Abstract
Duplicate genes tend to have a more variable expression program than singleton genes, which was thought to be an important way for the organism to respond and adapt to fluctuating environment. However, the underlying molecular mechanisms driving such expression variation remain largely unexplored. In this work, we first rigorously confirmed that duplicate genes indeed have higher gene expression variation than singleton genes in several aspects, i.e. responses to environmental perturbation, between-strain divergence, and expression noise. To investigate the underlying mechanism, we further analyzed a previously published expression dataset of yeast segregants produced from genetic crosses. We dissected the observed expression divergence between segregant strains into cis- and trans-variabilities, and demonstrated that trans-regulation effect can explain larger fraction of the expression variation than cis-regulation effect. This is true for both duplicate genes and singleton genes. In contrast, we found, between a pair of sister paralogs, cis-variability explains more of the expression divergence between the paralogs than trans-variability. We next investigated the presence of cis- and trans-features that are associated with elevated expression variations. For cis-acting regulation, duplicate genes have higher genetic diversity in their promoters and coding regions than singleton genes. For trans-acting regulation, duplicate and singleton genes are differentially regulated by chromatin regulators and transcription factors, and duplicate genes are more severely affected by the deletion of histone tails. These results showed that both cis-and trans-factors have great effect in causing the increased expression variation of duplicate genes, and explained the previously observed differences in transcription regulation between duplicate genes and singleton genes.
INTRODUCTION
Gene duplication is widely regarded as a primary contributor to the emergence of new genes, and an important substrate for adaptation during evolution (1–3). It was suggested that a majority of the genes are the result of past gene duplication events (4). Following a duplication event, the resultant gene pairs usually return to single copy (singleton) within a short evolutionary period; however, a small fraction of duplicates (paralogs) are fixed in the genome, usually as the result of dramatic functional divergence (5). The functional innovations required for post-duplication fixation are generally explained by two alternate scenarios: neo- and sub-functionalization. Neo-functionalization refers to the acquisition of novel, beneficial functions; whereas sub-functionalization describes partition of ancestral functions (1,6,7). In some cases, additional models such as dosage maintenance and protein stoichiometry are also in the play (8,9).
Appropriately regulated transcription processes are vital to ensure proper function of cellular pathways and the survival of an organism, which presumably have minimum tolerance for errors and fluctuations. However, as was first proposed over several decades ago (2), alteration (or innovation) in gene expressions is an essential way to generate biological novelty and diversity. The recent availability of microarray data in yeast and other organisms has provided mounting evidence that gene duplications have significantly increased the expression divergence between sister genes (9–14). Moreover, examinations of expression divergence with regard to the age of the paralogs revealed that many pairs had undergone dramatic expression divergence shortly after their ‘birth’ (5). With regard to their fitness effects, it was suggested that an evolved and maintained variation in expression profiles between extant duplicates can potentially facilitate certain cellular processes such as metamorphosis in yeast and fly (15,16), and stress response in Arabidopsis (17,18). It was also proposed that difference in expression levels can provide genetic robustness to the organisms through genetic buffering (16,19,20).
Recent advances in experimental determination of transcription factor (TF) binding sites (e.g. ChIP-chip and ChIP-seq) have made it possible to compare the promoter regions of duplicate genes, and identify sequence features that have potentially driven the variability of gene expression. Specifically, studies have attempted to elucidate whether the variation or divergence in gene expression is primarily the result of cis- (close to the gene in question) or trans- (far from the gene) regulatory elements (21–24). In favor of cis-regulation, accelerated divergence in regulatory elements (21), and dramatic turnover of translation start sites (23) associated with duplicate genes have both been observed. However, linkage analysis using strain of Saccharomyces cerevisiae (10,25,26), allowing dissection of cis- and trans-mutational effects contributing to gene expression divergence, found trans-variation to have a more profound effect within yeast species. A study by Leach et al. (22) compared the genetic divergence of duplicate and singleton genes, and found gene expression variation can be explained by both cis-divergence and trans-divergence. Furthermore, it was suggested that chromatin structure contributed significantly to the gene expression variation (11,27,28). Consequently, the exact influences of cis- and trans-regulatory adaptations on altered expression of yeast genes, regardless whether singletons or duplicates, remain unsettled.
In this study, we comprehensively investigated the genomic features that potentially influence gene expression variation in yeast, and compared the influence of cis- and trans-acting variations on gene expression of both duplicate and singleton genes. In addition, we also evaluated the contribution of histone modifications on the divergence of gene expression. The lessons learned here in yeast will provide insight on the evolution of duplicate genes in higher eukaryotes such as human.
MATERIALS AND METHODS
Identification of duplicate and singleton genes
Protein sequences of S. cerevisiae were downloaded from Saccharomyces Genome Database (SGD, http://www.yeastgenome.org/) in February 2010. All-against-all FASTA reciprocal searches were performed. Singleton genes were defined as proteins that did not have any hits with other proteins using an E-value cutoff of 0.1; a gene was designated as a duplicate gene if it met the following criteria: (i) it had a FASTA hit with E < 1E–10; (ii) the aligned sequence between the two homologs was longer than 50% for the longer protein; (iii) the sequence similarity between the two proteins was greater than I [I = 30% if L> 150 a.a. and I = 0.01n + 4.8L−0.32[1+exp(−L/1000)] if L < 150 a.a., where n = 6 and L was the length of alignments (29)] . In the end, we identified a total of 1591 duplicate genes and 2081 singleton genes, respectively. Whole-genome duplication (WGD) and small-scale duplication (SSD) gene pairs were obtained from Guan et al. (30). The rates of nonsynonymous substitutions (Ka) and synonymous substitutions (Ks) between duplicate genes were estimated using the YN00 model nested in PAML software (31). The ages of each SSD gene pairs were estimated based on Ks values, and the gene pairs were divided into young (0 < Ks < 0.5) or old (0.5 < Ks < 1.5) pairs.
Calculation of gene expression variation
Expression differences between two yeast strains (BY4716 and RM11-1a) were obtained from Brem et al. (10). The expression variation data measured under the perturbation and stress conditions was compiled from Tirosh et al. (32); these conditions included heat shock, oxidative stress, nitrogen starvation, DNA damage and carbon source switch. Stochastic expression noise data was from Newman et al. (33); distance to median (DM) values were used in our analysis. Mutational variance data from Landry et al. (14) was measured from expression variations among mutation accumulation lines. All expression variation data were normalized using Z-scores.
Sequence polymorphism data in S. cerevisiae
The yeast sequence polymorphism data was downloaded from Wellcome Trust Sanger Institute (ftp://ftp.sanger.ac.uk/pub/dmc/yeast/). The 5′ promoter sequence were defined as 500 base pairs regions upstream from the start codon, and the 3′ UTRs were defined as 100 base pairs region downstream from the stop codon. The core promoter region was defined as 200 base pairs upstream from the start codon. The genetic diversity was calculated in a moving window by counting the number of different nucleotide at all positions between all possible pairs of strains within a population of interest, and then dividing by the window size.
Cis- and trans-variability
The genotypes and expression profiles were previously determined for the S. cerevisiae laboratory strain (BY4716) and wild strain (RM11-1a), and the 112 segregants that were generated by crossing these two strains (10). We used the following procedures to dissect the total expression variation into trans- and cis-variability. For each yeast gene, we grouped the 112 segregants into two groups based on the genotype of the genetic markers located in the 10-kb cis-region upstream of the translation start sites, i.e. we grouped the strains based on from which parent (BY4716 or RM11-1a) the promoter sequence was inherited. For each group, we next calculated the standard deviation of the expression levels among the segregants, and designated it as trans-variability since the segregants in each group shared the same genotype in the upstream cis-region. For most of the genes, the trans-variability calculated from the two groups were very similar, therefore we only used the trans-variability calculated from those segregants descended from the BY4716 strain. For each gene, we also calculated the mean expression level in each of the two groups, and designated the difference between the two means as cis-variation. The percentages of cis- and trans-variability in the total expression variation were calculated by linear regression.
Chromatin and TF regulation effect
We used two previously published yeast expression compendiums, which measured the changes in gene expression after deleting or mutating chromatin regulators (CRs) (34) or TFs (35), respectively. The expression compendium of CR perturbations contains 188 expression profiles, corresponding to 60 unique CRs. The expression compendium of TF perturbations contains 269 profiles, each corresponding to a unique TF. For each gene i, the sensitivity score Sij represents the changes in gene expression level when a trans-acting factor j, either a CR or TF, is perturbed. For each gene i, its regulatory effect Si is defined as:
where n is the number of trans-acting factors that were perturbed. The regulatory effect Si is calculated separately for CRs and TFs. To examine the effects of trans-acting factor regulation between duplicate genes and singleton genes, we calculated Kolmogorov–Smirnov (K–S) test for each expression profile, and K–S scores are defined as –log10 (P-value).
Histone modification
Genome wide histone modification data was obtained from Pokholok et al. (36). They have measured histone methylation and acetylation (H3K9ac, H3K14ac, H4ac, H3K4me1, H3K4me2, H3K4me3, H3K36me3, H3K79me3) in yeast promoters using the ChIP-chip method. Among them, H3K4m3, H3K14ac and H4ac can activate gene expression, and H3K9ac can repress gene expression. In total the histone modification data were available for 5661 yeast genes.
RESULTS
Duplicate genes have higher expression variation than singleton genes
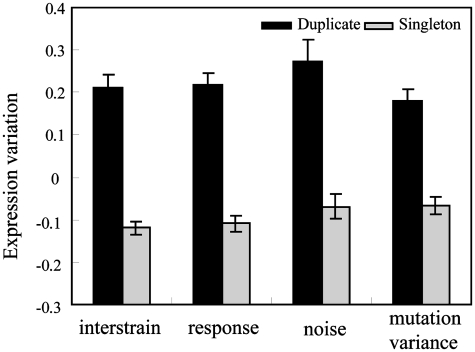
Variation in gene expression has been recognized to be an intrinsic property of simple or complex organisms (11); in budding yeast S. cerevisiae a number of experimental datasets are available to facilitate computational and comparative analysis. In this work, we are interested in comparing gene expression variation between duplicate genes and singleton genes and we considered four different types of expression variation: (i) divergence in expression level between orthologous genes in related yeast strains (10), (ii) variation of expression level under multiple environmental conditions (32), (iii) variation of gene expression following mutational accumulation (14), and (iv) stochastic noise in expression level among isogenic cells under the same condition (33). In the following, we discuss these four data types in more detail. (i) In Figure 1, we compared the expression divergence of orthologous genes from two S. cerevisiae isolates, the laboratory strain BY4716 and the wine strain RM11-1a (25). Clearly, the expression divergences between orthologs of duplicate genes are significantly higher than the divergences between orthologs of singleton genes (Wilcoxon rank sum test P-value <1e–10), which is consistent with what was observed previously (29). To examine potential enriched biological functions associated with duplicate genes that have higher expression variation, we next clustered yeast genes into functional groups based on the Gene Ontology (GO) annotations. We found genes that are involved in amino acid metabolism tend to have high expression variation. Given the previously proposed theory of backup compensation between duplicate genes, it is possible that duplicate genes are more likely to undergo regulatory changes, which would have made these genes more adaptable and variable in different strains or species. (ii) We next compared how the duplicate genes and singleton genes responded differently under environmental perturbations, using a previously generated data set (32). As shown in Figure 1, the duplicate genes show elevated changes in expression level in response to environmental stimuli, which suggested the functional roles of duplicate genes in stress response (Wilcoxon rank sum test P-value <1e–10). Moreover, functional groups involved in oxidoreductase activity tended to have high expression variation. (iii) Gene expression is subjected to stochastic noise, which influences most aspects of biological processes. In this study, we found that duplicate genes have higher noise than singleton genes (Wilcoxon rank sum test P-value = 4e−8) (Figure 1), and stress-related duplicate genes were found to be noisy too. (iv) Next, we analyzed a dataset which measured the response of gene expression to random mutations in mutation accumulation experiments (mutational variation) (14), and found that duplicate genes have higher gene expression variation among mutation lines (Wilcoxon rank sum test P-value <1e−10) (Figure 1), and genes encoding proteins localized in the plasma membrane have high expression variation. We next investigated whether the genes that were the most variable for each of these four types of expression variation were the same group of genes (11). For each type of variation, we selected the top 20% most variable genes and found that only environmental responsiveness and expression noise are highly correlated with each other (Fisher’s exact test, P-value <0.01), which agrees with previous findings that stress-related genes tend to have more noisy expression characteristics (33).
Figure 1.
Comparison of four types of expression variation between duplicate genes (dark grey bars) and singleton genes (light grey bars) in yeast. ‘Interstrain’ represents the expression divergence of orthologous genes between two S. cerevisiae isolates, the laboratory strain BM4716 and the wine isolate RM11-1a; ‘response’ represents changes in expression level in response to environmental stimuli; ‘noise’ represents fluctuation in expression level among isogenic cells; ‘mutation variance’ represents changes in expression level in mutation accumulation (MA) yeast lines.
However, it is possible that these observed trends were dominated by genes from a few selected functional categories. We next performed a series of control experiments to ensure that the higher expression variation of duplicate genes is a general trend across a wide range of genes. (i) Effect of TATA-box. We first removed those duplicate and singleton genes that contain a TATA-box in their promoter region since it is known that TATA-box containing genes tend to have higher expression variation and duplicate genes are enriched in TATA-boxes (32,37). The results persisted after removing TATA-box containing genes (Supplementary Table S1). (ii) Effect of tandem repeats. The presence of tandem repeats in the promoter regions has also been linked to higher gene expression variation (38). Indeed, we found the duplicate genes are more likely to have tandem repeats in their promoters than singletons, which partially explained their elevated expression variation (Chi-square test, P-value = 6e−7). After removing those tandem repeat containing genes from the duplicate and singleton genes, we observed that duplicate genes still had significantly higher expression variation than singleton genes (Supplementary Table S1). (iii) Effect of gene functions. It has been noted that biological function of genes can influence expression variation, for example, genes involved in stress response are known to have higher level of gene expression divergence and stochastic noise (32,39). To control for functional bias, we removed all the genes that contained the words ‘stress-related’ in their functional annotation (GO) and repeated our analysis. The results remained the same (Supplementary Table S1). (iv) Effect of expression level. We divided the yeast genes into highly expressed and lowly expressed groups by ranking their mRNA abundances (40) (Supplementary Table S1). Within each group, the duplicate genes still had higher expression variation than singleton genes.
To extend the analysis further, we next asked whether, in terms of expression variation, the sister paralogs in WGD resultant gene pairs were more divergent than SSD resultant gene pairs. We calculated expression variation similarity (Pearson correlation coefficient, PCC) between sister paralogs in each duplicate gene pair. Table 1 shows that, for inter-strain divergence, stochastic noise, and mutation variance, WGD resultant paralogs had lower noise similarity between themselves than SSD resultant paralogs (1000 permutation test, P-value < 1e−10). However, for expression responsiveness (under environmental perturbations), WGD resultant paralogs had higher noise similarity than SSD resultant paralogs (1000 permutation test, P-value < 1e−10). This is in accordance with the previously reported asymmetric partitioning of stress responses for WGD resultant paralogs in plants (41). To further validate whether the expression divergences between duplicate genes were coupled with evolutionary time, we used the rate of synonymous substitution, Ks, as a proxy for divergence time for duplicate gene pairs. We classified SSD resultant gene pairs into two different age groups (young and old SSD pairs) based on Ks values. Our results indicated that divergence in expression noise (after normalization for expression level) between duplicate paralogs increases over time, except for environmental responsiveness (Table 1).
Table 1.
Similarities in expression variation and genetic diversity between duplicated paralogs
| All duplicate pairs | SSD pairs | WGD pairs | Young SSD pairs | Old SSD pairs | Random pairs | |
|---|---|---|---|---|---|---|
| Inter strain | 0.47 | 0.49 | 0.43 | 0.51 | 0.44 | −0.03 |
| Responsiveness | 0.33 | 0.29 | 0.35 | 0.28 | 0.32 | 0.06 |
| Stochastic noise | 0.19 | 0.32 | 0.05 | 0.34 | 0.11 | 0.05 |
| Mutation variance | 0.24 | 0.31 | 0.17 | 0.36 | 0.21 | 0.02 |
| Trans-variability | 0.3 | 0.31 | 0.21 | 0.37 | 0.25 | −0.04 |
| Cis-variability | 0.15 | 0.17 | 0.13 | 0.19 | 0.14 | 0.02 |
| Total variability | 0.43 | 0.48 | 0.29 | 0.53 | 0.37 | −0.07 |
| Genetic diversity (codon) | 0.35 | 0.42 | 0.09 | 0.44 | 0.41 | 0.04 |
| Genetic diversity (promoter) | 0.15 | 0.21 | −0.02 | 0.26 | 0.16 | 0.01 |
| Genetic diversity (3′UTR) | 0.03 | 0.06 | −0.03 | 0.11 | 0.02 | 0.02 |
| Chromatin regulation effects | 0.42 | 0.49 | 0.28 | 0.54 | 0.33 | 0.01 |
| Transcription factor regulation effects | 0.18 | 0.17 | 0.18 | 0.19 | 0.14 | 0.03 |
| H3 perturbation | 0.21 | 0.25 | 0.13 | 0.29 | 0.16 | −0.01 |
| H4 perturbation | 0.28 | 0.31 | 0.25 | 0.26 | 0.21 | −0.01 |
PCCs were calculated between paralogs. All significant PCCs are highlighted with bold fonts.
In summary, these results demonstrated that duplicate genes indeed have a more variable and responsive expression program than the comparable singleton genes, and divergence in expression variation between sister paralogs may be coupled with evolutionary time. Next, we sought to determine how much of these differences between duplicates and singletons were caused by cis- or trans-regulatory effects, respectively.
The contribution of cis- and trans-variability
Having established that duplicate genes have higher expression variation than singleton genes, we next sought to determine how much of this was contributed by cis- or trans-acting factors (42). For this purpose, we used a dataset previously published by Brem and colleagues, who genetically crossed two yeast strains (BY4716 and RM11-1a) and measured the genotypes and expression profiles of 112 segregants (10). This dataset is ideal for our purpose since cis- and trans-variability can be ascertained by defining them as transcriptional variance between and within groups of segregants that share the same cis-genetic markers (see ‘Materials and methods’ section). We found that duplicate genes had both higher cis- and trans-variabilities than singleton genes (Wilcoxon rank sum test P-value = 1.4e−8 for cis-variability; P-value = 6.1e − 21 for trans-variability, respectively). For duplicate genes, the trans- and cis-variability could explain 67 and 30% of the total expression variability (measured from all 112 segregants), whereas it was 78 and 19% for singleton genes, respectively. Collectively these results indicated that trans-acting factors play a bigger role than cis- factors in causing gene expression variation for both duplicate genes and singleton genes. Trans-acting factors also had a slightly lower effect for duplicate genes than for singleton genes (67 versus 78%, Wilcoxon rank sum test P-value <1e − 6 with 1000 bootstraps).
We next compared the cis- and trans-variabilities between sister paralogs of duplicate gene pairs, aiming to explain how the divergence in expression variation between the paralogs can be explained by cis- and trans-factors. We calculated the expression variation similarity (Pearson correlation) between duplicate copies. Table 1 shows that trans-variabilities are more highly correlated between paralogs than cis-variabilities (R = 0.31 for trans-variability, and R = 0.15 for cis-variability, Wilcoxon rank sum test P-value = 0 with 1000 bootstraps), which suggested that cis-regulation underwent a more rapid divergence than trans-regulation for duplicate genes in the course of evolution. We next distinguished between paralogs created by different mechanisms, i.e. WGD versus SSD. Table 1 shows that the both cis- and trans-variability are slightly more conserved between SSD paralogs (R = 0.17 for cis-variability and R = 0.31 for trans-variability) than for WGD paralogs (R = 0.13 for cis-variability and R = 0.21 for trans-variability). Furthermore, the trans- and cis-variations are more conserved between young duplicates than older duplicates (Table 1).
Duplicate genes have higher level of cis-acting variation than singleton genes
In the above section we observed that cis-acting variations have a higher effect on duplicate genes than on singleton genes, we next attempted to find genomic features that could explain such discrepancy. Generally, cis-acting variation can be measured by allele-specific differential expression (ADE) by comparing the contribution of two alleles (43,44). In a recent study, genome-wide allele specific expression profiling was conducted in yeast and a number of genes were found to have significant allele imbalance (45). We found that these ADE genes are highly enriched in duplicate genes as 123 genes are duplicate genes and 114 are singleton genes (Fisher’s exact test P-value = 2.2e−6), respectively. These results suggested that duplicate genes indeed are more prone than singleton genes to be affected by cis-acting elements.
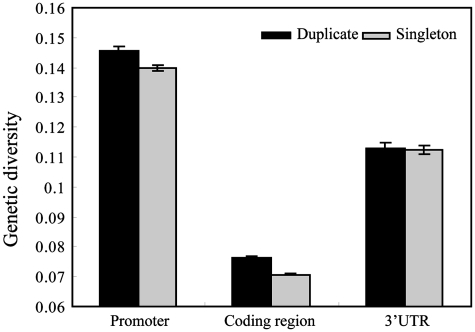
Having established the notion that duplicate genes have higher expression variation than singleton genes, we next asked whether this is because they have higher sequence divergence or heterogeneity in their regulatory regions. Our rationale is that the greater expression divergence and variation may be correlated or caused by accelerated evolution in the promoter and coding regions of the duplicate genes (10,25,42,46–48). We analyzed the previously published yeast population genomics data, which were measured from various ecologically and geographically diverse stains (49). It is generally assumed in practice that the density of polymorphisms in the promoter regions can be taken as an indicator for selective regulatory constraints (26). We calculated such genetic diversity in the coding and cis- regulatory regions (promoters and 3′UTR), and compared between duplicate and singleton genes. As shown in Figure 2, we found that duplicate genes had acquired a significant excess of polymorphisms in comparison to singleton genes (Wilcoxon rank sum test P-value = 2.3e−19 for the coding region, P-value = 1.4e−5 for promoters and P-value >0.05 for 3′UTR, respectively). Notably, this excess of polymorphism is most significant in the coding region, whereas the difference in 3′UTR is not statistically significant. This result suggests that the increased genetic diversities in the promoter region have significantly influenced gene expression variation of duplicate genes between related yeast species, whereas the higher polymorphism in the coding region likely reflects the relaxed constraints on coding sequence or synonymous substitutions. When we considered the core promoter region (200-bp upstream of start codon), we also found that duplicate genes have elevated genetic diversities (Supplementary Figure S1). We next examined the correlation of genetic diversity between sister paralogs; we found that the regulatory regions (promoter and 3′UTR) diverged more dramatically than protein coding regions (Table 1). Such elevated divergence in regulatory regions can cause expression divergence between sister paralogs, which has been theorized to provide genetic buffering to organisms and increase their tolerance to external perturbations (16). We also observed that SSD resultant paralogs are more similar to each other in the coding region and promoters than WGD pairs (Table 1). Moreover, comparing young and old duplicate gene pairs, we found that genetic diversity between duplicates is consistent with the evolutionary time (Table 1).
Figure 2.
Comparison of density of sequence polymorphisms in regulatory regions and coding regions. The duplicate genes (dark grey bars) have significantly higher polymorphism density in promoters than singleton genes (light grey bars).
Two types of trans-regulators: CRs and TFs
CRs and TFs are both important trans-acting factors that regulate gene transcription, and they are usually coupled and coordinated together (50). It was previously suggested that, in yeast, CRs may have a higher impact on expression variation than TFs (11). Here, we attempted to discern how these two types of trans-regulators influence the expression variation between duplicate genes and singleton genes.
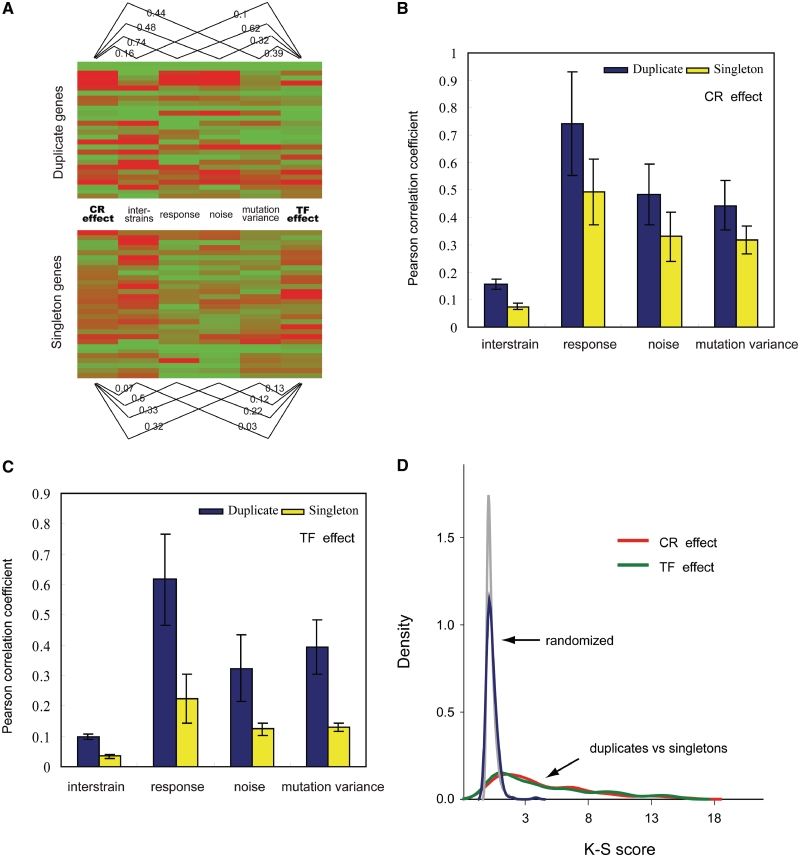
We first compiled two separate gene expression compendiums, which consist of genome-wide expression profiles, measured after either a CR or a TF was perturbed (34,35). Specifically, for each gene we calculated a TF and CR effect, which represent how much of a gene’s expression profile is influenced on average by TFs or by CRs (see ‘Materials and methods’ section). We then calculated genome-wide PCCs between the trans-regulatory effects (CR or TF) and each of the four types of expression variations (Figure 3A). Our rationale is that, if one type of expression variation is highly correlated with trans-regulation effects (either CR or TF effect), then it is taken as evidence that these trans-acting factors are more important in generating this type of expression variation. As shown in Figure 3B and C, we observed significantly positive correlations between the trans-regulation effects and all four types of expression variation, which suggested that the genes that have greater expression variation are also more likely to be sensitive to the deletions of TFs or CRs. Interestingly, these correlations are significantly higher for duplicate genes than for singleton genes, suggesting the duplicate genes are more likely to be influenced by the regulation of trans-acting factors.
Figure 3.
Comparison of the regulation effects of trans-acting factors between duplicate genes and singleton genes. (A) Pearson correlation coefficients between trans-regulation effects (CR and TF effect) and four categories of expression variations among duplicate and singleton genes, respectively. Note this is a schematic diagram showing only a few genes. (B) Effect of chromatin regulation (CR), measured as PCCs between level of variation and deletion effects. Duplicate genes are shown as blue bars and singleton genes are shown as yellow bars. (C) Effect of transcription factors (TF) regulation, measured as PCCs between level of variation and deletion effects. (B) and (C) show that duplicate genes are more sensitive to the perturbation of trans-acting factors than singleton genes. The error bars were 95% confidence intervals after 1000 bootstraps. (D) The distribution of K–S scores calculated between duplicate genes and singleton gene, comparing the regulatory effects of chromatin regulators (red) and transcription factors (green). The gray and blue curves are the distribution of K–S scores calculated between randomly chosen groups of genes: gray line (chromatin regulation effect), blue line (transcription factor regulation effect).
When comparing between sister paralogs in the same duplicated pair, we found that the sister paralogs shared more similar CR effects (Pearson correlation, R = 0.42) than TF effects (Pearson correlation, R = 0.18) (Table 1). This indicated that regulatory effects by TFs are more asymmetric between the paralogs than the effect by CRs, which is consistent with what was recently described (51). We also observed that the divergence in chromatin regulation effects is very different between WGD and SSD gene pairs (R = 0.28 for WGD genes and R = 0.49 for SSD genes, respectively). In contrast, the divergence in TF regulation effects is similar between WGD pairs (R = 0.18) and SSD gene pairs (R = 0.17). We also found that the divergence in both chromatin and TF regulation effects are coupled with evolutionary time (Table 1).
Next, we explored the influence of each CR or TF on gene expression variation on a genome-wide scale. For each CR or TF, we compared its regulation effect on the duplicate genes and singleton genes by calculating a K–S score between these two groups (see ‘Materials and methods’ section). K–S test is a non-parametric test that can distinguish whether two groups of samples have the same distribution. For the purpose of comparison, for each regulator, we also randomly picked two groups of yeast genes and calculated the K–S score between these random control groups. Figure 3D shows that indeed the K–S scores of CRs and TFs are significantly shifted to the right side, indicating that duplicate genes and singleton genes were distinctly regulated by CRs and TFs. A list of the 10 CRs and TFs with highest K–S scores were shown in Supplementary Table S2, respectively. However, we did not find any significant difference between the two different types of trans-factors, i.e. CRs versus TFs.
Previously we have shown that both TFs and chromatin structure have contributed to the expression divergence of paralogs (51). In eukaryotes, CRs help to conduct chromatin assembly and organization, and even affect the interaction between histones and DNA. Kim et al. (37) also reported that recent duplicates in yeast are highly enriched in occupied proximal nucleosome (OPN) genes. We analyzed the previously published promoter nucleosome depleted region (PNDR) scores which measured the nucleosome occupancy in the promoters (52), but did not find any significant difference between duplicate genes and singleton genes (P-value = 0.1, Wilcoxon rank sum test). We next examined other epigenetic regulatory effects that could have potentially contributed to the differences between duplicate and singleton genes.
The influence of epigenetic regulation
The N-terminal tails of eukaryotic histones are subjected to numerous post-translational modifications, such as methylation, acetylation, phosphorylation, ubiquitination and ADP-ribosylation (53,54). Histone modifications can change regional chromatin status, thus affect gene regulation and play a central role in gene expression variation (27,55). However, very little work has been done to explore how the expression variation of duplicate genes were influenced by histone modifications (56).
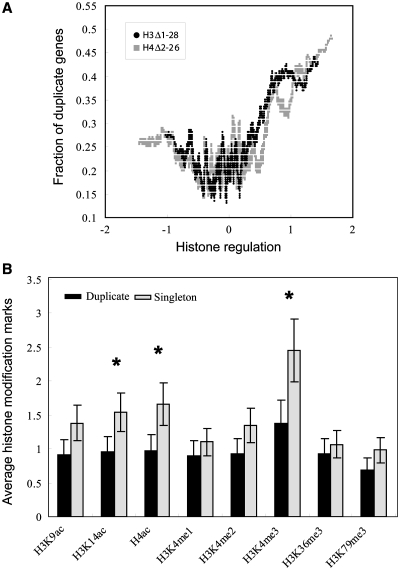
In a previously published genome-wide study, the N-termini of H3 and H4 histone genes were removed, and their effects on the expression level of yeast genes were measured (57). Using this data set, we asked how the duplicate and singleton genes were influenced differently by such perturbation. In Figure 4A, on the x-axis, we sorted yeast genes according to the changes in their expression level upon histone tail deletion; on the y-axis, we showed the fraction of genes in each sliding window of 200 ordered genes that are duplicate genes. It is clear that duplicate genes are more sensitive to the perturbation of histone tails than singleton genes are (Wilcoxon rank sum test P-value = 1.3e−7 for H3Δ1–28 and P-value = 4.2e−9 for H4Δ2–26, respectively), as the duplicate genes are enriched at the left and right ends of the distribution. This indicates that the expression levels of duplicate genes are more significantly affected by histone tail deletions than singleton genes are. Furthermore, the fraction of genes that were duplicated is much higher on the right end of the curve, suggesting that duplicate genes are under more severe chromatin mediated expression repression (since deletion of the histone tails increased their expression). We next asked whether the divergence in expression noise under histone perturbations is different between WGD and SSD paralogs. Table 1 shows that, for both H3 and H4 perturbations, WGD resultant paralogs had higher divergence in expression noise than SGD resultant paralogs. For example, in H3 perturbation experiments, WGD paralogs had correlation of 0.13 and SSD paralogs had correlation of 0.25.
Figure 4.
Comparison of epigenetic effects on the expression variation of duplicate and singleton genes. (A) Impact of histone tail deletion on gene expression variation between duplicate genes and singleton genes. We sorted the genes by expression changes resulting from histone deletion (x-axis); y-axis represents the fraction of genes which are duplicate in each sliding window of 200 sorted genes. (B) Effect of histone modifications on duplicate genes (dark grey bars) and singleton genes (light grey bars). The average values of histone modification marks are shown. *statistically significant (Wilcoxon rank sum test, see text).
Genome-wide histone modification profiles in yeast have been produced using the ChIP-chip platform (36), which can be used to examine the global effect of epigenetic processes influencing gene expression variations. We collected a compendium of such histone modification data (see ‘Materials and methods’ section), and subsequently compared their distributions between duplicate genes and singleton genes. Some of these histone modifications are highly associated with expression levels, i.e. H3K4m3, H3K14ac and H4ac are associated with transcriptional activation, whereas H3K9ac is generally associated with transcriptional repression. Figure 4B showed that, on average, duplicate genes (dark grey bars) have lower density of histone modification marks than singleton genes (light grey bars), especially for H3K4m3, H3K14ac and H4ac, for which the differences are statistically significant between duplicates and singletons (Wilcoxon rank sum test P-value <1e−4). Notably all of these three marks are activating histone marks. This is consistent with our observation (see above) that duplicate genes are under more severe chromatin mediated expression repression, which might be introduced by the lower level of such expression activating histone modifications in duplicate genes. Such a lower level of histone modifications of duplicate genes might play an important role in the dramatic expression variation of duplicate genes.
We next investigated the similarities in histone modifications between sister paralogs and found that all of the eight histone modification marks exhibited asymmetric distributions between sister paralogs (Table 2). Notably, we found the WGD resultant paralogs have higher level of divergence in their histone modifications than SSD paralogs, suggesting a potential distinct evolutionary trajectory for paralogs arisen from different duplication mechanisms. Furthermore, we found that the divergence in histone modifications between duplicate gene pairs (SSD only) is proportional to the divergence time. This, in addition to the divergence in trans-regulation, may partially account for the divergence in gene expression between the paralogs.
Table 2.
Similarities in histone modifications between duplicate pairs
| All duplicate pairs | SSD pairs | WGD pairs | Young duplicate pairs | Old duplicate pairs | Random pairs | |
|---|---|---|---|---|---|---|
| H3K9ac | 0.35 | 0.31 | 0.43 | 0.37 | 0.29 | 0.04 |
| H3K14ac | 0.49 | 0.41 | 0.69 | 0.44 | 0.4 | −0.02 |
| H4ac | 0.39 | 0.35 | 0.47 | 0.41 | 0.32 | 0.01 |
| H3K4me1 | 0.21 | 0.15 | 0.36 | 0.22 | 0.13 | 0.02 |
| H3K4me2 | 0.21 | 0.19 | 0.25 | 0.23 | 0.16 | 0.05 |
| H3K4me3 | 0.52 | 0.45 | 0.69 | 0.55 | 0.43 | 0.02 |
| H3K36me3 | 0.24 | 0.18 | 0.31 | 0.27 | 0.14 | −0.02 |
| H3K79me3 | 0.29 | 0.26 | 0.37 | 0.31 | 0.22 | 0.03 |
PCCs were calculated between paralogs. All significant PCCs are highlighted with bold fonts.
DISCUSSION
Gene duplications and the subsequent divergences in sequence, expression and interactions are considered as one of the major driving forces for the evolution of phenotypic complexity. Previous studies in yeast, fly and Arabidopsis have observed that, in comparison to singleton genes, duplicate genes usually undergo faster divergence in expression profile, and have higher expression divergence within and between species (17,21,29,58). This has been proposed to be either the results of relaxed selective constraints on expression levels, or alternatively the results of positive selection for genetic backup or functional innovation (i.e. subfunctionalization and neofunctionalization). In this study, we further presented evidence that duplicate genes in yeast also have higher level of expression variations, either in response to environmental and genetic perturbations or as the results of intrinsic stochastic fluctuations.
The molecular mechanisms of transcriptional regulation are intrinsically complicated, even for simple organisms such as S. cerevisiae. It is widely recognized that, in addition to changes in protein sequences, gene regulation effects and the subsequent changes in gene expression profiles play a pivotal role for biological phenotypic variation (59). Similarly, variation in gene expression, manifested either at the population level in the form of response to perturbations or inter-strain divergence, or at the single cell level in the form of stochastic noise, can offer clues on the regulatory mechanisms. In this work we comprehensively analyzed several types of expression variation among yeast genes, and dissected the molecular mechanisms into cis- and trans-effects. Our results showed that trans-effects play a much bigger role in generating expression variations than cis-effects, i.e. 67 versus 30% for duplicate genes and 78 versus 19% for singleton genes. This observation is significant since most of the current efforts in analyzing the evolution of duplicate genes have focused on the divergence in the cis-regulatory regions, e.g. divergence in TF binding sites and chromatin state (51). In contrast, very little work has been done to elucidate the contribution of trans-factors in the divergence of expression profiles between duplicated paralogs. Our work underscored the importance of these trans-factors, and paved the way for future studies.
Nevertheless we still observed that cis-factors played a significantly bigger role in generating expression noise for duplicate genes than for singleton genes. Indeed, it was previously demonstrated that duplicate genes exhibited a dramatic acceleration of regulatory evolution (21,60,61), and the start codon of duplicate genes underwent dramatic turnovers (23). The divergence in regulatory regions can be analyzed in the conceptual framework of subfunctionalization versus neofunctionalization, which was often invoked in analyzing function evolutions (6). A recent work reported that the TF binding sites in the promoter regions of duplicate genes often undergo asymmetric evolution, which was taken as an evidence for neofunctionalizations (62). Finding examples of subfunctionalization is more difficult since it requires knowledge of the regulatory regions of the ancestral gene before duplication events. The concept of neofunctionalization and subfunctionalization does not directly apply to expression variations, however after comparing expression variations between duplicate pairs, we indeed found that in the majority of the duplicated pairs, one sister paralog has higher level of variation than the other paralog (Supplementary Figure S2). This asymmetric expression program between paralogs has been previously taken as evidence to support the notion of ‘transcriptional reprogramming’, i.e. genetic backup was provided predominantly by paralogs that are expressed dissimilarly in most growth conditions (16).
As we know, trans-variability results from either changes in a trans-factor’s responsiveness to upstream signals, or changes in the factor’s ability to bind cis-regulatory sites. Our work suggests that duplicate genes are highly regulated by histone modifications and require integral histone N-termini for proper regulation, particularly for repression. This might be the reason why duplicate genes tend to lack activating histone modifications (H3K4m3, H3K14ac and H4ac). A recent work has shown that segmentally duplicated regions in the human genome often undergo asymmetric histone modifications, which was suggested to be the result of genome ‘pseudogenization’, i.e. the divergence in histone marks were the result of silencing of previously active genes (56). Our observation in yeast provided an alternate view of such asymmetric histone modifications, as they likely have a role in the functional divergence and innovation of duplicate genes.
In conclusion, we have shown that duplicate genes tend to have higher expression variation than singleton genes, which may have a pivotal role in generating phenotypic plasticity and complexity. Taking advantage of a population genomics data set which linked genetic markers to expression profiles, we were able to dissect the total variation into cis- and trans-regulation effects, and explained mechanistically why duplicate genes have higher expression variations. Our findings also highlighted the importance of trans-acting factors on duplicate gene expression variation, and proposed that epigenetic modifications played important role in expression divergence and variation. With increasing amount of expression and genomics data becoming available for other organisms, we expect our work will pave the way for more in-depth analysis on the duplicate genes in other organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funded by a Team Grant from the Canadian Institutes of Health Research (CIHR MOP#82940). Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Gabe Musso for his insightful comments. They would like to thank the Associate Editor and three referees for their constructive comments that have significantly improved the quality of this article.
REFERENCES
- 1.Conant GC, Wolfe KH. Turning a hobby into a job: How duplicated genes find new functions. Nature Rev. Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. Evolution by Gene Duplication. New York, USA: Springer-Verlag; 1970. [Google Scholar]
- 3.Zhang JZ. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. [Google Scholar]
- 4.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 5.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 6.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc. Royal Soc. London Ser. B-Biol. Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 8.Birchler JA, Veitia RA. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006;7:R13. doi: 10.1186/gb-2006-7-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Natl Acad. Sci. USA. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi JK, Kim YJ. Intrinsic variability of gene expression encoded in nucleosome positioning sequences. Nat. Genet. 2009;41:498–503. doi: 10.1038/ng.319. [DOI] [PubMed] [Google Scholar]
- 12.Duarte JM, Cui LY, Wall PK, Zhang Q, Zhang XH, Leebens-Mack J, Ma H, Altman N, dePamphilis CW. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- 13.Ganko EW, Meyers BC, Vision TJ. Divergence in expression between duplicated genes in Arabidopsis. Mol. Biol. Evol. 2007;24:2298–2309. doi: 10.1093/molbev/msm158. [DOI] [PubMed] [Google Scholar]
- 14.Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science. 2007;317:118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- 15.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 16.Kafri R, Bar-Even A, Pilpel Y. Transcription control reprogramming in genetic backup circuits. Nat. Genet. 2005;37:295–299. doi: 10.1038/ng1523. [DOI] [PubMed] [Google Scholar]
- 17.Ha M, Kim ED, Chen ZJ. Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc. Natl Acad. Sci. USA. 2009;106:2295–2300. doi: 10.1073/pnas.0807350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha M, Li WH, Chen ZJ. External factors accelerate expression divergence between duplicate genes. Trends Genet. 2007;23:162–166. doi: 10.1016/j.tig.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 20.Musso G, Costanzo M, Huangfu MQ, Smith AM, Paw J, Luis BJS, Boone C, Giaever G, Nislow C, Emili A, et al. The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res. 2008;18:1092–1099. doi: 10.1101/gr.076174.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu X, Zhang Z, Huang W. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc. Natl Acad. Sci. USA. 2005;102:707–712. doi: 10.1073/pnas.0409186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leach LJ, Zhang Z, Lu CQ, Kearsey MJ, Luo ZW. The role of Cis-regulatory motifs and genetical control of expression in the divergence of yeast duplicate genes. Mol. Biol. Evol. 2007;24:2556–2565. doi: 10.1093/molbev/msm188. [DOI] [PubMed] [Google Scholar]
- 23.Park C, Makova KD. Coding region structural heterogeneity and turnover of transcription start sites contribute to divergence in expression between duplicate genes. Genome Biol. 2009;10:R10. doi: 10.1186/gb-2009-10-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang ZQ, Gu JY, Gu X. How much expression divergence after yeast gene duplication could be explained by regulatory motif evolution? Trends Genet. 2004;20:403–407. doi: 10.1016/j.tig.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Brem RB, Storey JD, Whittle J, Kruglyak L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 27.Choi JK, Kim YJ. Epigenetic regulation and the variability of gene expression. Nat. Genet. 2008;40:141–147. doi: 10.1038/ng.2007.58. [DOI] [PubMed] [Google Scholar]
- 28.Tirosh I, Weinberger A, Bezalel D, Kaganovich M, Barkai N. On the relation between promoter divergence and gene expression evolution. Mol. Syst. Biol. 2008;4:159. doi: 10.1038/msb4100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Z, Rifkin SA, White KP, Li WH. Duplicate genes increase gene expression diversity within and between species. Nat. Genet. 2004;36:577–579. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- 30.Guan Y, Dunham MJ, Troyanskaya OG. Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics. 2007;175:933–943. doi: 10.1534/genetics.106.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 32.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat. Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 33.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 34.Steinfeld I, Shamir R, Kupiec M. A genome-wide analysis in Saccharomyces cerevisiae demonstrates the influence of chromatin modifiers on transcription. Nat. Genet. 2007;39:303–309. doi: 10.1038/ng1965. [DOI] [PubMed] [Google Scholar]
- 35.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 36.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 37.Kim Y, Lee JH, Babbitt GA. The enrichment of TATA box and the scarcity of depleted proximal nucleosome in the promoters of duplicated yeast genes. J. Mol. Evol. 2010;70:69–73. doi: 10.1007/s00239-009-9309-3. [DOI] [PubMed] [Google Scholar]
- 38.Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 41.Zou C, Lehti-Shiu MD, Thomashow M, Shiu SH. Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet. 2009;5:e1000581. doi: 10.1371/journal.pgen.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams RB, Chan EK, Cowley MJ, Little PF. The influence of genetic variation on gene expression. Genome Res. 2007;17:1707–1716. doi: 10.1101/gr.6981507. [DOI] [PubMed] [Google Scholar]
- 43.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gagneur J, Sinha H, Perocchi F, Bourgon R, Huber W, Steinmetz LM. Genome-wide allele- and strand-specific expression profiling. Mol. Syst. Biol. 2009;5:274. doi: 10.1038/msb.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osada N, Kohn MH, Wu CI. Genomic inferences of the cis-regulatory nucleotide polymorphisms underlying gene expression differences between Drosophila melanogaster mating races. Mol. Biol. Evol. 2006;23:1585–1591. doi: 10.1093/molbev/msl023. [DOI] [PubMed] [Google Scholar]
- 47.Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat. Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- 48.Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nature Genetics. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 49.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komili S, Silver PA. Coupling and coordination in gene expression processes: a systems biology view. Nature Rev. Genet. 2008;9:38–48. doi: 10.1038/nrg2223. [DOI] [PubMed] [Google Scholar]
- 51.Li J, Yuan Z, Zhang Z. Revisiting the contribution of cis-elements to expression divergence between duplicated genes: the role of chromatin structure. Mol. Biol. Evol. 2010;27:1461–1466. doi: 10.1093/molbev/msq041. [DOI] [PubMed] [Google Scholar]
- 52.Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat. Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Higgs DR, Vernimmen D, Hughes J, Gibbons R. Using genomics to study how chromatin influences gene expression. Ann. Rev. Genomics and Human Genet. 2007;8:299–325. doi: 10.1146/annurev.genom.8.080706.092323. [DOI] [PubMed] [Google Scholar]
- 56.Zheng DY. Asymmetric histone modifications between the original and derived loci of human segmental duplications. Genome Biol. 2008;9:R105. doi: 10.1186/gb-2008-9-7-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabet N, Tong F, Madigan JP, Volo S, Smith MM, Morse RH. Global and specific transcriptional repression by the histone H3 amino terminus in yeast. Proc. Natl Acad. Sci. USA. 2003;100:4084–4089. doi: 10.1073/pnas.0637524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li WH, Yang J, Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–607. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 59.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 60.Gu ZL, Nicolae D, Lu HHS, Li WH. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 2002;18:609–613. doi: 10.1016/s0168-9525(02)02837-8. [DOI] [PubMed] [Google Scholar]
- 61.Castillo-Davis CI, Hartl DL, Achaz G. Cis-regulatory and protein evolution in orthologous and duplicate genes. Genome Res. 2004;14:1530–1536. doi: 10.1101/gr.2662504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tirosh I, Barkai N. Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol. 2007;8:R50. doi: 10.1186/gb-2007-8-4-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.