Abstract
Initiation of translation of the full-length messenger RNA of HIV-1, which generates the viral structural proteins and enzymes, is cap-dependent but can also use an internal ribosome entry site (IRES) located in the 5′ untranslated region. Our aim was to define, through a mutational analysis, regions of HIV-1 IRES that are important for its activity. A dual-luciferase reporter construct where the Renilla luciferase (Rluc) translation is cap-dependent while the firefly luciferase (Fluc) translation depends on HIV-1 IRES was used. The Fluc/Rluc ratio was measured in lysates of Jurkat T cells transfected with the dual-luciferase plasmid bearing either the wild-type or a mutated IRES. Deletions or mutations in three regions decreased the IRES activity but deletion or mutations of a stem-loop preceding the primer binding site increased the IRES activity. The wild-type IRES activity, but not that of an IRES with a mutated stem-loop, was increased when cells were treated with agents that induce oxidative stress. Such stress is known to be caused by HIV-1 infection and we propose that this stem-loop is involved in a switch that stimulates the IRES activity in cells infected with HIV-1, supporting the suggestion that the IRES activity is up-regulated in the course of HIV-1 replication cycle.
INTRODUCTION
Initiation of translation of most eukaryotic cellular messenger RNAs (mRNAs) occurs by a cap-dependent mechanism that requires ribosomal scanning of their 5′UTR. The 40S ribosomal subunit bearing the initiator tRNA, Met-tRNAMeti, interacts with the initiation factors bound at the cap (m7GpppG) at the 5′ end of the mRNA, and then scans the mRNA in the 5′–3′ direction until it encounters an initiation codon in an appropriate context. The 60S ribosomal subunit joins the 40S subunit and translation of the mRNA begins (1–3). However, several groups of viruses and a minority of cellular mRNAs initiate translation in a cap-independent manner, using internal ribosome entry sites (IRESes) (3,4,5–8). IRESes are structured RNA regions that are able to directly recruit the 40S ribosomal subunit at or near an initiation codon. This mode of initiation usually requires the participation of some canonical initiation factors and of host cell factors called IRES trans-acting factors (ITAFs). The structure, the molecular mechanism and the requirement for ITAFs of different IRESes vary greatly, reflecting their functional diversity (9–11). IRESes were first discovered in picornaviruses (12,13), and have since been found in several other groups of viruses, including retroviruses (14) such as simian immunodeficiency virus (15,16), human immunodeficiency virus type 1 (HIV-1) (17,18), human immunodeficiency virus type 2 (19,20), murine leukemia virus (21,22), Rous sarcoma virus (23), feline immunodeficiency virus (FIV) (24) and mouse mammary tumor virus (25).
Translation of HIV-1 full-length mRNA produces the precursor of structural proteins, Gag (p55), and, through a programmed −1 ribosomal frameshift (26,27), it produces the precursor of viral enzymes, Gag-Pol (p220). This full-length RNA also serves as a genomic RNA that is encapsidated in viral particles as a dimer. The HIV-1 full-length mRNA is capped and polyadenylated, but the long structured 5′UTR of this mRNA could interfere with ribosomal scanning from the cap and decrease the efficiency of the cap-dependent translation initiation (28,29). Indeed, the 5′UTR of HIV-1 RNA contains several structural elements that are required for the replication cycle (30), such as the transactivation response domain (TAR), the poly(A) hairpin, the primer binding site (PBS), the dimerization initiation site (DIS), the major splicing donor site (SD) and the packaging signal (ψ) (see Figure 1A). Various models for the 5′UTR region of HIV-1 full-length RNA encompassing the IRES have been proposed, based on phylogenetic, chemical mapping and mutagenesis approaches (31). Among them, Huthoff and Berkhout (32) had described two mutually exclusive conformations for the 5′UTR of HIV-1 RNA, the branched multiple hairpin (BMH) and the long-distance interaction (LDI). Recently, using the powerful SHAPE (selective 2′hydroxyl acylation analyzed by primer extension) technology, the Weeks group found that the 5′UTR of HIV-1 full-length mRNA exists inside and outside the virus particles in a single predominant conformation, which we refer to as the Weeks conformation (see Figure 1A) (31,33). The BMH conformation has several similarities with the Weeks conformation, but the existence of the LDI conformation, which has never been detected in infected cells or in viral particles, has been challenged (31,34).
Figure 1.
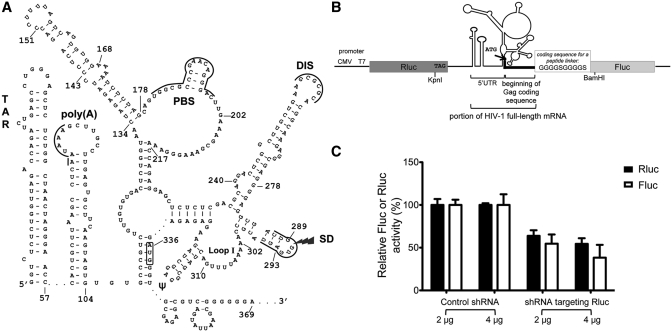
The 5′UTR of HIV-1 RNA that encompasses the IRES. (A) Representation of the Weeks conformation of the 5′UTR region of HIV-1 full-length RNA. This representation is adapted from Wilkinson et al. (31). The regulatory motifs are indicated: the TAR structure (nt 1–57), the polyadenylation signal [poly(A)], the primer binding site (PBS), the dimerization initiation site (DIS), the major splice donor site (SD) and the packaging signal (ψ) (nt 312–325). The AUG initiator codon is boxed. (B) Representation of the dual-luciferase reporter, plasmid pFRT-dual-IRES-HIV. This plasmid contains the Rluc and the Fluc coding sequences, under control of a CMV and a T7 promoter, and separated by the 5′UTR region of HIV-1 full-length RNA (nt 1–369 based on pLAI). Translation of Rluc is cap-dependent while translation of Fluc depends on the 5′UTR IRES of HI1 RNA. The initiator codon for the Fluc coding sequence is within the 5′UTR IRES. An oligonucleotide coding for a peptide linker was inserted between the 30 nt from the Gag coding sequence that are present in the construct and the beginning of the Fluc coding sequence. (C) Effect of RNA interference against Rluc on the expression of Rluc and Fluc. Jurkat T cells were cotransfected with the dual-luciferase reporter and either pBS-U6-RLi that expresses a shRNA targeting Rluc or pBS-U6-ApaI that expresses a control shRNA. Jurkat T cells lysates were assayed for Fluc and Rluc activities 48 h post-transfection and these activities were normalized for the protein content. A value of 100% is ascribed to the Rluc and Fluc activity with the control plasmid. Results are the mean ± SEM of three independent experiments.
Translation studies in reticulocyte lysates showed that Gag (and implicitly Gag-Pol) can be expressed both by cap- and IRES-dependent modes but that the cap-dependent mode is predominant (35). The full-length mRNA of HIV-1 group M subtype B, the group and type that predominates in infected patients of the Western World, contains an IRES in its 5′UTR (18). This IRES generates Gag (p55), initiating translation at the same initiation codon as the classical cap-dependent initiation. Another IRES was also found in the Gag coding sequence (17,20), which generates, in addition to Gag, a shorter isoform of Gag (p40). Brasey et al. (18), using deletion mutants, delimited the boundaries of HIV-1 5′UTR IRES from nucleotides (nt) 104 to 336. They also showed that the portion encompassing nt 104–289 has an IRES activity very near that of the 104–336 segment. The 104–289 segment is present in all the RNA transcripts from HIV-1, suggesting that all these transcripts could be translated by an IRES-dependent mechanism in addition to the classical cap-dependent mechanism [see also ref. (29)]. This is supported by recent studies from Charnay et al. (36), showing that the mRNA coding for Tat can be translated by an IRES-dependent mechanism. Two proteins were found to modulate the activity of the 5′UTR IRES of HIV-1: the human embryonic-lethal abnormal vision (ELAV)-like protein HuR down-regulates this IRES efficiency (37), whereas the heterogenous nuclear ribonucleoprotein (hnRNP) A1 up-regulates its efficiency (38). Interestingly, it was shown that infection with HIV-1 promotes relocalization of hnRNP A1 in the cytoplasm and enhances its expression, which then increases HIV-1 5′UTR IRES activity. It was also observed that the La autoantigen binds to the 5′UTR of HIV-1 full-length RNA (39,40) although a direct effect of La on the HIV-1 5′UTR IRES activity as an ITAF remains to be proven.
Although the use of the 5′UTR IRES of HIV-1 during viral infection is still questioned, several studies support the suggestion that this IRES benefits the virus during its replication in infected cells. The 5′UTR IRES is activated in cell extracts that were blocked in the G2/M phase of the cell cycle, a phase where cap-dependent translation is decreased (18). The IRES-dependent translation of HIV-1 full-length mRNA could be useful to ensure viral replication when the viral protein Vpr induces G2 cell cycle arrest (41,42). Recently, Castello et al. (43) showed that HIV-1 protease cleaves the canonical initiation factors eIF4GI and PABP, which inhibits cap-dependent translation in HeLa extracts, but not translation of HIV-1 full-length mRNA. Stimulation of a viral IRES by viral infection has also been described for FIV whose RNA contains a dormant IRES that is activated by FIV infection and by cellular stress (24). It is well-known that infection of HIV-1 causes oxidative and endoplasmic reticulum stresses (44–47) and such stresses could influence the activity of the HIV-1 IRES.
In this study, we investigated the activity of the 5′UTR IRES of HIV-1, using site-directed mutagenesis. We also investigated the effect of various stress conditions on this IRES activity. We found that deleting an unstructured region downstream of the PBS (nt 202–217), mutating 3 nt (nt 240–242) in a 4-nt bulge in the DIS hairpin or mutating a stretch of four A (nt 302–305) in loop I downstream of the SD hairpin, decrease the IRES activity. In contrast, deleting or mutating a stem-loop upstream of the PBS (nt 134–178) increases the IRES activity. A similar increase is observed when the activity of the wild-type IRES is assessed in cells treated with agents that induce oxidative stress. The relationship between the mutational analysis and the IRES activity is discussed. A model is proposed to account for the effect of the 134–178 stem-loop on the IRES activity in relationship with the oxidative stress caused by infection with HIV-1.
MATERIALS AND METHODS
Plasmid construction
To measure the IRES activity in the 5′UTR of HIV-1 group M subtype B full-length mRNA, we used a dual-luciferase reporter, pFRT-dual-IRES-HIV. This plasmid is derived from pcDNA5FRT (Invitrogen) where the KpnI and BamHI restriction sites of the original polycloning site were deleted to facilitate subsequent cloning of mutant IRESes. Our reporter pFRT-dual-IRES-HIV contains the 5′UTR region of HIV-1 originating from pLAI, a vector expressing a molecular clone of HIV-1 group M subtype B proviral DNA (48), inserted between the coding sequences of the Renilla luciferase (Rluc) and the firefly luciferase (Fluc). Expression of these genes is under control of a CMV promoter followed by a T7 promoter. The corresponding mRNA contains both the Rluc and the Fluc coding sequences. Rluc translation is cap-dependent and Fluc translation depends on HIV-1 5′UTR IRES. The initiator codon for Fluc expression is within the IRES and the context of the AUG (30 nt from the Gag precursor) was included in our constructs. Since the presence of these additional amino acids could affect Fluc activity, a sequence coding for a peptide linker (GGGGSGGGGS) was inserted by PCR before the Fluc coding sequence. The mutant IRESes were made by PCR amplification with four primers (49). Mutants were named according to the position where the mutation/deletion starts in the 5′UTR of HIV-1 full-length mRNA according to pLAI. The details and the primers used for all these cloning experiments are found in the Supplementary Materials and Methods section.
Jurkat T cells transfection
Jurkat T cells (CD4+ T cells) were maintained in RPMI 1640 medium (Gibco) supplemented with 10% (v/v) FBS (Wisent). All transfections were made by electroporation, using the NeonTM transfection system (Invitrogen) according to the manufacturer’s instructions. The conditions for electroporation were one pulse of 1150 V for 40 ms. Transfections were performed in 24-well plates containing 1.5 × 105 Jurkat T cells in 1 ml of complete medium without antibiotics. Briefly, 1 µg of DNA was mixed with 1.5 × 105 Jurkat T cells in 10 µl of R buffer supplied by the manufacturer. For assays with shRNA, co-transfections with shRNA-encoding plasmids were performed in 6-well plates containing 2.0 × 106 Jurkat T cells in 5 ml of complete medium without antibiotics. Two micrograms of dual-luciferase reporter were mixed with 2 µg of the plasmid encoding either control shRNA (pBS-U6-ApaI) or shRNA targeting Rluc (pBS-U6-RLi), a gift from Ivan Shatsky. Each mix was added to 2.0 × 106 Jurkat T cells in 100 µl of R buffer. Transfections for treatments with chemical agents were performed in 6-well plates containing 2 ml of complete medium without antibiotics. Four µg of dual-luciferase reporter were mixed with 1.0 × 106 Jurkat T cells in 10 µl of R buffer. Twenty-four hours post-transfection, transfected cells were pooled and splitted in 24-well plates (500 µl per well) and the chemical agents (H2O2 at final concentrations of 5 and 10 µM, TBHQ at final concentrations of 150 and 300 nM, thapsigargin at a final concentration of 450 nM and DFX at final concentrations of 250 and 500 µM; control: DMSO 0.25%) were added in a final volume of 1 ml per well for 4–8 h, as indicated in the legend to Figure 3.
Figure 3.
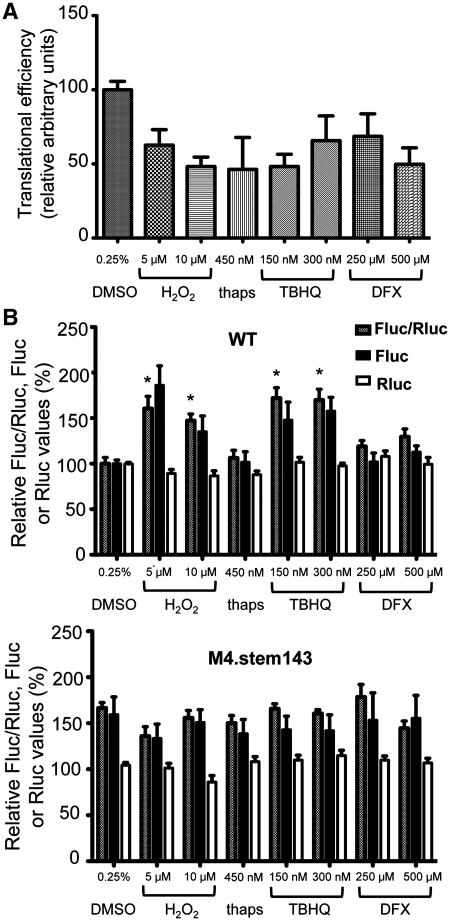
HIV-1 5′UTR IRES activity is increased in presence of agents that induce oxidative stress. Jurkat T cells were exposed to different agents [DMSO: control (6 h), H2O2: hydrogen peroxide (4 h), thaps: thapsigargin (6 h), TBHQ: tert-butylhydroquinone (6 h) and DFX: deferoxamine (8 h)]. (A) The cap-dependent translation is decreased following exposure to the different agents, as shown by metabolic labeling with [35S]methionine and measurement of the radioactivity incorporated into trichloroacetic acid-precipitable material. Radioactive counts were normalized for total protein content. A value of 100% was arbitrarily ascribed to the control sample (DMSO). (B) The activity of WT and M4.stem143 mutant IRES was assessed in cell lysates as described in the legend to Figure 2. Results show the Fluc/Rluc ratio and the Fluc and Rluc activities. A value of 100% was arbitrarily ascribed to WT. Results are the mean ± SEM of three independent experiments. The asterisk indicates the treatments that significantly increase the activity of the WT IRES (P < 0.05). The activity of the mutant IRES was not significantly altered by exposure to the different agents.
Jurkat T cells harvesting and luciferase assays
Transfected Jurkat T cells were harvested 24 h post-transfection, centrifuged at 200 g for 5 min, washed with PBS and lysed in 100 µl of Cell Passive Lysis Buffer (Promega). Cell lysates were frozen at −80°C until being used. Prior to luciferase assays, cell lysates were thawed and centrifuged 2 min at 1300 g to remove cell debris. The Fluc and the Rluc activities were assayed using a Dual-Luciferase Reporter Assay System kit (Promega). The IRES activity was monitored as a Fluc/Rluc ratio where the Fluc activity is the readout for IRES-dependent translation while Rluc expression depends upon cap-dependent translation. Fluc and Rluc activities were measured as relative light units with a TriathlerTM multilabel tester (Hidex Oy). One-way ANOVA with Bonferroni’s multiple comparison test was performed using GraphPad Prism version 5.00 for Mac OS X.
Metabolic labeling
Briefly, Jurkat T cells were transfected with pFRT-dual-IRES-HIV and were incubated with different stress-inducing agents for the appropriate amount of time, as described above. The medium was replaced with a methionine-free medium supplemented with 10% dialyzed FBS for 30 min, [35S]methionine (150–225 μCi/ml; 1Ci = 37 GBq) (Perkin-Elmer) was next added to the medium, and the cells were incubated for 15 min. Cells were collected by centrifugation, washed two times and lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS). Radiolabeled proteins were isolated by trichloroacetic acid precipitation on Whatman 3 MM paper. The amount of radioactivity was determined by scintillation counting, and the counts were normalized to protein concentration, which had been determined, using the DC Protein Assay (Bio-Rad).
Sequence alignment
Ninety-seven 5′UTR regions of HIV-1 group M subtype B sequences were obtained for the 134–178 stem-loop, 129 5′UTR regions of HIV-1 group M subtype B sequences were obtained for the 202–217 region, 142 5′UTR regions of HIV-1 group M subtype B sequences were obtained for the 240–242 bulge and 149 5′UTR regions of HIV-1 group M subtype B sequences were obtained for the 302-305 segment. These sequences were aligned, using the HIV sequence database (QuickAlign) of the Los Alamos National Laboratory available at the following URL : http://www.hiv.lanl.gov/.
RESULTS
The IRES located in the 5′UTR of HIV-1 RNA is functional in Jurkat T cells
To perform a mutational analysis of the IRES located in the 5′UTR of HIV-1 full-length mRNA, a standard dual-luciferase reporter system was used (Figure 1B), where the Rluc translation is cap-dependent whereas the Fluc translation depends on the HIV-1 5′UTR IRES. This reporter was similar to the reporter described by Brasey et al. (18), who demonstrated the existence of the HIV-1 5′UTR IRES. The intercistronic region contains the complete 5′UTR, including the TAR and poly(A) stem-loops. It is well known that the presence of a thermodynamically stable stem-loop such as TAR interferes with readthrough, ribosomal scanning and reinitiation that could occur after Rluc termination of translation (36,50). Moreover, the Rluc coding sequence is terminated by three stop codons, which ensures efficient termination of translation. The initiation codon for Fluc is located within the HIV-1 5′UTR IRES. The 30 nt of HIV-1 RNA that follow this initiator codon and encode the beginning of the sequence of HIV-1 Gag were included in our construct, in order to maintain the integrity of the IRES. A sequence coding for a peptide linker was added between these 30 nt and the beginning of the Fluc coding sequence to avoid any interference of HIV-1 5′UTR with Fluc folding and activity. Jurkat T cells, a CD4+ T-cell line, were co-transfected with the dual-luciferase reporter and a reporter coding for a shRNA targeting the Rluc coding sequence (51). In the presence of a shRNA targeting Rluc, but not of a control shRNA, Rluc and Fluc expressions were decreased proportionally by ∼50%, confirming that Fluc is not expressed from a cryptic promoter or by spurious splicing (Figure 1C). The HIV-1 5′UTR IRES activity had been previously studied in HeLa cells (18,38). We worked with Jurkat T cells since they are closely related to the natural target cells of HIV-1. We observed here that the HIV-1 IRES is functional in Jurkat T cells, being ∼5-fold more active than a control construct where the 5′UTR was inserted in the opposite direction between the Rluc and Fluc coding sequences, and than a control construct where there is no IRES between the Rluc and Fluc coding sequences (data not shown). However, HIV-1 5′UTR IRES is weakly active, being ∼5-fold less efficient than the well-characterized HCV IRES (data not shown), an IRES known to be very active in a variety of cell lines (52).
Deleting two different regions has an opposite effect on the HIV-1 5′UTR IRES activity
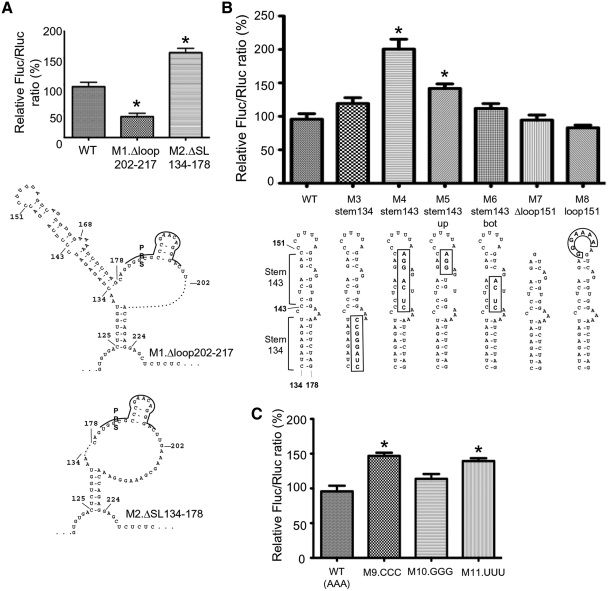
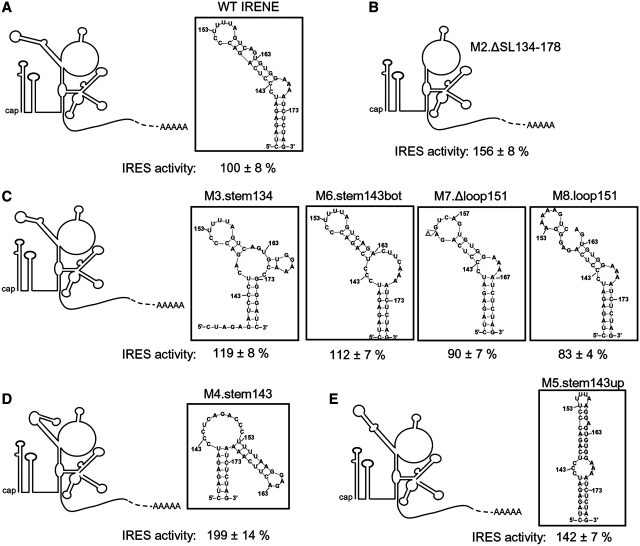
We investigated which are the regions of the 5′UTR of HIV-1 that are important for the IRES activity. Two deletion mutants were constructed. In mutant M1.Δloop202–217, 16 nt in the unstructured loop portion following the PBS were deleted while in mutant M2.ΔSL134–178, the irregular stem-loop encompassing nt 134–178, which has no known function to date, was deleted. The IRES activity was decreased to 39 ± 6% of the wild-type IRES activity with M1.Δloop202–217, but increased to 160 ± 8% with M2.ΔSL134–178 (Figure 2A). These results suggest that the region encompassing nt 202–217 contains a positive determinant of the 5′UTR IRES activity. In contrast, the 134–178 stem-loop acts as a negative determinant of this IRES activity and was named IRENE (for IRES negative element). Both regions are highly conserved among group M subtype B natural variants (see Supplementary Results section and Supplementary Figures S1A and B).
Figure 2.
HIV-1 5′UTR IRES activity is modulated by two RNA regions. (A) Two deletion mutations, represented in the Weeks conformation, were made in HIV-1 5′UTR IRES and their effect on the IRES activity is shown. (B) Mutations of the upper part of the stem-loop 134–178, named IRENE, increase the IRES activity. The mutations are indicated in boxes. Note that this figure describes the mutations that were introduced but not the structure adopted by the mutant stem-loops (see Figure 5). (C) Replacement of the 3A bulge of IRENE by 3C or 3U but not by 3G increases the IRES efficiency. The IRES activities for the mutants were assessed in lysates from Jurkat T cells transfected with the corresponding dual-luciferase plasmids. The Fluc/Rluc ratio obtained with the wild-type pFRT-dual-IRES-HIV (WT) was arbitrarily set at 100%. Results are the mean ± SEM of six independent experiments. The asterisk indicates mutants that were significantly different from WT (P < 0.05).
Disruption of the upper portion of IRENE stimulates the HIV-1 5′UTR IRES activity
To further investigate which portions of IRENE are involved in the control of HIV-1 5′UTR IRES activity, different mutations were made (described in Figure 2B and C). IRENE is formed by two stems separated by a 3A internal bulge on one side and a C bulge on the other side. The upper stem is itself interrupted by an AG bulge. We first disrupted either the lower stem (M3.stem134) or the upper stem (M4.stem143). We also made two mutants of the upper stem, M5.stem143up and M6.stem143bot, where either the top or the bottom part of the upper stem was disrupted, (see details in Figure 2). Finally, we deleted the 7-pyrimidine loop that caps this stem-loop (M7.Δloop151) or substituted the pyrimidines of this capping loop with purines (M8.loop151). Mutating the lower stem of IRENE did not influence the IRES activity (see M3.stem134) while disruption of the upper stem increased the IRES activity, as seen with M4.stem143 (200 ± 15%) (Figure 2B). An increase was also observed, although to a lesser extent, when only the top part of the upper stem was disrupted (M5.stem143up, 142 ± 7%), an effect similar to that obtained with M2.ΔSL134–178. Deleting or substituting the capping loop (M7.Δloop151 and M8.loop151) had no effect. Therefore, our results suggest that the negative determinant for the 5′UTR IRES activity seems to be located in the upper part of IRENE, but that the capping loop is not involved in this effect. We also investigated whether the 3A bulge that separates the upper from the lower stem in IRENE has an effect on the 5′UTR IRES activity. The 3A were substituted with either 3C, 3G or 3U. It was found that when the bulge is made of pyrimidines, the IRES efficiency is significantly higher (M9.CCC: 147 ± 5% and M11.UUU: 139 ± 5%) than when the bulge is formed by purines (WT with the 3A bulge: 100 ± 8% and M10.GGG: 114 ± 7%) (Figure 2C). Sequence alignment showed that IRENE is highly conserved in group M subtype B natural variants as mentioned in the above section. Most of the differences are located in the upper 7-pyrimidine loop, which does not influence the IRES activity. Twenty-one of the 97 variants analyzed have mutations in the upper stem, but only five of these mutations disrupt a base-pair. The 3A bulge is conserved in all these variants except three (see details in Supplementary Results section and Supplementary Figure S1A).
Oxidative stress increases the activity of wild-type HIV-1 5′UTR IRES, but not that of the M4.stem143 mutant
We investigated whether the activity of the 5′UTR IRES could be altered by a treatment with chemical agents causing a variety of stresses. We used hydrogen peroxide (H2O2) and tert-butylhydroquinone (TBHQ) to induce oxidative stress, thapsigargin to induce an endoplasmic reticulum (ER) stress and deferoxamine (DFX), which is a hypoxia mimetic (Figure 3). We exposed to these agents either the wild-type IRES or the M4.stem143 mutant, where IRENE is altered and whose activity was increased compared to the wild-type. It is known that a treatment with these stress-inducing agents causes a decrease in cap-dependent initiation (53). This was verified in our assays, using a [35S]methionine labeling and it was observed that global translation, that reflects cap-dependent translation, was decreased by ∼2-fold under the different stress conditions investigated (Figure 3A). We measured the Fluc/RLuc ratio under these conditions. Rluc cannot be used to assess transient changes in cap-dependent initiation (54) because of its stability (half-life superior to 50 h) and its activity remained unchanged when the cells were exposed to the various stresses (see Figure 3B). We observed that wild-type HIV-1 IRES activity, but not that of the M4.stem143 mutant, was increased ∼2-fold in presence of H2O2 and TBHQ, which induce oxidative stress. No change in the IRES activity was seen with agents that induce either an ER stress or hypoxia. These results therefore suggest that the effect of mutating IRENE can be reproduced by an exposure of the wild-type IRES to oxidative stress.
The HIV-1 5′UTR IRES activity is decreased by mutations located in the DIS hairpin or in loop I downstream of the SD hairpin
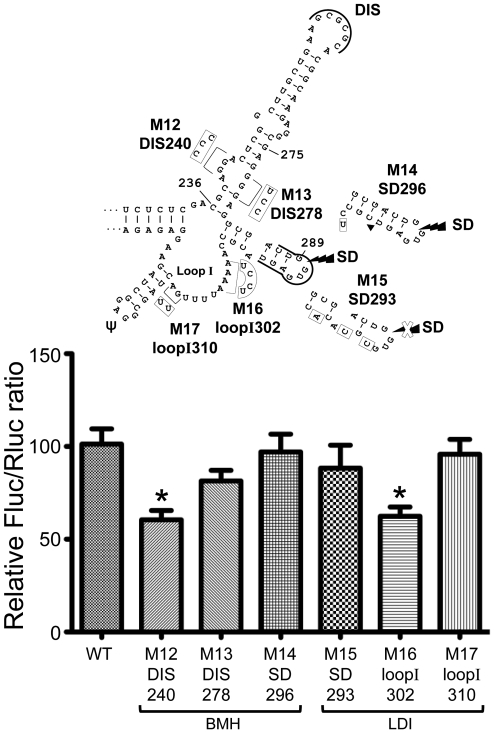
To further characterize the regulation of the HIV-1 5′UTR IRES activity, we also investigated substitution mutations that were found by Abbink et al. (55) to influence the BMH-LDI switch (see Introduction section) (Figure 4). Two of the six mutations investigated, M12.DIS240 and M16.loopI302, influenced the 5′UTR IRES activity, decreasing its value to ∼60% of wild-type efficiency. These mutations consisted, respectively, of a substitution of three purines with pyrimidines in a 4-nt bulge in the bottom part of the DIS hairpin and in a substitution of four A with pyrimidines in the beginning of loop I, downstream of the SD hairpin. When the purines directly opposite to the 4-nt bulge in the DIS hairpin were substituted with pyrimidines (M13.DIS278), when the two purines located at the end of loop I were substituted with pyrimidines (M17.loopI310) or when the SD hairpin was mutated (M14.SD296 and M15.SD293), there was no effect on the IRES activity. It was also observed that, in M15.SD293, the sequence of the splicing donor site in the 5′UTR of HIV-1 RNA is mutated, but the expression of Fluc in this construct is not affected (data not shown), in line with the fact that Fluc expression results from a genuine IRES activity and not from a splicing event (see above). The 4-nt bulge in the DIS hairpin is highly conserved, but the A stretch in loop I shows some variability among natural variants of HIV-1 group M subtype B (see Supplementary Results section and Supplementary Figure S1C and D).
Figure 4.
HIV-1 5′UTR IRES activity is decreased when nt 240–242 or 302–305 are mutated. The mutants studied here were previously found by Abbink et al. (55) to favor the BMH or the LDI conformation of the HIV-1 5′UTR (see the text). Mutations (boxes) or a deletion (filled triangle) are represented in the Weeks conformation and the conformation they favor is indicated. The WT and mutant IRES efficiencies were assessed in lysates from Jurkat T cells as described in the legend to Figure 2.
M12 was shown by Abbink et al. (55) to favor the BMH conformation whereas M16 favored the LDI conformation. Both mutants had a similar effect on the IRES activity, which does not support an involvement of the proposed BMH-LDI switch in the IRES activity. Abbink et al. had already observed that such a switch did not influence translation of the HIV-1 full-length mRNA, using conditions where cap-dependent translation predominates. Our results indicate that this putative switch has also no impact on the IRES activity but they reveal two additional regions that can influence HIV-1 IRES activity.
DISCUSSION
Our aim was to obtain insights into the regulation of the activity of the IRES located in the 5′UTR of HIV-1 full-length mRNA through a mutational analysis. In this study, we describe RNA regions important for the activity of the HIV-1 5′UTR IRES. We found three positive determinants (nt 202–217, 240–242 and 302–305) whose presence increases the IRES activity ∼2-fold and one negative determinant, a stem-loop encompassing nt 134–178 called IRENE, which decreases the IRES activity ∼2-fold. We also found that oxidative stress increases the wild-type IRES activity ∼2-fold but not that of the M4.stem143 mutant IRES with a mutated IRENE, whose activity is ∼2-fold that of the wildtype. This is the first study showing that oxidative stress activates the 5′UTR IRES of HIV-1.
Deleting the 202–217 portion of the PBS loop or mutating the 240–242 bulge of the DIS hairpin or the 302–305 portion of loop I could remove the binding site for an unknown cellular factor whose binding stimulates the IRES activity, or could result in a conformational change that promotes a less active IRES conformation. The mutated/deleted nt in all these three mutants are very reactive according to Wilkinson et al. (31), which implies that they are well exposed and free to interact with a factor. We observed that mutating the nt opposite to the 240–242 bulge in the DIS hairpin or mutating the last 2 nt of loop I did not change the IRES activity. This suggests that the decrease in the IRES activity obtained when nt 240–242 or 302–305 are mutated rather results from an effect localized to a well-defined region of the IRES than from a conformational change in the DIS hairpin or in loop I. It thus favors the possibility that nt 240–242 and 302–305 could be parts of the binding site for an ITAF. For the mutant with the deleted PBS loop, both a conformational change and the loss of the binding site for an ITAF could influence the IRES activity.
The effect of IRENE on the IRES activity is more complex. Our results show that the IRES activity is weaker in the presence of IRENE than in its absence, but that mutations in the upper stem of IRENE increase the IRES activity. Moreover, inducing oxidative stress increases the activity of the wild-type IRES but not that of the M4.stem143 mutant, which is more active than the wild-type. Figure 5 shows the secondary structure of wild-type and mutated IRENE, determined by the standard mfold algorithm. The secondary structure of IRENE is quasi-identical in the different structures that were proposed for the 5′UTR of HIV-1 full-length RNA (31) and the structure of IRENE determined by mfold is identical to that found in the Weeks conformation that we used throughout this study. The structure of IRENE for the M3.stem134, M6.stem143bot, M7.Δloop151 and M8.loop151 mutants is very similar to that of the wild-type (Figure 5A and C), conserving the same orientation of the bend induced by the 3A bulge separating IRENE in two parts. These four mutants have an IRES activity comparable to that of the wild-type IRES and it could be suggested that the wild-type IRES and these mutants adopt a weakly active conformation, which could be stabilized by the binding of a negative factor. As to the mutants with an increased IRES activity due to either the deletion or a mutation in the upper stem of IRENE (M2.ΔSL134-178, M4.stem143, M5.stem143up), they have a different conformation for IRENE, but this is consistent with the fact that a variety of IRES structures can recognize the ribosome (3). Finally, substituting the 3A bulge with 3G in IRENE did not change the IRES activity while replacing the 3A with pyrimidines increased the IRES activity ∼1.5-fold (see Figure 2C). These 3A are poorly reactive according to Wilkinson et al. (31) and could thus be involved in RNA–RNA contacts with other parts of the IRES, which could contribute to a weakly active conformation of the IRES. Such contacts would be maintained when the 3A are replaced by 3G but not by pyrimidines.
Figure 5.
The structure of IRENE influences the activity of HIV-1 5′UTR IRES. (A) The secondary structure (boxed) of WT IRENE. The presence of the 3A bulge causes a bend between the upper and the lower parts of this stem-loop. (B) In the absence of IRENE (M2.ΔSL134–178), the IRES activity is increased 1.5-fold. (C) With the M3.stem134, M6.stem143bot, M7.Δloop151 and M8.loop151 mutants, the IRES activity is unchanged and the secondary structure of IRENE (boxed) is very similar to that of WT. (D) In the M4.stem143 mutant, the structure of IRENE is drastically changed and the IRES activity is increased 2-fold. (E) The M5.stem143up mutation moderately changes the structure of IRENE (boxed), and the IRES activity is increased 1.5-fold. The secondary structure of WT or mutant IRENE was predicted by the mfold algorithm (version 3.2; http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi) (60,61).
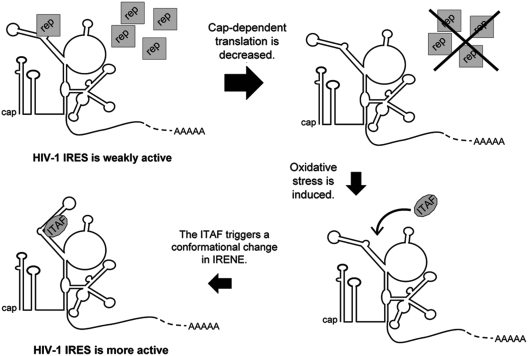
As mentioned in the ‘Introduction’ section, HIV-1 IRES is activated during viral infection (38). Oxidative stress, one of the pleiotropic effects caused by HIV infection, increases the activity of the wild-type IRES but not of the M4.stem143 mutant. Since the effect of oxidative stress is not additive to that of the M4 mutation, this result strongly suggests that oxidative stress increases the IRES activity via an effect on IRENE. A model can be proposed to describe the role of IRENE and oxidative stress in the increase in HIV-1 IRES activity, which is illustrated in Figure 6. The IRES could initially adopt a weakly active conformation that is stabilized by the binding of a repressor cellular protein to IRENE, as suggested above. In the course of the viral infection, a decrease in global protein synthesis would decrease the level of repressor. This could make IRENE available for interacting with a stimulatory ITAF that could act as a chaperone and help the IRES to adopt a more active conformation, such as that we artificially induced by mutating the upper stem of IRENE (Figure 5D). The ITAF could be a cellular protein which would be relocalized in the cytoplasm or whose expression would be induced by oxidative stress following viral infection. This hypothetical model thus suggests that IRENE controls the IRES activity through the conformation it adopts. Although this model is attractive, it is presently speculative and relies strongly on the structures of IRENE obtained with the folding algorithm. Further study of the effect of IRENE on HIV-1 IRES activity requires a characterization of the proteins that interact with this structure.
Figure 6.
A hypothetical model that accounts for the effects of IRENE and oxidative stress on HIV-1 5′UTR IRES activity. The model proposes that WT IRENE maintains the IRES in a weakly active conformation, which is stabilized by the binding of a repressor cellular protein (rep). In the course of HIV-1 infection, a decrease in global protein synthesis would decrease the level of this repressor. IRENE could then interact with a stimulatory ITAF, whose expression or relocalization was induced by oxidative stress. Binding of this ITAF causes a conformational change and promotes a more active IRES conformation (see the text).
It could be argued that the effect of mutating IRENE or inducing oxidative stress is modest. It is however worth recalling that an increase of ∼2-fold in the IRES activity of the foot-and-mouth disease virus was related to its hypervirulence in BHK (baby hamster kidney) −21 cells (56) and that an increase of ∼2-fold in the IRES activity of c-myc-RNA was linked to the occurrence of multiple myeloma (57). Also, microRNAs have powerful physiological effects although their effects on translation of individual targets are often moderate, being in the range of 1.5–2-fold (58,59).
In conclusion, our results suggest that the control of the HIV-1 5′UTR IRES activity is a complex event that likely combines conformational changes and the effect of cellular repressors and activators that bind to this IRES. Our results identified IRENE, a stem-loop that could play a role in the activation of this IRES upon viral infection in response to oxidative stress. More work is needed to understand the mechanism of stimulation of HIV-1 IRES by oxidative stress via IRENE and the regulation of this IRES during the virus replication cycle. The IRES-dependent translation of HIV-1 full-length mRNA must be tightly regulated during viral infection so as to favor viral replication. It will be important to characterize when this IRES is activated and utilized during the virus replication cycle and to identify the proteins which are involved in this utilization.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Canadian Institutes of Health Research (CIHR) to L.B.-G., G.F., N.H. K.G. acknowledges a studentship from CIHR, N.H. acknowledges a CIHR New Investigator fellowship and G.F. acknowledges a FRSQ Senior fellowship. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Ivan Shatsky from Moscow State University for the generous gift of pBS-U6-RLi and pBS-U6-Apa1. They also thank Johanie Charbonneau, Pascal Chartrand, Dominic Dulude, Alexey Karetnikov and Stephen Michnick for stimulating discussions and for critical reading of this manuscript.
REFERENCES
- 1.Pestova TV, Lorsh JR, Hellen CUT. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 2007. pp. 87–128. [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Lastra M, Rivas A, Barria MI. Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol. Res. 2005;38:121–146. doi: 10.4067/s0716-97602005000200003. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 6.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 7.Doudna J, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Hershey JW, Mathews MB, Sonenberg N, editors. Translational control in Biology and Medicine. New York: CSH Press; 2007. pp. 129–153. [Google Scholar]
- 8.Hellen CU. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta. 2009;1789:558–570. doi: 10.1016/j.bbagrm.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta. 2009;1789:542–557. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta. 2009;1789:518–528. doi: 10.1016/j.bbagrm.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 14.Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology. 2009;6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohlmann T, Lopez-Lastra M, Darlix JL. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson MG, Rue SM, Clements JE, Barber SA. An internal ribosome entry site promotes translation of a novel SIV Pr55(Gag) isoform. Virology. 2006;349:325–334. doi: 10.1016/j.virol.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 17.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbreteau CH, Weill L, Decimo D, Prevot D, Darlix JL, Sargueil B, Ohlmann T. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 20.Weill L, James L, Ulryck N, Chamond N, Herbreteau CH, Ohlmann T, Sargueil B. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010;38:1367–1381. doi: 10.1093/nar/gkp1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vagner S, Waysbort A, Marenda M, Gensac MC, Amalric F, Prats AC. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J. Biol. Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 22.Berlioz C, Darlix JL. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deffaud C, Darlix JL. Rous sarcoma virus translation revisited: characterization of an internal ribosome entry segment in the 5′ leader of the genomic RNA. J. Virol. 2000;74:11581–11588. doi: 10.1128/jvi.74.24.11581-11588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camerini V, Decimo D, Balvay L, Pistello M, Bendinelli M, Darlix JL, Ohlmann T. A dormant internal ribosome entry site controls translation of feline immunodeficiency virus. J. Virol. 2008;82:3574–3583. doi: 10.1128/JVI.02038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallejos M, Ramdohr P, Valiente-Echeverria F, Tapia K, Rodriguez FE, Lowy F, Huidobro-Toro JP, Dangerfield JA, Lopez-Lastra M. The 5'-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation. Nucleic Acids Res. 2010;38:618–632. doi: 10.1093/nar/gkp890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brierley I, Dos Ramos FJ. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brakier-Gingras L, Dulude D. Programmed -1 ribosomal frameshift in the human immunodeficiency virus of type 1. In: Atkins J, Gesteland R, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. New York: Springer; 2010. pp. 175–192. [Google Scholar]
- 28.Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yilmaz A, Bolinger C, Boris-Lawrie K. Retrovirus translation initiation: Issues and hypotheses derived from study of HIV-1. Curr. HIV Res. 2006;4:131–139. doi: 10.2174/157016206776055039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 35.Ricci EP, Soto Rifo R, Herbreteau CH, Decimo D, Ohlmann T. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem. Soc. Trans. 2008;36:690–693. doi: 10.1042/BST0360690. [DOI] [PubMed] [Google Scholar]
- 36.Charnay N, Ivanyi-Nagy R, Soto-Rifo R, Ohlmann T, Lopez-Lastra M, Darlix JL. Mechanism of HIV-1 Tat RNA translation and its activation by the Tat protein. Retrovirology. 2009;6:74. doi: 10.1186/1742-4690-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivas-Aravena A, Ramdohr P, Vallejos M, Valiente-Echeverria F, Dormoy-Raclet V, Rodriguez F, Pino K, Holzmann C, Huidobro-Toro JP, Gallouzi IE, et al. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology. 2009;392:178–185. doi: 10.1016/j.virol.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 38.Monette A, Ajamian L, Lopez-Lastra M, Mouland AJ. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J. Biol. Chem. 2009;284:31350–31362. doi: 10.1074/jbc.M109.048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waysbort A, Bonnal S, Audigier S, Esteve JP, Prats AC. Pyrimidine tract binding protein and La autoantigen interact differently with the 5' untranslated regions of lentiviruses and oncoretrovirus mRNAs. FEBS Lett. 2001;490:54–58. doi: 10.1016/s0014-5793(01)02137-8. [DOI] [PubMed] [Google Scholar]
- 40.Chang YN, Kenan DJ, Keene JD, Gatignol A, Jeang KT. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davy C, Doorbar J. G2/M cell cycle arrest in the life cycle of viruses. Virology. 2007;368:219–226. doi: 10.1016/j.virol.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen JL, Planelles V. The role of Vpr in HIV-1 pathogenesis. Curr. HIV Res. 2005;3:43–51. doi: 10.2174/1570162052772988. [DOI] [PubMed] [Google Scholar]
- 43.Castello A, Franco D, Moral-Lopez P, Berlanga JJ, Alvarez E, Wimmer E, Carrasco L. HIV- 1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One. 2009;4:e7997. doi: 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pyo CW, Lee SH, Choi SY. Oxidative stress induces PKR-dependent apoptosis via IFN-gamma activation signaling in Jurkat T cells. Biochem. Biophys. Res. Commun. 2008;377:1001–1006. doi: 10.1016/j.bbrc.2008.10.103. [DOI] [PubMed] [Google Scholar]
- 45.Deshmane SL, Mukerjee R, Fan S, Del Valle L, Michiels C, Sweet T, Rom I, Khalili K, Rappaport J, Amini S, et al. Activation of the oxidative stress pathway by HIV-1 Vpr leads to induction of hypoxia-inducible factor 1alpha expression. J. Biol. Chem. 2009;284:11364–11373. doi: 10.1074/jbc.M809266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindl KA, Akay C, Wang Y, White MG, Jordan-Sciutto KL. Expression of the endoplasmic reticulum stress response marker, BiP, in the central nervous system of HIV-positive individuals. Neuropathol. Appl. Neurobiol. 2007;33:658–669. doi: 10.1111/j.1365-2990.2007.00866.x. [DOI] [PubMed] [Google Scholar]
- 47.Peruzzi F. The multiple functions of HIV-1 Tat: proliferation versus apoptosis. Front Biosci. 2006;11:708–717. doi: 10.2741/1829. [DOI] [PubMed] [Google Scholar]
- 48.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 49.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 50.Venkatesan A, Sharma R, Dasgupta A. Cell cycle regulation of hepatitis C and encephalomyocarditis virus internal ribosome entry site-mediated translation in human embryonic kidney 293 cells. Virus Res. 2003;94:85–95. doi: 10.1016/s0168-1702(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 51.Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borman AM, Le Mercier P, Girard M, Kean KM. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 54.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 55.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Salas E, Saiz JC, Davila M, Belsham GJ, Domingo E. A single nucleotide substitution in the internal ribosome entry site of foot-and-mouth disease virus leads to enhanced cap-independent translation in vivo. J. Virol. 1993;67:3748–3755. doi: 10.1128/jvi.67.7.3748-3755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, Ralston SH, Helfrich MH, Willis AE. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- 58.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 60.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 61.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.