Abstract
Plasmids are autonomously replicating extrachromosomal DNA molecules that often impart key phenotypes to their bacterial hosts. Plasmids are abundant in marine bacteria, but there is scant knowledge of the mechanisms that control their replication in these hosts. Here, we identified and characterized the factors governing replication of a new family of plasmids from marine bacteria, typified by the virulence-linked plasmid pB1067 of Vibrio nigripulchritudo. Members of this family are prevalent among, yet restricted to, the Vibrionaceae. Unlike almost all plasmid families characterized to date, the ori regions of these plasmids do not encode a Rep protein to initiate DNA replication; instead, the ori regions encode two partially complementary RNAs. The smaller, termed RNA I, is ∼68-nt long and functions as a negative regulator and the key determinant of plasmid incompatibility. This Marine RNA-based (MRB) plasmid family is the first characterized family of replicons derived from marine bacteria. Only one other plasmid family (the ColE1 family) has previously been reported to rely on RNA-mediated replication initiation. However, since the sequences and structures of MRB RNA I transcripts are not related to those of ColE1 replicons, these two families of RNA-dependent replicons likely arose by convergent evolution.
INTRODUCTION
Plasmids are autonomously replicating extrachromosomal DNA molecules that often impart key phenotypes, such as toxin production, to their bacterial hosts (1). Replication of bacterial plasmids, like that of chromosomes, is subject to stringent regulatory mechanisms that promote their maintenance at a consistent copy number within the cell (2,3) These processes enable initiation of plasmid DNA synthesis and prevent ‘runaway’ replication, which can be toxic to the host bacterium. Furthermore, regulation of copy number helps to ensure that there are sufficient plasmids for segregation to daughter cells. Typically, the factors that enable and govern plasmid replication are encoded within a single region of the replicon, adjacent or close to the cis-acting origin of replication (ori) (2). Thus, the origin region is usually necessary and sufficient for propagation of the element in the appropriate host. Plasmid replication is not truly autonomous, in that chromosome-encoded factors, such as DNA polymerase, also play an essential role. However, initiation of replication—the step at which the process is always regulated—is dependent upon sequences, and the factors that they encode, contained within the origin region.
Plasmids are widespread within the bacterial kingdom, and there is considerable diversity among their origin sequences. Despite this heterogeneity, it is possible to broadly classify them into two distinct categories, based upon whether initiation of synthesis of plasmid DNA depends upon a plasmid encoded replication protein (Rep) or RNA species (3). The vast majority of characterized plasmids, including plasmids that replicate by very distinct processes (e.g. theta versus rolling circle replication), encode a Rep protein, and they have evolved a variety of mechanisms to control Rep expression and/or activity (2,3). To our knowledge, RNA-mediated plasmid replication initiation has only been observed for ColE1 and related plasmids from the Enterobacteriaceae (2). Replication of ColE1-family plasmids is dependent upon transcription of an origin-proximal RNA (RNA II) that can, under permissive conditions, serve as a primer for plasmid DNA synthesis (4). The activity of RNA II is negatively regulated by a small (110 nt) antisense RNA (RNA I), which is complementary to the 5′-end of RNA II (5,6). Modulation of RNA II secondary structure by RNA I, which is itself folded into a complex structure, inactivates RNA II, and thereby prevents initiation of replication (7,8).
The regulatory factors and processes that govern plasmid copy number control also inhibit stable co-existence of multiple plasmids with identical origins (9). Two plasmids that share origin regions are generally ‘incompatible’; cross-talk between their oris and regulators typically results in loss of one of the two replicons. In contrast, multiple plasmids subject to distinct regulatory processes are compatible and can readily be maintained within a bacterial lineage.
Besides encoding activities that enable their replication and maintenance, plasmids carry highly diverse gene repertoires that impart a vast array of phenotypes to their hosts (1). Since plasmids are often transmissible, they can enable dissemination of these phenotypes, such as antibiotic resistance, within bacterial populations. Although plasmids have been detected within a broad cross-section of bacterial species, most well characterized plasmids originated within species with clinical relevance for human populations. Mechanistic analyses have also been restricted to plasmids that can reside within bacterial species amenable to culture, which excludes, among others, the majority of marine bacterial species. However, sequence analyses have demonstrated that marine isolates contain abundant and diverse plasmid replicons that in many cases appear to be unrelated to previously described incompatibility groups (10).
We recently reported that an 11-kb plasmid, pSFn1 (now referred to as pB1067) is necessary for the virulence of Vibrio nigripulchritudo, a shrimp pathogen (11). This plasmid is highly similar to pAK1 (13.5 kb), which was isolated from the coral pathogen V. shiloni (12). However, sequence analyses did not reveal homology between pB1067 (or pAK1) and a known Rep protein or incompatibility group, suggesting that replication of this plasmid might be governed by novel sequences (11).
In this study, we identified the pB1067 ori and characterized the process by which pB1067 replication is regulated. We found that a 500-bp fragment of pB1067 is sufficient to mediate autonomous replication in V. nigripulchritudo and several other species of Vibrionaceae. The 500 bp pB1067 fragment that enables replication does not contain any recognizable protein coding sequences, but does encode two RNAs, which are transcribed from opposite strands. The smaller RNA (RNA I; 68 nt) is fully complementary to the larger transcript (RNA II) and functions as a negative regulator. We identified sequences homologous to this RNA I sequence within 24 additional plasmids isolated from at least six other Vibrionaceae species and confirmed that they are subject to a similar regulatory paradigm, leading us to define a new group of replicons: the Marine RNA-based (MRB) plasmid family. Despite the extensive homology of their ori regions, these replicons are compatible, due to differences between RNA I sequences at sites that appear to mediate RNA I: RNA II interactions. The sequences and structures of MRB RNA I transcripts are not related to those of ColE1 replicons, suggesting that these two families of RNA-dependent replicons arose by convergent evolution.
MATERIALS AND METHODS
Bacterial strains, plasmids and culture conditions
Bacterial strains and plasmids used in this study are described in Supplementary Tables S1 and S2. Escherichia coli strains DH5α λpir and β2163 were used respectively for DNA cloning and to mobilize DNA. Vibrio cholerae strain O395 was used for analyses of replication by mutant plasmid oris. V. cholerae and E. coli strains were grown in Luria-Bertani (LB) at 30 or 37°C. Other Vibrionaceae strains were grown in LB-NaCl 0.5 M at 30°C. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml chloramphenicol (Cm), 5 or 20 μg/ml kanamycin (Km), 25 μg/ml spectinomycin (Spec), 50 μg/ml. Diaminopimelate (DAP) and l-arabinose were added when necessary to final concentrations of 0.3 mM and 0.02%, respectively.
Molecular biology techniques
Plasmid DNA was isolated using spin miniprep kits (Qiagen). Matings between E. coli (β2163) and Vibrionaceae were performed at 30°C as described earlier (13). Nucleotide substitutions were introduced by PCR assembly as described earlier (14). RNA isolation and northern blotting were performed using standard laboratory protocols.
Identification of the pB1067 ori region
DNA amplified from pB1067 was partially digested with Sau3AI, and fragments of 1–2 kb were ligated to pSW25T (15), and then transformed into DH5α λpir. DNA from more than 100 SpecR transformants was pooled, introduced into β2163 donor cells, and transferred to V. nigripulchritudo strain SFn118 via conjugation. Inserts from eight randomly selected SpecR colonies were sequenced.
OriVn700 random mutagenesis library
Random mutagenesis was performed using the Diversify PCR Random Mutagenesis Kit according to the manufacturer’s instructions (Clontech). The pB1067 ori was amplified using Titanium Taq DNA polymerase in the presence of 160 mM MnSO4 and 40 mM dGTP, ligated to Placgfp, and transformed into DH5α λpir. Plasmid DNA from 1000 pooled transformants was introduced into V. cholerae strain O395 containing PBADRNA I-Vn. Fluorescence relative to OD600 was determined for 200 randomly selected colonies after growth in LB supplemented with Spec, Cm and arabinose. Plasmid DNA was isolated from 30 clones; ori sequence was determined for the nine clones yielding the highest pOriVn700- Placgfp DNA and fluorescence.
RNA analyses
RNA was purified from log phase cultures unless otherwise noted, using Trizol (Invitrogen) followed by DNase I (Ambion) treatment. For northern analyses, RNA was electrophoresed on acrylamide gels, transferred to Bright Star Plus nylon membranes (Ambion), and hybridized to 32P-labelled in vitro transcribed riboprobes in ULTRAhyb (Ambion) at 68°C. For primer extension analyses, RNA was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) and 32P-labelled primers, and electrophoresed on sequencing gels. For 3′-RACE analyses, RNA was polyadenylated with PAP (Ambion) prior to reverse transcription with a primer containing a 3′-polyd(T) tail and subsequent amplification with nested primers.
RNA I was transcribed in vitro, labelled and purified largely as described (16). RNAse T1 (Ambion) structural probing was performed according to the manufacturer’s instructions.
Bioinformatic analyses
GenBank was searched for sequences similar to the pB1067 ori region using BLASTN (17). The ori sequences were aligned using Seaview and phylogenetic trees were built using the Phylo-win program (18) applied to Neighbour Joining method and Kimura’s 2-parameter distances. Reliability of topologies was assessed by the bootstrap method with 1000 replicates. Plasmid alignments were performed using Mauve (19) and rendered graphically using PlasmiG: a Python-based sequence map and alignment drawing tool for plasmids (Van der Auwera, G., manuscript in preparation), with identity scoring by the BLAST algorithm bl2seq (20). RNA structure was investigated using RNAalifold (http://rna.tbi.univie.ac.at/cgi-bin/RNAalifold.cgi) and RNAshapes (http://bibiserv.techfak.uni-bielefeld.de/rnashapes/submission.html).
RESULTS
Identification of the pB1067 ori region
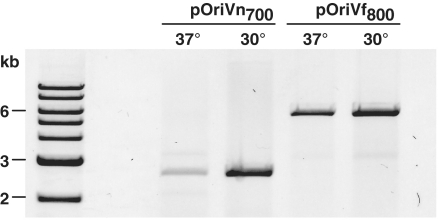
In order to define the sequences within pB1067 that mediate its replication, a library of pB1067 fragments was generated in the suicide vector pSW25T and screened for plasmids that replicated autonomously in V. nigripulchritudo. All sequenced plasmids shared at least a 700-bp sequence that corresponds to base pairs 2700–3400 of pB1067 (accession number: NC_010733). A pSW25T derivative containing only these 700 bp (pOriVn700) replicated autonomously in all six Vibrio strains tested as well as one Photobacterium and two Enterovibrio strains (Supplementary Figure S1), confirming that base pairs 2700–3400 of pB1067 are sufficient to mediate replication. Unexpectedly, plasmid DNA could not be detected in the small SpecR colonies obtained following transfer of pOriVn700 to pir- E. coli at 37°C. However, the plasmid was visible when the E. coli exconjugants were selected and cultured at 30°C, although it was still far less abundant than in most Vibrionaceae strains (Supplementary Figure S1). These data suggested (i) that replication of pB1067 may be aided by Vibrionaceae-specific factors, and ii) that this replicon might be temperature sensitive. Additional studies have confirmed the latter hypothesis: the copy number of pOriVn700 in V. cholerae is significantly greater when cells are cultured at 30°C rather than 37°C (Figure 1). Furthermore, growth at 42°C for 40 generations resulted in plasmid curing, suggesting that this temperature completely inhibits plasmid replication. The basis for this thermoregulation is currently unknown.
Figure 1.
The effect of temperature on replication of pOriVn700 and pOriVf800. Plasmid DNA was isolated from V. cholerae transformed with the indicated ori constructs and cultured in the presence of antibiotics at 37 or 30°C, digested, and electrophoresed.
The pB1067 ori region lacks open reading frames
Bioinformatic analysis of the pB1067 origin region identified relatively few features. No coding regions are predicted between 2700 and 3400 bp (11) suggesting that propagation of this replicon is not dependent upon a Rep protein. Consistent with this hypothesis, the region lacks direct repeat sequences (iterons), which frequently are the binding sites for Rep proteins. The origin region was found to contain an inverted repeat flanked by runs of As and Ts at the 5′- and 3′-ends respectively, a structure that often functions as a Rho-independent transcriptional terminator. Collectively, these findings suggested that replication of pB1067 might depend upon the activity of RNA(s), as has been described for ColE1-family replicons.
pB1067 belongs to a new family of replicons
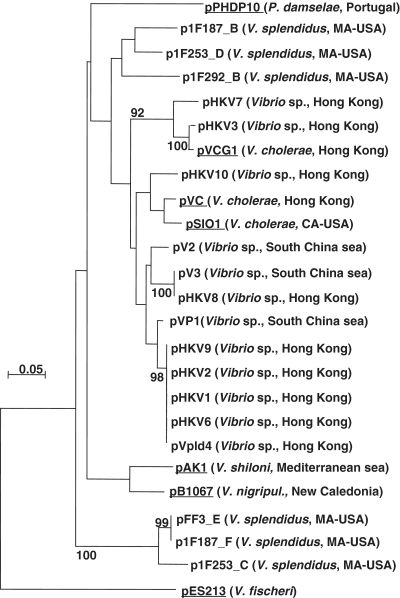
BLASTN analyses were performed using the 700-bp sequence in order to identify related genetic elements. These analyses revealed that a 488-bp region (2813–3301) within this sequence had high identity (54–94%) to 11 other plasmids and 12 partial plasmids sequences (Figure 2), all of which had been obtained from Vibrio or Photobacterium, although in numerous cases the precise identity of the species had not been determined. In total, a minimum of seven distinct species harbored plasmids containing sequences related to the pB1067 ori. Phylogenetic trees based on the homologous sequences indicated that closely related sequences were often, but not always, isolated from a single geographic region (Figure 2). We observed that several V. splendidus isolates (1F187 and 1F253) yielded two plasmids containing sequence homologous to the pB1067 ori. In both cases, the two plasmids belong to distinct clusters within the phylogenetic tree. Additionally, little homology was found among most plasmids outside of the putative ori sequence; in fact, no region (or gene) other than the putative ori was conserved among all seven plasmids for which complete and annotated sequence is available (Supplementary Figure S2). The most extensive homology was that previously described for pB1067 and pAK1. Collectively, these analyses suggest that the pB1067 ori is (i) representative of a new family of plasmid origins that is widespread among, yet limited to, Vibrionaceae and (ii) found in plasmids that encode highly diverse functions.
Figure 2.
Phylogenetic tree of plasmids based on sequences homologous to the pB1067 ori region. The tree was built (‘Materials and Methods’ section) based on sequences aligned using Seaview. Branch lengths are drawn to scale and are proportional to the number of nucleotide changes. Numbers at the nodes represent the percentage value given by bootstrap analysis of 1000 replicates. The bacterial host and the geographic origin are noted in parentheses. Fully annotated plasmids are underlined; comparisons of their complete sequences are presented in Supplementary Figure S2.
The region of homology in most plasmids containing sequences similar to the pB1067 origin region corresponded to the full 488-bp region noted above. However, in one plasmid, pES213, which was isolated from V. fischeri (21) homology was evident for a more limited sequence (nucleotides 3138–3248), a 110-bp region that includes the inverted repeat noted above. The homologous V. fischeri sequence is contained within an 863-bp fragment previously described as sufficient to mediate plasmid replication (21). Rather than constructing additional pES213 derivatives, we used a previously described pES213-derived plasmid, pVSV104 (22), for our studies, although for simplicity it is referred to within our work as pOriVf800. We found that replication of pOriVf800 is also thermosensitive in V. cholerae (Figure 1), although the effect of temperature is less striking that with pOriVn700.
Definition of a mimimal pB1067 ori
Given the observation that not all of the 700-bp fragment within pOriVn700 is conserved among related replicons, we explored whether a smaller fragment might be sufficient to enable plasmid replication. A construct (pOriVn500) consisting of 488 bp (nucleotides 2813–3301 of pB1067) ligated directly to a CmR cassette was found to replicate within V. cholerae (Figure 3). Similarly, a plasmid (pOriVs500) containing ∼500 bp from the comparable region of pAK1 could replicate autonomously (Figure 3). In contrast, smaller fragments derived from this region of pB1067 did not give rise to functional replicons. Mutational analyses also suggest that the ∼500 bp region is necessary and sufficient for replication. The majority (8 of 10) of 4–6 bp deletions within pOriVn700 that altered conserved regions of its 488 bp core abolished the autonomous replication capacity of the resulting plasmids (Supplementary Figure S3). In contrast, a deletion outside of this core region did not impair replication. Interestingly, the two core region deletions that did not prevent replication (which disrupted the inverted repeat) instead significantly increased the abundance of plasmid DNA, suggesting that the inverted repeat/transcription terminator may be important for negative regulation of plasmid replication.
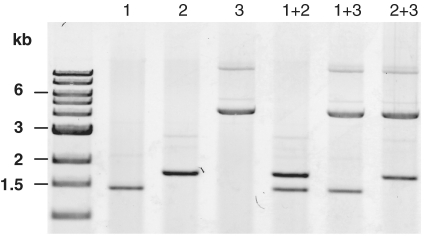
Figure 3.
Related replicons derived from plasmids of V. nigripulchritudo, V. shiloni and V. fischeri are compatible. Plasmid DNA was isolated from V. cholerae O395 transformed with pOriVn500 (1), pOriVs500 (2), and pOriVf800 (3), either singly or in pairs. DNA was digested and electrophoresed.
Plasmids containing related origins are compatible
As the pB1067-related origin sequences are quite similar, we explored whether derivatives of the related plasmids could be maintained together or whether they were incompatible replicons. Derivatives of plasmids isolated from V. nigripulchritudo, V. shiloni and V. fischeri (plasmids pOriVn500, pOriVs500 and pOri-Vf800, respectively) were introduced either singly or in pairs into V. cholerae, and their abundance was assessed in the resulting transformants. Notably, the level of plasmid DNA in strains containing two plasmids was almost identical to that detected in strains containing a single plasmid (Figure 3). This observation suggests that the small differences between the sequences of these origins are sufficient to prevent interference between their regulatory processes. The observed compatibility of these replicons is also consistent with the observation that two V. splendidus isolates were found to contain two distinct pB1067-related plasmids (Figure 2). However, it is important to note that plasmids from this family that utilize identical origin sequences cannot be stably maintained together. When pOriVn700 was transferred to V. nigripulchritudo strain SFn1 that contains pB1067, 10/10 SpecR colonies lost pB1067 (23).
The pB1067-ori is transcriptionally active
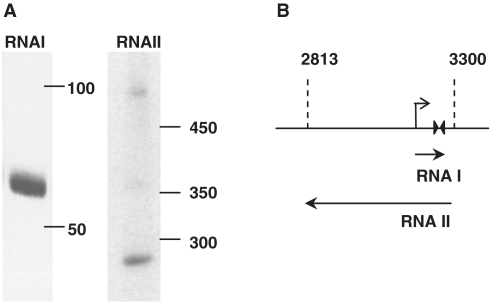
Although the pB1067 ori region does not appear to encode proteins to mediate plasmid replication, northern blot analyses revealed that RNA is generated from both strands of the origin region. Abundant transcripts of ∼70 nt that corresponded to the forward strand of the origin were detected (Figure 4A). Transcripts corresponding to the reverse strand were also observed, although they were less abundant than forward transcripts, longer, and more heterogenous in size (∼250–500 nt; Figure 4A). By analogy to the ColE1 replication system [reviewed in (24,25)], the small RNA has been named RNA I, and the larger RNA II.
Figure 4.
Detection and mapping of transcripts from the pB1067 ori region. (A) Northern blots of RNA from V. nigripulchritudo SFn1 were probed with riboprobes complementary to the forward (left) and reverse (right) strands of the 700-bp region that contains the pB1067 ori. (B) The location of sequences encoding RNA I and RNA II (arrows) are shown relative to the 700-bp origin region (horizontal line). Transcript termini were identified using primer extension and 3′-RACE assays and by the presence of a predicted transcriptional terminator (paired triangles). The positions and coordinates of the minimal 500-bp origin region are shown with dotted lines. The predicted promoter for RNA I is shown as a bent arrow; no consensus promoter for RNA II was identified.
Additional analyses were performed to identify the boundaries of these transcripts. The start of RNA I was mapped with northern blotting and primer extension to nucleotide 3181, using RNA either from V. nigripulchritudo SFn1 or V. cholerae containing pOriVn500 (Supplementary Figure S4A). Appropriately positioned −10 and −35 consensus promoter sequences can be detected upstream of the apparent +1 site. Based on the presence of the inverted repeat/terminator at position 3214–3242, the RNA I transcript is expected to be 68 nt in length, and in vitro transcription and northern blotting of these 68 nt confirmed that the length of the predicted transcript matches that produced in vivo from pB1067 (Supplementary Figure S4B). Notably, the position of RNA I and its promoter corresponds precisely to that of the 110-bp region that is conserved between pB1067, the V. fischeri plasmid pES213, and the 23 homologous Vibrionaceae plasmids described earlier.
The termini of RNA II were also mapped, using primer extension and 3′-RACE. The transcript was found to terminate at position 2820, which is near the boundary of the 500-bp sequence with homology to most of the related origin sequences. Primer extension analyses using RNA from V. nigripulchritudo SFn1 identified a +1 for this transcript at position 3285 of pB1067 (Supplementary Figure S4A); thus, RNA II contains sequences complementary to the entire RNA I transcript (Figure 4B). The full length RNA II is expected to be at least 465 nt, which is consistent with the length of the longer transcripts observed on northern blots of SFn1 RNA. Our results do not rule out the possibility that the primary transcript is slightly longer, particularly since, unlike for RNA I, a consensus promoter sequence that might drive expression of RNA II has not been identified. The shorter transcripts detected on northern blots likely result from RNA processing.
RNA I is a negative regulator of replication
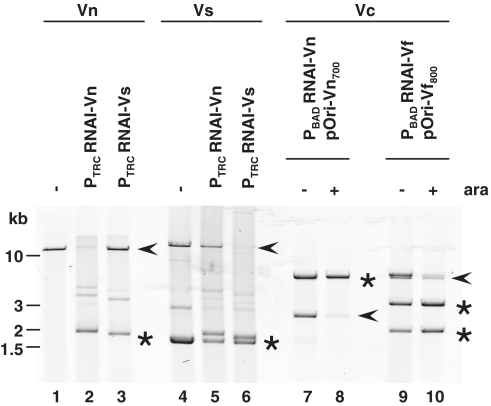
Small RNAs have been found to regulate numerous processes, including plasmid replication (26). We therefore explored the consequences of RNA I overexpression upon replication of pB1067 and related replicons. Constructs were generated for expression of putative RNA I transcripts (based on conserved promoter and terminator sequences) from pAK1 and pES213 as well that of pB1067. First, plasmids enabling constitutive expression of RNA I from pB1067 (PTRCRNA I-Vn) and pAK1 (PTRCRNA I-Vs) were introduced into strains containing the natural replicons. The presence of the RNA I-Vn expression construct dramatically reduced the abundance of pB1067 in V. nigripulchritudo, whereas the PTRCRNA I-Vs construct had no effect (Figure 5, lanes 1–3). Similarly, the abundance of pAK1 in V. shiloni was only markedly reduced by expression of RNA I-Vs (Figure 5, lanes 4–6). Neither plasmid had any effect on the abundance of pES213 in V. fischeri (Supplementary Figure S5). Subsequently, we assessed the effect of origin-specific RNA I upon the abundance of pOriVn700 and pOriVf800 in V. cholerae. The levels of both plasmids were markedly reduced following induction (using PBAD and arabinose) of their cognate RNA I transcripts (Figure 5, lanes 7 versus 8 and 9 versus 10). Collectively, these data suggest that RNA I transcripts encoded within pB1067 and the related replicons pAK1 and pES213 all function as negative regulators, and that the regulatory effect of each is specific for the plasmid from which it originates. We presume that the specificity of these RNAs regulators accounts for the compatibility of the replicons from which they are derived.
Figure 5.
The effect of exogenous RNA I expression upon plasmid abundance. Exogenous RNA I was constitutively expressed from pPTRCRNA I-Vn and pPTRCRNA I-Vs or transcribed in the presence of arabinose (ara) from PBADRNA I-Vn and PBADRNA I-Vf. Expression constructs were introduced into V. nigripulchritudo SFn1 (contains pB1067; lanes 1–3), V. shiloni AK1 (contains pAK1 and an unrelated ∼1.8-kb plasmid; lanes 4–6), or into V. cholerae strain O395 containing pOriVn700 (lanes 7, 8) or pOriVf800 (lanes 9, 10). Plasmid DNA from these strains was digested and electrophoresed on an agarose gel. pB1067-related replicons are marked with arrowheads, and RNA I expression constructs are marked with stars. Digestion of pPBADRNA I-Vf in lanes 9 and 10 yields two bands.
Folded RNA I’s contain two stem loops
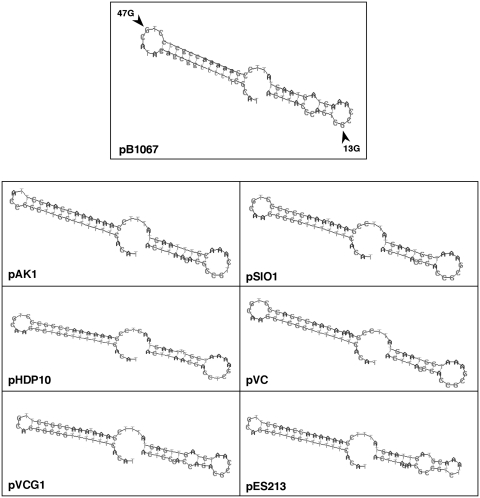
The secondary structures of regulatory RNAs are often critical determinants of their activities (26). We therefore investigated the secondary structure of RNA I, using both predictive and experimental approaches. Using primary alignments of RNA I sequences, coupled with the RNAalifold (http://rna.tbi.univie.ac.at/cgi-bin/RNAalifold.cgi) and RNAshapes (http://bibiserv.techfak.uni-bielefeld.de/rnashapes/submission.html) analysis software, we predicted that RNA I-Vn and related regulatory RNA molecules all would fold into a structure comprised of two stem loops with a single-stranded 3′-tail (Figure 6). The two stem loops are separated by a single-stranded region of 4–6 nt. Structural probing of in vitro transcribed RNA I-Vn, using RNAse T1, confirmed that it does adopt the predicted structure. RNAse T1, which only cleaves RNA at single-stranded Gs, cleaved non-denatured RNA I-Vn at nucleotide 13 and 47, which are the only G residues predicted to lie within single-stranded regions, and did not cleave at Gs expected to lie within double-stranded stems (Supplementary Figure S6).
Figure 6.
Predicted structures of RNA I transcripts. Structural predictions for putative RNA I transcripts from the indicated pB1067-related plasmids were generated using RNAalifold and RNAshapes. Single-stranded Gs identified through structural probing of RNA I from pB1067 (Supplementary Figure S6) are indicated with arrowheads.
The 5′-loop of RNA I is necessary but not sufficient for repression
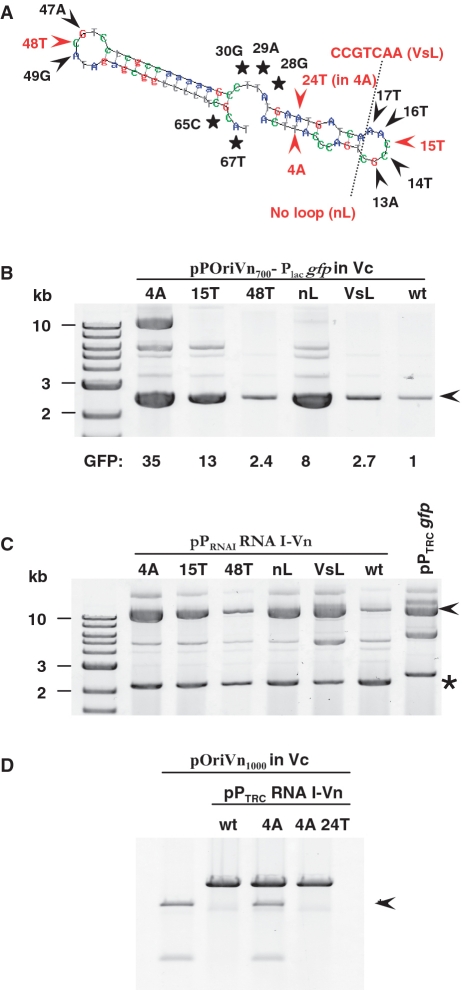
RNA I’s function as a negative regulator is likely to be a consequence of interactions between RNA I and RNA II. All of RNA I could theoretically pair with RNA II; however, the analysis of RNA I structure suggests that only a subset of its nucleotides are likely to promote initial binding to RNA II, since many are also capable of forming intramolecular pairs (i.e. form stems). Such an interaction between loops has been termed a ‘kissing complex’ (27,28). To identify key interactions underlying RNA I-mediated repression, we generated a library of origin region mutants and screened for mutants that were less subject to repression. Mutants were generated in a GFP-marked derivative of pSW25T, which enabled fluorescence-based screening of mutants and facilitated identification of unrepressed replicons. In addition, screening was done in a strain containing an exogenous source of RNA I (PBADRNA I-Vn), in order to minimize detection of mutants in which RNA I had been inactivated. Of 300 randomly selected mutants, 30 initially yielded fluorescence more than two times that of the wild-type control (Supplementary Figure S7), and after rescreening, plasmids from nine clones with fluorescence more than four times greater than that of the wild-type were selected for sequence analysis. Sequencing results confirmed that the region of overlap between RNA I and RNA II is pivotal in maintaining pB1067 copy number. All nine plasmids contained at least one mutation within this region, and four of nine mutants differed from the wild-type solely by a single nucleotide change within the region of overlap. The latter finding strongly suggests that a rate-limiting interaction occurs between folded forms of RNA I and RNA II, as single nucleotide changes could dramatically alter the thermodynamics of such pairing, whereas a ‘zippered’ interaction between antisense sequences should not be significantly influenced by a single nucleotide change (6).
Although RNA I appears to fold into a structure containing several single-stranded regions (Figure 6), mutants isolated from the screen only disrupted one of these, namely the 5′-loop (Supplementary Figure S7A). To determine whether other single-stranded regions were also important for pB1067 regulation, we generated a series of additional constructs containing mutations at other sites within RNA I, and assessed their effects upon plasmid regulation, which was monitored both by visualizing plasmid DNA and by measuring plasmid-derived GFP fluorescence. Mutations were introduced that modified the 3′-loop, the central single-stranded region, and additional positions within the 5′-loop. Unexpectedly, all of the five mutations that altered the central loop or the 3′-tail (Figure 7A, black stars) rendered the resulting plasmid non-functional in V. cholerae (i.e. SpecR transformants could never be isolated), although normal plasmid yields were obtained from pir+ E. coli. The sites of these mutations thus appear to be critical for plasmid replication and/or regulation, although their precise contribution has not been assessed. Most mutations that altered the 3′- and 5′-loops (Figure 7A, black arrows) had relatively subtle effects; they resulted in an increase of 2- to 3-fold in each replicon’s fluorescence and copy number, although it should be noted that some of these mutations would permit alternate base pairing and hence their consequences might understate the importance of the base of interest. Replacement of the 5′-loop with the putative 5′-loop sequence from pAK1 (VsL) resulted in a similar increase (Figure 7B). However, a few mutations dramatically increased fluorescence and replicon abundance. Alteration of RNA I nucleotide 15 (C15T) resulted in a 13-fold increase in fluorescence, while deletion of the 5′-loop sequences (nL) caused an 8-fold increase (Figure 7B). These results, like those of the mutagenesis, suggest that the 5′-loop may be the most important element in mediating an RNA I:RNA II interaction. Consistent with this hypothesis, a mutation within the 5′-stem sequence (T4A), which is expected to disrupt folding (and therefore loop availability), also caused a significant (35-fold) increase in plasmid abundance (Figure 7B).
Figure 7.
Mutations within RNA I disrupt repression and alter plasmid replication. (A) The secondary structure for RNA I-Vn is shown (Supplementary Figure S6). Mutations that prevented replication of plasmids dependent upon the pB1067 ori region are labelled with black stars. Mutations characterized in subsequent analyses are labelled in red. In (B) and (D) plasmid DNA was isolated from V. cholerae transformants and in (C) from V. nigripulchritudo exconjugants and then electrophoresed on agarose gels. Mutations within RNA I are noted above each gel; numbering is based on the position within RNA I as shown in (A). In (B) plasmid DNA is that of wt pOriVn700-Placgfp and its derivatives; relative fluorescence (GFP) is shown underneath. In (C), plasmid DNA is that of wt pB1067 (arrowhead) and constructs for expression of wt RNA I (pPRNA IRNA I-Vn) and its derivatives (star). No RNA I is encoded by the control plasmid, pPTRCgfp. In (D) the pB1067-related replicon (arrowhead) is pOriVn1000, which was used instead of pOriVn700GFP because it confers resistance to ampicillin rather than spectinomycin. RNA I is expressed from wt and mutant variants of pPTRCRNA I-Vn.
To confirm that the mutations described alter the activity of RNA I (rather than that of RNA II), expression constructs were generated for production of the mutant regulatory RNAs in trans. These constructs, which utilized the endogenous RNA I promoter, were transferred to V. nigripulchritudo SFn1, so that their effect on replication of pB1067 could be monitored. Compared to a vector control (pPTRCgfp), the expression of wt RNA I caused a readily detectable reduction in the amount of pB1067 (Figure 7C). Repression was also evident (although slightly less) when the C48T (3′-loop) variant of RNA I was expressed, consistent with the relatively minor effect of this mutation in the previous assays. However, no repression was observed due to expression of RNA I containing the T4A, C15T, nL and VsL mutations, indicating that RNA I containing these mutations is largely inactive against a wt RNA II (Figure 7C). (Note that in the previous assay, the target of RNA I containing the VsL mutation was a mutant version of RNA II). Notably, the VsL expression construct also did not alter replication of pAK1 in V. shiloni (Supplementary Figure S7B), providing further evidence that the 5′-loop of RNA I is necessary but not sufficient for RNA I-mediated repression.
Finally, in order to confirm the importance of RNA I’s structure for its activity as a regulator, we assessed whether a change expected to restore wild-type structure to the T4A mutant RNA I (namely A24T) also restored its activity. Strikingly, the activity of the doubly mutant RNA I (T4A A24T) was equivalent to that produced from the wt expression construct (Figure 7D). This result provides strong evidence that the T4A mutation inactivates RNA I by disrupting formation of a hairpin essential for presentation of a single-stranded binding loop, and confirms that the nucleotide at position 4 does not directly bind to RNA II.
DISCUSSION
We have identified and characterized the regulatory controls governing replication of a new family of plasmids. These plasmids, typified by the virulence-linked plasmid pB1067 of V. nigripulchritudo, are prevalent among, yet restricted to, members of the Vibrionaceae family. Unlike almost all plasmid families characterized to date, the ori regions of these plasmids do not encode a Rep protein to initiate DNA replication. Instead, the ori regions give rise to two RNAs, which are transcribed from opposite strands and contain complementary sequences. The smaller of these RNAs, termed RNA I, is ∼68-nt long, and functions as a negative regulator and the key determinant of plasmid incompatibility. Our data suggests that RNA I regulates the activity of the longer RNA (RNA II) via formation of a ‘kissing complex’ between single-stranded loops in each molecule, rather than through ‘zippering’ of antisense sequences. We have named this family, which is the first characterized family of replicons derived from marine bacteria, the Marine RNA-based (MRB) plasmid family. Only one other plasmid family (the ColE1 family) has previously been reported to rely on RNA-mediated replication initiation.
The MRB family consists of plasmids isolated from at least six distinct Vibrio species as well as from a Photobacterium. These plasmids are linked by the presence of a ∼110-bp sequence containing an inverted repeat/rho-independent terminator, which encodes RNA I and its promoter. Although the presence of this conserved sequence in a few of these plasmids has been previously noted (10,21,29,30), its role as a regulator was not known. Most fully sequenced members of the MRB family contain additional homologous sequences, which, together with the RNA I-encoding region, comprise the ∼500-bp minimal ori for these plasmids. These additional sequences encode the remainder of RNA II. One member of the family, the V. fisheri plasmid pES213 (21), lacks homology outside of the 110-bp core; however, northern blot analyses revealed that a larger RNA, encoded opposite to pES213′s RNA I, is also encoded by this plasmid’s ori region (Supplementary Figure S8). This observation suggests that pES213 replication likely depends upon a mechanism similar to that of pB1067 and other MRB family members, despite the limited scope of their homology.
Among previously characterized plasmid oris the only ones that lack a Rep protein and instead rely upon RNA-mediated replication initiation (as MRB plasmids appear to do) are plasmids of the ColE1 family. The ori regions of these well-studied replicons from the Enterobacteriaceae also yield two complementary transcripts. As observed for MRB RNA I, the smaller (∼110 nt) ColE1 RNA I transcript is a negative regulator and the determinant of plasmid incompatibility. Subtle nucleotide changes within RNA I molecules from both families can result in loss of regulatory activity and the emergence of a new incompatibility group (6). Additionally, for both plasmids, RNA I’s regulatory activity appears to be mediated (at least within the initial and rate limiting step) by interactions between single-stranded loops within highly structured RNAs (a ‘kissing complex’) rather than by extensive pairing of complementary sequences, although we do not exclude the possibility that such zipped complexes are ultimately formed. However, despite the many similarities exhibited by these regulatory elements, there is no similarity between either the primary sequences or the folded structures of MRB and ColE1 RNA Is (e.g. folded ColE1 RNA I has three, rather than two, stem loops), suggesting that they arose via convergent evolution. As the mechanism of action of RNA II from MRB plasmids has yet to be characterized, it remains to be seen whether the RNA II components of these replicons have likewise evolved very similar roles.
All 25 MRB family members have been isolated from members of the Vibrionaceae, suggesting that these plasmids may face constraints that limit their host range. It seems possible that MRB replication is dependent upon or enhanced by a species-specific factor, as has previously been described for several other replicons (31–33). Such a factor might be required for expression of RNA I and/or RNA II, might mediate their interaction, e.g. an RNA chaperone, or might be needed for the activity of RNA II, e.g. by cleaving the RNA to enable priming of DNA synthesis. Additional experimental characterization of the MRB host range, coupled with comparative genomic analyses, should enable identification of gene candidates that may underlie MRB host specificity. Differential expression of such a host factor at different temperatures may also account for our observation that MRB replication is influenced by temperature. Alternatively, it is possible that host restriction and temperature sensitivity are unrelated phenomena and that temperature-dependent variability in the efficiency of RNA I-mediated repression, or some other key step of replication control, independently accounts for the temperature sensitivity of MRB replicons.
Our analyses suggest that MRB and ColE1 family plasmids differ in the extent of their heterogeneity. No gene other than that encoding RNA I is conserved among all members of the MRB family, and few other genes are conserved even among a subset of members. In contrast, there appears to be more conservation among ColE1 family members, both with respect to the ori region (in which RNA II is routinely conserved as well as RNA I) and sequences outside it. For example, many ColE1 family plasmids encode an accessory regulator of replication, Rop, which promotes RNA I:RNA II interaction and thereby suppresses replication (25). No such factor appears to be present among MRB plasmids. The heterogeneity in MRB plasmid gene content suggests that there has been abundant recombination between ancestral plasmid(s) and additional genetic elements; coupled with the prevalence of these plasmids, it raises the possibility that they have been key agents of horizontal gene transfer among Vibrio sp.
MRB replicons may serve as the basis for a new set of vectors and genetic tools for study of marine bacteria, particularly the Vibrionaceae. The majority of vectors that are widely used in microbiological research utilize ColE1 family replication origins; however, at least some of these vectors (e.g. those containing the p15A ori) can be unstable in Vibrios (22). Broad host range vectors have also proved unsatisfactory in some Vibrios; consequently, a need for Vibrio-specific or Vibrio-optimized vectors has been proposed (22). Our characterization of MRB family replication control and our demonstration that numerous MRB family replicons and derivatives are compatible is a significant advance towards this goal. Through use of plasmids with subtly different MRB-derived ori regions, it should be possible to co-express proteins of interest and to vary their expression levels via modulation of plasmid copy number. Additionally, the ease with which new MRB compatibility groups can be developed suggests that there is a potentially limitless supply of such reagents. Finally, our observation that MRB replicons display temperature sensitivity raises the possibility that they might have additional utility as conditional replicons, which are currently unavailable in Vibrios.
NOTE ADDED IN PROOF
While this manuscript was under review, a bioinformatic analysis of MRB family replicons was published by Pan, Leung, and Gu in J. Microbiol. Biotechnol.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ifremer; National Institutes of Health (grant number R37-AI-42347); Howard Hughes Medical Institute. Funding for open access charge: HHMI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Martin Polz and Hong Xue for graciously providing unpublished sequences of plasmids derived from V. splendidus isolates and Eric Stabb for V. fischeri strains and plasmids. The authors are grateful for help with bioinformatics provided by Cynthia Sharma, Géraldine Van der Auwera and Stephane Duigou.
REFERENCES
- 1.Funnell BE, Phillips GJ, editors. Plasmid Biology. Washington, D.C: ASM press; 2004. [Google Scholar]
- 2.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kues U, Stahl U. Replication of plasmids in gram-negative bacteria. Microbiol Rev. 1989;53:491–516. doi: 10.1128/mr.53.4.491-516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh T, Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. USA. 1980;77:2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomizawa J, Itoh T, Selzer G, Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc. Natl. Acad. Sci. USA. 1981;78:1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacatena RM, Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981;294:623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- 7.Masukata H, Tomizawa J. Effects of point mutations on formation and structure of the RNA primer for ColE1 DNA replication. Cell. 1984;36:513–522. doi: 10.1016/0092-8674(84)90244-7. [DOI] [PubMed] [Google Scholar]
- 8.Masukata H, Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986;44:125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- 9.Novick RP. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobecky PA, Hazen TH. Horizontal gene transfer and mobile genetic elements in marine systems. Methods Mol. Biol. 2009;532:435–453. doi: 10.1007/978-1-60327-853-9_25. [DOI] [PubMed] [Google Scholar]
- 11.Reynaud Y, Saulnier D, Mazel D, Goarant C, Le Roux F. Correlation between detection of a plasmid and high-level virulence of Vibrio nigripulchritudo, a pathogen of the shrimp Litopenaeus stylirostris. Appl. Environ. Microbiol. 2008;74:3038–3047. doi: 10.1128/AEM.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg E, Falkovitz L. The Vibrio shiloi/Oculina patagonica model system of coral bleaching. Annu. Rev. Microbiol. 2004;58:143–159. doi: 10.1146/annurev.micro.58.030603.123610. [DOI] [PubMed] [Google Scholar]
- 13.Le Roux F, Binesse J, Saulnier D, Mazel D. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 2007;73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto-Mashimo C, Guerout AM, Mazel D. A new family of conditional replicating plasmids and their cognate Escherichia coli host strains. Res. Microbiol. 2004;155:455–461. doi: 10.1016/j.resmic.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marliere P, Mazel D. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 2005;156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 19.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 21.Dunn AK, Martin MO, Stabb EV. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid. 2005;54:114–134. doi: 10.1016/j.plasmid.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ. Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Roux F, Labreuche Y, Davis BM, Iqbal N, Mangenot S, Goarant C, Mazel D, Waldor MK. Environ. Microbiol. in press doi: 10.1111/j.1462-2920.2010.02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988;55:929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 25.Camps M. Modulation of ColE1-like plasmid replication for recombinant gene expression. Recent Pat. DNA Gene Seq. 2010;4:58–73. doi: 10.2174/187221510790410822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerhart E, Wagner H. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 27.Wagner EG, Brantl S. Kissing and RNA stability in antisense control of plasmid replication. Trends Biochem. Sci. 1998;23:451–454. doi: 10.1016/s0968-0004(98)01322-x. [DOI] [PubMed] [Google Scholar]
- 28.Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984;38:861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- 29.Purdy A, Rohwer F, Edwards R, Azam F, Bartlett DH. A glimpse into the expanded genome content of Vibrio cholerae through identification of genes present in environmental strains. J Bacteriol. 2005;187:2992–3001. doi: 10.1128/JB.187.9.2992-3001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang R, Wang Y, Leung PC, Gu JD. pVC, a small cryptic plasmid from the environmental isolate of Vibrio cholerae MP-1. J. Microbiol. 2007;45:193–198. [PubMed] [Google Scholar]
- 31.Chattoraj DK. Role of molecular chaperones in the initiation of plasmid DNA replication. Genet. Eng. 1995;17:81–98. [PubMed] [Google Scholar]
- 32.Maestro B, Sanz JM, Faelen M, Couturier M, Diaz-Orejas R, Fernandez-Tresguerres E. Modulation of pPS10 host range by DnaA. Mol Microbiol. 2002;46:223–234. doi: 10.1046/j.1365-2958.2002.03155.x. [DOI] [PubMed] [Google Scholar]
- 33.Wada C, Imai M, Yura T. Host control of plasmid replication: requirement for the sigma factor sigma 32 in transcription of mini-F replication initiator gene. Proc. Natl. Acad. Sci. USA. 1987;84:8849–8853. doi: 10.1073/pnas.84.24.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.