CO2.
DNA fingerprinting
PCR was used to amplify the variable numbers of tandem repeats (VNTR) of the D1S80 locus on human chromosome 1. Genomic DNAs were isolated with a DNA isolation kit (Roche) and 50 ng of the DNAs were amplified by PCR with primers 5′-GAAACTGGCCTCCAAACACT GCCCGCCG-3′ and 5′-GTCTTGTTGGAGATGCACGT GCCCCTTGC-3′ (50 pmol each). PCR was performed as follows: one cycle at 94°C for 4 min and then 30 cycles through 94°C for 30 s, 68°C for 30 s and 72°C for 2 min, using the Expand High Fidelity PCR kit (Roche). The amplified DNAs were further fractionized by 2.5% agarose gel electrophoresis.
Statistic analysis
Any change over 75% of signal intensity in the analyses of immunostaining, western blotting and northern blotting was considered as a positive result, which in turn was analyzed and presented as mean ± SE. Statistical analysis of data was performed by one-way ANOVA. When main effects were significant, the Dunnett’s post-hoc test was used to identify the groups that differed significantly from the controls. For pairwise comparison between two treatment groups, the two-tailed student t-test was used. For experiments involving more than two treatment groups, ANOVA was performed followed by a post hoc multiple range test. Probability values of P < 0.05 was considered significant. All P-values were determined from two-tailed tests.
RESULTS
Mir-302-induced SCR is a dose-dependent mechanism involving AOF2 suppression and Oct3/4–Sox2–Nanog co-activation
Mir-302-mediated gene silencing is a dose-dependent reaction due to its mismatched targeting. Following an increase of Dox concentration up to 10 µM, we observed that transcription of mir-302 and its RGFP marker gene was proportionally elevated, while expressions of melanocytic markers tyrosinase-related protein 1 (TRP1) and keratin 16 were reduced in all transfected cells (Figure 1C). Accordingly, core reprogramming factors Oct3/4, Sox2 and Nanog were all strongly stimulated by a threshold of Dox ≥7.5 µM, indicating a dose-dependent correlation between mir-302 concentration and Oct3/4–Sox2–Nanog co-activation. This concurrent Oct3/4–Sox2–Nanog gene activation is an essential step for iPS cell induction (2,3). Time-course measurement of reprogramming hHFCs to mirPS cells further showed that the Oct3/4–Sox2–Nanog co-activation was most prominent after Days 5–6, the time frame required for initiating SCR (Figure 1D). Almost no cell division was detected during the first 3–4 days after treatment of ≥7.5 µM Dox. In this dose-dependent SCR process (Figure 1C), we discovered three critical mir-302 concentrations. First, at the treatment of 5 µM Dox, the induced mir-302 level was closely similar to that found in hES H1 and H9 cells, but not sufficient to reprogram hHFCs. Second, at 7.5 µM Dox, mir-302 concentration was raised to over 30% higher than that of H1 and H9 cells and began inducing co-expression of the core reprogramming factors Oct3/4–Sox2–Nanog and Lin28 (Figure 1E). Last, at 10 µM Dox, the stimulated mir-302 concentration was optimal for reprogramming hHFCs to mirPS cells and forming embryoid bodies. Treatment of >12 µM Dox caused severe cell cycle arrest at the G0/G1 phase, which ultimately hindered mirPS cell formation. Fluorescence-activated cell sorter (FACS) analysis further confirmed that 91% of the transfected cells treated with 10 µM Dox expressed over twice more Oct3/4 than those treated with only 5 µM Dox (Figure 1F). For the first time, this study established the threshold concentration of a hES-specific miRNA for SCR. The mechanism underlying this SCR process is consistent with the reported iPS cell induction using ectopic transfection of Oct3/4–Sox2–Nanog–Lin28 factors (2).
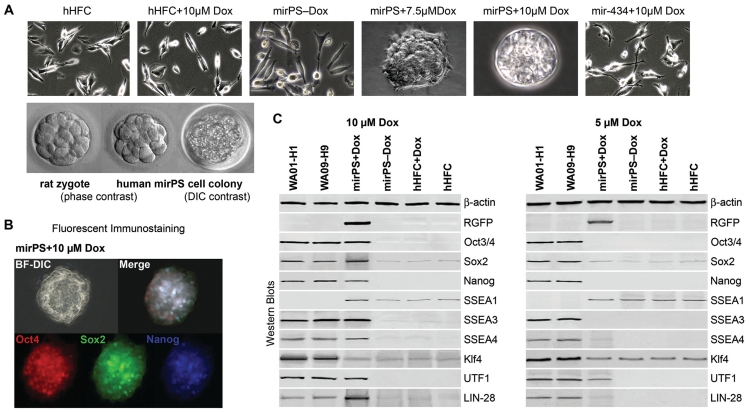
Following the increase of Dox-induced mir-302 expression, we also observed a significant change of mirPS cell colony morphology from hES-cell-like 2D aggregations (at 7.5 µM Dox) to embryoid body (EB)-like three-dimensional colonies (at 10 µM Dox), which highly resembled early mammalian zygotes (Figure 2A). After cultivated on a low attachment (overhanging) surface, these EB-like colonies formed sphere-like EBs strongly and homogeneously expressing pluripotent Oct3/4–Sox2–Nanog markers as well as other standard hES markers (Figure 2B and C), indicating that their undifferentiated status was well maintained by the induced mir-302. A similar undifferentiated status is often observed in mammalian zygotes before the morula stage, but rarely in three/four-factor-induced iPS cell colonies. This is probably due to a lower mir-302 level expressed in both human iPS and late blastocyst-derived hES cells (9). From morphology and hES-specific gene expression, there was no significant difference between mirPS cell colonies and mirPS cell-derived EBs. Accompanying the Oct3/4–Sox2–Nanog co-activation, we also found a proportional loss of multiple epigenetic regulator expression, including AOF1, AOF2, MECP1-p66 and MECP2 (Figure 1C). Western blot analysis showed that hHFCs expressed abundant AOF2 and MECP1/2 but scarce AOF1, suggesting that silencing of AOF2 plays a major role in hHFC reprogramming. Deficiency of either AOF1 or AOF2 is sufficient to cause global demethylation (13,14). Our previous studies have also demonstrated that mir-302 mediated MECP1/2 silencing enhances global demethylation (6,15). Therefore, mir-302 induces SCR through a combinatory mechanism involving both Oct3/4–Sox2–Nanog co-activation and AOF1/2 suppression, while further MECP1/2 silencing enhances the result of AOF1/2-associated global demethylation.
Figure 2.
Morphological and genetic changes of mirPS cell properties in response to different mir-302 concentrations. (A) Morphological changes of mirPS cell colonies in response to Dox-induced mir-302 expression. Mir-434-expressing cells generated by electroporating hHFCs with pTet-On-tTS-miR434-5p were served as a negative control. A morphological comparison between a morula-staged rat embryo and a mirPS cell colony at 32–64 cell stage was shown in the lower panel. (B) Fluorescent microscope examination showing strong and homogeneous expression of the core pluripotent stem cell markers Oct3/4, Sox2 and Nanog in a mirPS-derived embryoid body. BF-DIC, bright field with differential interference contrast. (C) Western blotting confirming the expression patterns of major hES-specific marker proteins in completely (10 µM Dox) and partially (5 µM Dox) reprogrammed mirPS cells compared to those found in hES H1 and H9 cells (n = 4, P < 0.01). Mir-302 expression was proportional to Dox concentration.
Mir-302 silences AOF2 to trigger DNMT1 deficiency and global demethylation
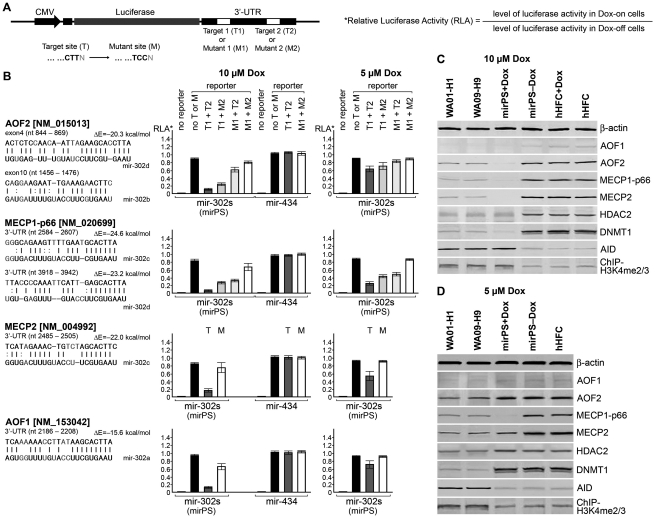
To validate the interactions between mir-302 and its epigenetic targets, we used a luciferase 3′-untranslated region (3′-UTR) reporter assay (Figure 3A), which showed that treatments with various mir-302 concentrations resulted in different epigenetic patterns in mirPS cells. In the presence of 10 µM Dox, mir-302 effectively bound to the target sites of AOF1, AOF2, MECP1-p66 and MECP2 mRNAs and successfully silenced over 80% of the reporter luciferase expression in all targets (Figure 3B). Suppression of the real target genes in mirPS cells was also confirmed by western blot analyses, consistent with the results of the luciferase 3′-UTR reporter assay (Figure 3C). Accordingly, we detected a significant decrease of DNMT1 (loss over 87 ± 3% of the level in hHFCs) and increase of H3K4 di-/tri-methylation (H3K4me2/3; gain 103 ± 17%) in response to the silencing of AOF2 (loss 93 ± 2%) by mir-302. Previous studies have demonstrated that AOF2 is required for stabilizing DNA methyltransferase 1 (DNMT1) and preserving its activity on the maintenance of global DNA methylation (14), whereas active global demethylation can promote Oct3/4–Nanog activation in early mouse embryos and mouse–human fused heterokaryons (17,18). Conceivably, the deficiency of DNMT1 caused mirPS cell genomes to be susceptible to a certain demethylation activity. This demethylation effect was further enhanced by co-suppression of MECP1/2 (loss 96 ± 1% and 81 ± 5%, respectively) (6) and eventually led to global demethylation and Oct3/4–Nanog activation (Figure 4A and B). On the flipside, a lower mir-302 concentration induced by 5 µM Dox failed to trigger any significant silencing effect on either the target sites of the reporter gene or the targeted epigenetic genes, except MECP1-p66, indicating that mir-302-induced global demethylation is dose dependent and requires co-suppression of AOF1/2 and MECP1/2 (Figure 3B and D).
Figure 3.
Analysis of mir-302-induced silencing effects on targeted epigenetic genes and global demethylation. (A) Constructs of the luciferase 3′-UTR reporter genes, which carry either two normal (T1 + T2) or two mutant (M1 + M2), or a mixture of both (T1 + M2 or M1+T2), mir-302 target sites in the 3′-UTR. The mutant sites contained a mismatched TCC motif in place of the uniform 3′-CTT end of the normal target sites. (B) Effects of Dox-induced mir-302 on the luciferase expression (n = 5, P < 0.01). (C) and (D) Western blot analyses showing the changes of major mir-302-targeted epigenetic gene expression induced by high (10 µM Dox) and low (5 µM Dox) mir-302 concentrations in mirPS cells compared to those found in hES H1 and H9 cells (n = 4, P < 0.01).
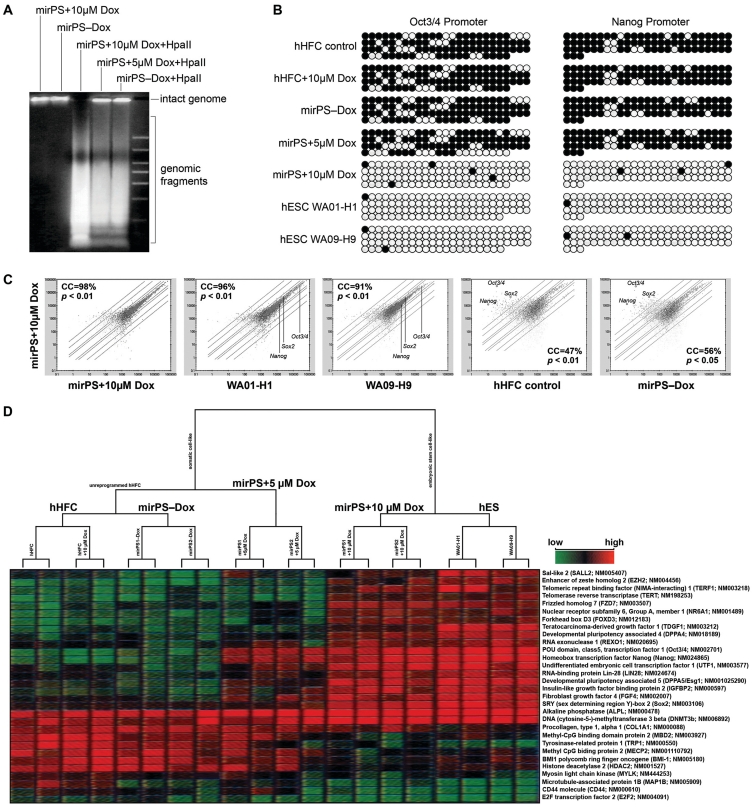
Figure 4.
Mir-302-induced genomic DNA demethylation and global gene expression alteration. (A) HpaII cleavage showing the vast loss of global CpG methylation, identified by increased presence of smaller DNA fragments, at a genome-wide scale in mirPS cells treated with 10 µM but not 5 µM Dox. (B) Bisulfite DNA sequencing in the promoter regions of Oct3/4 and Nanog. Methylation maps show the most frequently observed patterns. Black and white circles indicate the methylated and unmethylated cytosine sites, respectively. (C) Analysis of global gene expression patterns before and after global demethylation, using human genome GeneChip U133 plus 2.0 arrays (Affymetrix; n = 3, P < 0.01–0.05). (D) Microarray hierarchical clustering of the top 30 most differentially expressed hES-specific genes and epigenetic regulators between somatic hHFCs, reprogrammed mirPS-hHFCs (mirPS+10 µM Dox) and hES H1/H9 cells.
Methylation site-sensitive HpaII digestion assays confirmed that mirPS cell genomes isolated from the group treated with 10 µM Dox underwent global demethylation (Figure 4A). When further assessing the methylation patterns of Oct3/4 and Nanog promoters with bisulfite DNA sequencing, we observed that both promoters were almost completely demethylated in a fashion resembling hES H1 and H9 cells (Figure 4B). Similar global demethylation patterns have also been found in iPS cells (2,3). In contrast, neither global demethylation nor SCR was observed in the transfected cells treated with only 5 µM of Dox (Figures 1C, 4A and B). We subsequently evaluated this global demethylation effect in over 47 000 human gene expression patterns using microarray analyses and revealed that approximately half of the transcriptome expression in mirPS cells was changed from a somatic hHFC mode to a uniform hES-like expression pattern sharing over 91% similarity to that of H1/H9 cells (Figure 4C). Hierarchical clustering of the top 30 most differentially expressed hES-specific genes and epigenetic regulators in microarrays further showed an extremely high correlation between reprogrammed mirPS and hES H1/H9 cells (Figure 4D). Thus, we conclude that mir-302 regulates the epigenetic reprogramming of genomic methylation patterns through co-suppression of AOF2 and DNMT1 during SCR.
Mir-302-induced SCR is reversible and dependent on AOF2–DNMT1 suppression
After SCR completion (5–6 days post-transfection of mir-302), reprogrammed mirPS cells were visually distinguished by their sphere-shape morphology and expression of red fluorescent RGFP protein (Supplementary Figure S3A), and further selected by G418 antibiotics to ensure their purity. Consistent with our previous reports (6,15), each individual mirPS cell could grow into a homogeneous EB (Supplementary Figure S3B) and form teratoma-like tissue cysts (Figure 5C) in immunocompromised SCID-beige mice, containing various differentiated tissues derived from all three embryonic germ layers, ectoderm, mesoderm and definitive endoderm (Figure 5D). These results confirmed the hES cell-like pluripotency of mirPS cells.
Figure 5.
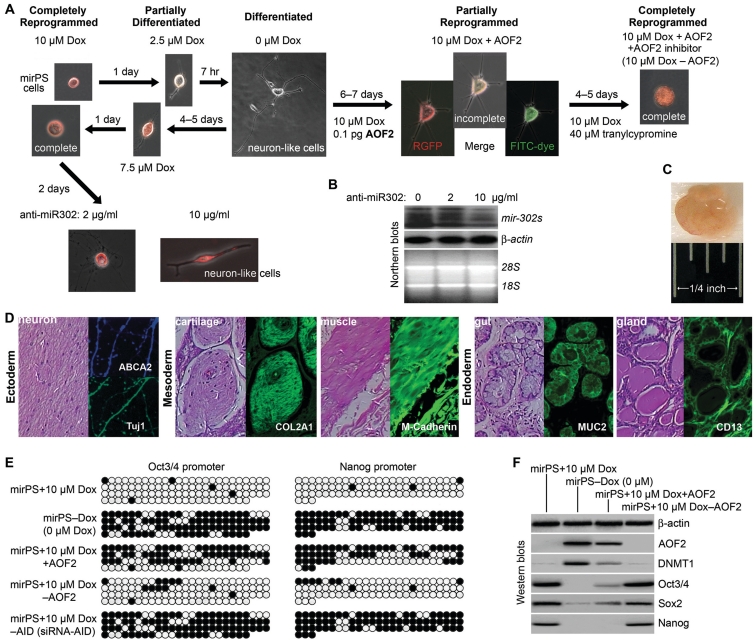
Reversible SCR process regulated by mir-302–mediated AOF2 silencing. (A) Reversible interchange between hES-like mirPS and neuron-like differentiated cells as a result of mir-302 and AOF2 counteraction. (B) Northern blots showing the mir-302 knockdown (∼65%) by 10 µg/ml anti-mir-302 LNA–DNA oligonucleotides. (C) Typical morphology of a mirPS EB-derived teratoma-like tissue cyst (2.5 weeks post-implantation). (D) Pluripotent differentiation of mirPS-hHFCs into teratoma-like cysts containing various tissues derived from all three embryonic germ layers. Immunostaining the cross-sections of mirPS EB-derived tissue cysts showed the positive results of specific differentiation markers, including neuronal β-tubulin isotype-III (Tuj1; green) and/or ATP-binding cassette sub-family A member 2 (ABCA2; blue), cartilage collagen type II α-1 (COL2A1; green), muscle-associated M-cadherin (green), gut-epithelium-related mucin 2 (MUC2; green), and glandular epithelium-related CD13 (green). Photographs were taken with Nikon TE2000 microscopic system at ×200 magnification. (E) Global demethylation patterns in Oct3/4 and Nanog promoters corresponding to the result of (A). (F) Changes of Oct3/4, Sox2, Nanog, AOF2 and DNMT1 expression in response to the results of (A) and (E).
Next, we evaluated the role of mir-302–targeted AOF2 silencing in the process of SCR using recombinant AOF2 protein and its inhibitor tranylcypromine (Figure 5A). Under conditions of 10 µM Dox stimulation, the success rate of complete SCR approached 100% (477/480). After that, the reprogrammed mirPS cells could be cultivated to over 26–28 passages under our feeder-free cultural condition in the absence of Dox and GSK inhibitor (‘Materials and Methods’ section), indicating the completion of SCR. However, when a GSK inhibitor was presented in the cultural medium without sufficient Dox stimulation, majority of mirPS cells differentiated into neuron-like cells. Given that glycogen synthase kinase 3 (GSK3) is a major gatekeeper for embryonic neural induction (19), this result suggests that GSK inhibitor can induce the ectodermic differentiation of mirPS cells. Notably, these differentiated cells could be reprogrammed back to mirPS cells after re-supplementation of ≥7.5 µM Dox in the cultural medium. The same neuronal differentiation could also be triggered by treatment of 10 µg/ml anti-mir-302 LNA–DNA oligonucleotides in mirPS cells (Figure 5A and B), indicating the critical role of mir-302 concentration in maintaining pluripotent cell stemness. Alternatively, while being microinjected with recombinant AOF2 into the cell nuclei, the differentiated cells failed to be reprogrammed back to mirPS cells even after 10 µM Dox stimulation, demonstrating the inhibitory role of AOF2 in the mechanism of mir-302-induced SCR. Further treatment of tranylcypromine removed the blockade of AOF2 in the process of SCR and, in conjunction with the stimulation of 10 µM Dox, could again reprogram the differentiated cells to mirPS cells with all hES-like properties. Microinjection of blank buffer did not cause any effect in all tests. Therefore, SCR is a reversible mechanism dependent on mir-302–mediated AOF2 silencing.
Following this reversible SCR process, we measured the corresponding changes of global demethylation, AOF2–DNMT1 co-suppression and Oct3/4–Sox2–Nanog co-activation in all tested mirPS and differentiated cells. Bisulfite DNA sequencing assays revealed that, under the GSK inhibitor influence, the global demethylation of Oct3/4 and Nanog promoters were observed only in mirPS cells treated with ≥7.5 µM Dox (mirPS+10 µM Dox) and the re-reprogrammed cells co-treated with 10 µM Dox, AOF2 and tranylcypromine (mirPS+10 µM Dox–AOF2), whereas the differentiated cells treated with 10 µM Dox and AOF2 (mirPS+10 µM Dox+AOF2) failed to complete the demethylation (Figure 5E). These resulting global demethylation patterns perfectly matched the morphological changes observed in Figure 5A. In addition, western blot assays also showed that the simultaneous AOF2–DNMT1 co-suppression and Oct3/4–Sox2–Nanog co-activation only occurred in the mirPS+10 µM Dox and mirPS+10 µM Dox–AOF2 cells but not in mirPS+10 µM Dox+AOF2 cells (Figure 5F), confirming that mir-302–targeted AOF2 silencing is required for global demethylation during SCR. In accordance to this counteraction between mir-302 and AOF2, we identified that SCR is a reversible process.
Global genomic DNA demethylation does not require any nuclear effector
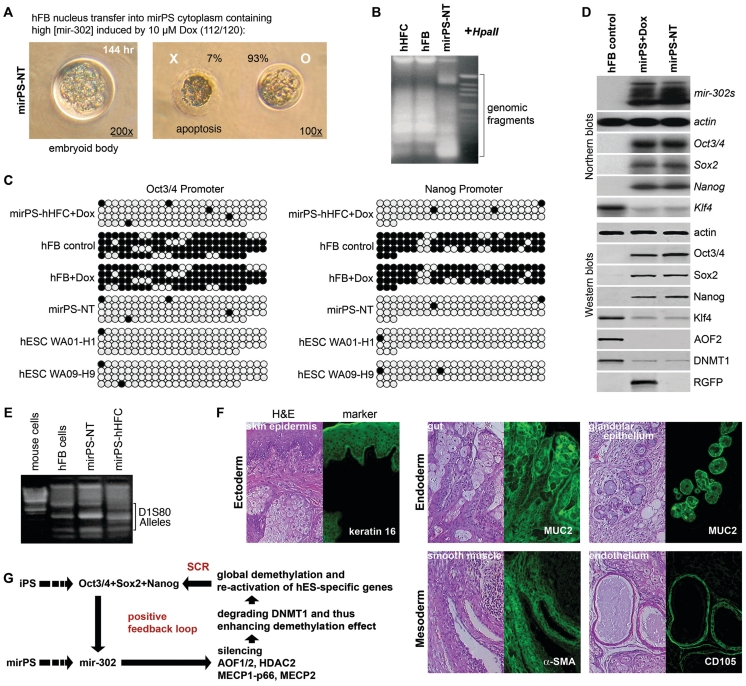
Mir-302 is a cytoplasmic effector. To further rule out the possible involvement of any nuclear factor from the mir-302–mediated global demethylation, we transferred human adult fibroblast (hFB) nuclei into the cytoplasm of mirPS cells pre-treated with 10 µM Dox. Most (93%) of the hybrid cells successfully formed mirPS-like iPS cells and embryoid bodies (Figure 6A). When cytoplasm derived from the mirPS cells pre-treated with <7.5 µM Dox, neither mirPS-like cell nor embryoid body was developed. Conversely, transfer of mirPS nuclei into hFB cytoplasm also failed to form any viable cells. Thus, the reprogramming ability of mirPS cells is preserved in the cytoplasm rather than nucleus. These hFB nucleus-transferred mirPS (mirPS-NT) cells preserved all the same traits as mirPS cells in terms of global demethylation (Figure 6B and C), Oct3/4–Sox2–Nanog co-activation and AOF2–DNMT1 co-suppression (Figure 6D), and in vivo pluripotency (Figure 6F and Supplementary Figure 4). Using DNA fingerprinting in human D1S80 alleles, the mirPS-NT-derived teratoma cysts were confirmed to be originated from human cell sources rather than mouse cells (Figure 6E). In view of these hES-like properties inherent in mirPS-NT cells, we conclude that mir-302 is able to not only induce SCR but also preserve this reprogramming ability in the cytoplasm of mirPS cells.
Figure 6.
Assessment of the reprogramming ability preserved in mirPS cytoplasm. (A) Embryoid body derived from a single mirPS–hFB hybrid (mirPS-NT) cell after limiting dilution. Scale bars = 100 µm. (B) HpaII cleavage showing global demethylation (smaller DNA fragments) in reprogrammed mirPS-NT cells compared to that in the somatic hHFC and hFB cells. (C) Bisulfite DNA sequencing in the Oct3/4 and Nanog promoters, showing CpG methylation maps. Black and white circles indicate the methylated and unmethylated cytosine sites, respectively. (D) Northern and western blot analyses of mir-302, AOF2–DNMT1 and Oct3/4–Sox2–Nanog expression patterns in mirPS-NT cells compared to those in the somatic hFB and reprogrammed mirPS (mirPS+Dox) cells. (E) DNA fingerprinting of the variable numbers of tandem repeats (VNTRs) in the D1S80 locus of human genome, confirming that mirPS-NT, mirPS-hHFC and hFB cells all originated from the same human source not mouse. (F) Pluripotent tissue types found in teratoma-like cysts derived from mirPS-NT cells. Markers (green): keratin 14, actin α-2 in smooth muscle aorta (a-SMS, ACTA2), CD105 and MUC2. H&E, histological staining with hematoxylin and eosin. BF, bright field. (G) Proposed mechanism of mir-302–induced SCR.
DISCUSSION
Our previous study established that mir-302 functions not only to improve the efficiency of SCR but also to enhance the stemness and pluripotency of the reprogrammed somatic cells (6,15). In this study, we further revealed the mechanism involved (Figure 6G), showing that mir-302 significantly decreases AOF2 and DNMT1 activities and, in conjunction with the co-suppression of MECP1/2, results in global genomic DNA demethylation and H3K4 modification. Subsequently, these epigenetic reprogramming events induce hES-specific gene expression, in particular Oct3/4, Sox2 and Nanog, which in turn further stimulates mir-302 expression, and so on to form a positive feedback cycle essential for maintaining SCR. When compared to the previous three/four-factor reprogramming methods, our approach adopts a novel entry point in the same mechanistic cycle to complete SCR. Also, without introducing the oncogenic Klf4 and c-Myc genes, this new approach likely provides safer in vivo applications. From this novel SCR mechanism, we learned that the effector(s) responsible for reprogramming genomic epigenetics resides in the mirPS cytoplasm and can enter somatic cell nuclei after NT because mir-302 is a cytoplasmic effector. Thus, our findings may also help in clarifying the mechanism of somatic cell NT, addressing the possible role of mir-302 in nuclear reprogramming.
Through deciphering the interactions between mir-302 and its targeted epigenetic genes, we elucidate the intricate gene regulation circuitry involved in epigenetic reprogramming during SCR. Global demethylation naturally occurs in two developmental periods—at the initial stage of gametogenesis and during early embryogenesis up until the morula stage (20). The mir-302 expression pattern in human embryos has not been studied due to ethical and legal conflicts. However, its homologous counterpart mir-291/294/295 in mice was found to present most abundantly during these two developmental periods (21). Previous studies in mice have also shown that deficiency of AOF2 and DNMT1 activities occurs most significantly during the same embryogenesis period of time (14). To this, our finding of mir-302-targeted AOF2 silencing connects the relationship of all these events and further extends the presence of this epigenetic reprogramming mechanism in iPS cells during SCR. In this extended model, cell stemness, analogous to a fountain of youth, is inherent in our somatic cells and can be activated and regulated by mir-302, providing a feasible means to rejuvenate cell stemness and pluripotency through mimicking the natural reprogramming pathway. However, our current work only reveals its partial functionality. To fully utilize this profound mechanism of mir-302, further studies are still needed to realize its potential in stem cell research and regenerative medicine.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The WJWU and LYNN institutional fund for regenerative medicine and biobanking development.
Conflict of interest statement. None declared.
REFERENCES
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 4.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin SL, Chang D, Chang-Lin S, Lin CH, Wu DTS, Chen DT, Ying SY. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puca AA, Daly MJ, Brewster SJ, Matise TC, Barrett J, Shea-Drinkwater M, Kang S, Joyce E, Nicoli J, Benson E, et al. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc. Natl Acad. Sci. USA. 2000;98:10505–10508. doi: 10.1073/pnas.181337598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 12.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 15.Lin SL, Ying SY. Role of mir-302 microRNA family in stem cell pluripotency and renewal. In: Ying SY, editor. Current Perspectives in MicroRNAs. NY: Springer Publishers press; 2008. pp. 167–185. [Google Scholar]
- 16.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papadimou E, Moiana A, Goffredo D, Koch P, Bertuzzi S, Brüstle O, Cattaneo E, Conti L. p66(ShcA) adaptor molecule accelerates ES cell neural induction. Mol. Cell. Neurosci. 2009;41:74–84. doi: 10.1016/j.mcn.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 21.Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.