Abstract
Previous studies have led to a model in which the promoter-specific recognition of prokaryotic transcription initiation factor, sigma (σ), is core dependent. Most σ functions were studied on the basis of this tenet. Here, we provide in vitro evidence demonstrating that the intact Bacillus subtilis primary sigma, σA, by itself, is able to interact specifically with promoter deoxyribonucleic acid (DNA), albeit with low sequence selectivity. The core-independent promoter-specific interaction of the σA is −10 specific. However, the promoter −10 specific interaction is unable to allow the σA to discern the optimal promoter spacing. To fulfill this goal, the σA requires assistance from core RNA polymerase (RNAP). The ability of σ, by itself, to interact specifically with promoter might introduce a critical new dimension of study in prokaryotic σ function.
INTRODUCTION
The bacterial RNA polymerase holoenzyme (holo RNAP) is composed of the catalytic core RNAP, α2ββ′ and the transcription initiation factor, sigma (σ). The functions of σ have been studied since 1969 (1). It is now clear that σ confers upon holo RNAP the ability to recognize and melt promoter deoxyribonucleic acid (DNA) during transcription initiation (2–6).
The bacterial σ factors are classified into the σ70 and σ54 families (7). The σ70 family members in all bacteria can be subdivided into four groups. The Group 1, or primary, σs are responsible for the transcription of house-keeping genes and are essential for viability, whereas the Groups 2–4, or alternative, σs are required for more specialized functions such as stress responses. The primary σs have four conserved regions (Regions 1–4) with each having 2–4 subregions, whereas the alternative σs of Groups 3–4 lack Region 1 (7–9). Regions 2.4, 3.0 and 4.2 are thought to interact with the −10, extended −10 (or TG motif) and −35 promoter elements, respectively, according to the results of genetic and biochemical analyses of σ in the context of holo RNAP (10–15). The idea for direct promoter interaction of σ independent of core RNAP has been put to the test (16–21); however, it was only observed for a group 3 σ, the σD of Bacillus subtilis (17). Thus, it was generally believed that the primary σ has to associate with core RNAP prior to interaction with promoter DNA.
The inability of primary σ, by itself, to bind promoter DNA was once ascribed to the auto-inhibition of σ through direct interaction between Regions 1.1 and 4.2 of the σ (22,23). However, the studies of a Thermotoga maritima σ revealed that this inhibition is owing to an indirect steric and/or electrostatic mechanism (24) or to the close proximity of the negatively charged Region 1.1 to the positively charged promoter recognition domains of σ, which makes σ adopt a compact structure incompatible with DNA binding (25). Multiple switches are thought to be built into σ to alleviate the auto-inhibition (26). They include the interactions between the flap domain of the β-subunit of core RNAP and Region 4 of the σ (27,28) as well as between the coiled coil domain of the β′-subunit and Region 2.2 of the σ (29). Although the switch mechanism may explain why σ is unable to bind promoter DNA in a core-independent manner, direct evidence supporting the hypothesis that σ, by itself, is unable to bind promoter DNA is still lacking. Actually, the failure to detect the core-independent promoter-specific interaction of σ could be attributed to the use of a σ devoid of promoter DNA-binding conformation or the use of an inappropriate condition for assaying the binding interaction. Since how σ recognizes promoter DNA would affect how we elucidate the functional mechanisms of σ and core RNAP during transcription initiation, we re-investigated this issue.
Besides the recognition of −10 element, TG motif and −35 element by σ, the spacer DNA, which separates the −10 and −35 elements and is important for a promoter to function, is also proposed to be recognized by σ (30). In general, it is the length of the spacer DNA (spacing), rather than the sequence (except the TG motif), which is highly homologous (31–33). Usually, the 17 ± 1 bp spacing confers the optimal activity on promoters recognized by the primary σ (34–39). The main roles of the spacer DNA are thought to hold a more favorable orientation of the −10 element relative to the −35 element for initial binding of RNAP and for facilitating subsequent formation of a competent RNAP–DNA open complex (40–46). However, the mechanism leading to the recognition of optimal promoter spacing by RNAP remains unclear. Using a series of glutathione S-transferase (GST)-fused partial σ70 polypeptides, which exhibit specific binding to either one or both of the −10 and −35 elements of tac promoter, for competition assay, it was inferred that the σ70, by itself is a molecular ruler, which is able to discern the optimal spacing of the tac promoter (23,30). In this study, we also re-examined the inference using the B. subtilis σA.
Here, we provide in vitro evidence demonstrating that the house-keeping B. subtilis σA is able to bind specifically to the −10 element of promoters independent of core RNAP. However, the σA is unable to discern the optimal promoter spacing. To fulfill this goal, it requires the assistance from core RNAP.
MATERIALS AND METHODS
Overproduction and purification of σA
Methods used for overproduction and purification of σA in inclusion bodies are similar to those reported previously (47). The cell lysate was centrifuged at 7500g to harvest the σA-containing inclusion bodies that were then denatured with TEDG buffer [10 mM tris–hydrochloric acid (HCl), pH 7.9, 0.1 mM ethylenediaminetetraacetic (EDTA), 0.1 mM dithiothreitol (DTT), 0.2 mM phenylmethanesulfonylfluoride (PMSF), and 5% glycerol] supplemented with 200 mM NaCl and 7 M guanidine–HCl (Gn–HCl), and refolded through drop-by-drop dilution of the denatured protein with the TEDG buffer containing 200 mM NaCl until the concentration of Gn–HCl was ∼55 mM. The refolded σA was centrifuged at 7500g to remove unfolded proteins. The soluble σA in the supernatant was then precipitated with 40% (w/v) ammonium sulfate and resuspended in TEDG buffer containing 200 mM NaCl before further purification with the molecular sieving columns, Superdex HR-200 and Superdex SD-75.
To overproduce soluble σA, Escherichia coli BL21 (DE3)/pCD2 or E. coli BL21 (DE3)/pCD2M, which is able to express the wild-type (Wt) σA or the mutant (Mu) σA with Gln-196-Ala and Thr-199-Ala substitutions, respectively, was grown at 37°C to an OD550 of 0.5 before being transferred to 16°C. After incubation of the culture at 16°C for 10 min, isopropyl β-D-1-thiogalactopyranoside (IPTG) (final concentration 0.4 mM) was added to induce the expression of σA. Fifty minutes later, rifampicin (final concentration 100 µg/ml) was added into the culture which was further incubated at 16°C for 12 h. The E. coli cells were harvested and disrupted with a microfluidizer in TEDG buffer supplemented with 100 mM NaCl and then centrifuged at 7500g. The soluble σA in the supernatant was absorbed with 20 g of diethylaminoethyl cellulose (DEAE)-cellulose pre-equilibrated with TEDG buffer containing 100 mM NaCl. The DEAE cellulose was packed into a column and a NaCl gradient from 0.1 to 0.45 M was applied. The major σA-containing fractions were pooled and the σA were precipitated with 40% (w/v) ammonium sulfate and resuspended in TEDG buffer containing 200 mM NaCl before further purification with the molecular sieving columns, Superdex HR-200 and Superdex SD-75.
Both the refolded and soluble σA purified had homogeneities higher than 95% and were stored in storage buffer (20 mM Tris–HCl, pH 7.9, 10 mM MgCl2, 1 mM EDTA, 200 mM KCl, 0.1 mM DTT, 0.2 mM PMSF and 50% glycerol).
Mutagenesis of the Gln-196 and Thr-199 in the Region 2.4 of σA
The sigA gene with Gln-196-Ala and Thr-199-Ala substitutions was constructed using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene). The plasmid, pCD2, which contains the full-length sigA gene, was used as the DNA template for mutagenesis. The two primers used for codon replacement are as follows: sigA-196-199A-F: 5′-GCTACGTGGTGGATCAGAGCGGCGATTGCACGCGCCATTGCCGATC-3′ and sigA-196-199A-R: 5′-GATCGGCAATGGCGCGTGCAATCGCCGCTCTGATCCACCACGTAGC-3′. The plasmid containing the correct mutant sigA gene was named pCD2M.
Construction of the spacing variants of the B. subtilis trnS promoter DNA
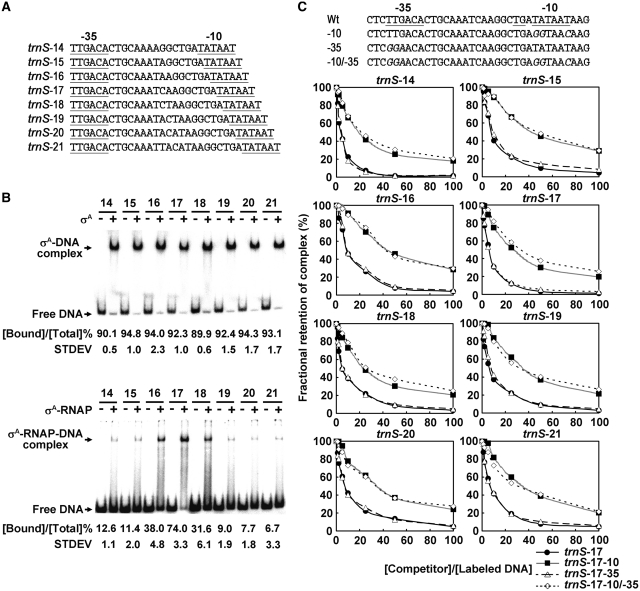
The trnS spacing variants with a 14- to 21-bp spacing (Figure 6A) were created by the QuikChange® Site-Directed Mutagenesis Kit (Stratagene). The templates and primer pairs used for the construction were shown in Supplementary Table S1. All of the resultant spacing variants were sequenced to be correct and were named as trnS-X, in which X indicates the length of spacer ranging from 14 to 21 bp.
Figure 6.
The promoter −10 specific interaction of σA is unable to allow the σA to discern the optimal promoter spacing in vitro. (A) The spacing variants of the B. subtilis trnS promoter used in this study. The highly conserved −10 and −35 elements of the trnS promoter are underlined. (B) The binding of σA (top panel) and σA-RNAP (bottom panel) to the trnS spacing variants. In EMSA, 10 µM σA or 50 nM σA-RNAP was used. The numbers above the horizontal lines are the spacer lengths of the trnS spacing variants. The plus and minus signs denote the addition and omission of σA or σA-RNAP, respectively. The values shown under each panel are the percentages of trnS spacing variant bound by σA or σA-RNAP ([Bound DNA]/[Total DNA] %). Each value was the average of triplicate measurements with a standard deviation (STDEV) shown. (C) Upper panel: the Wt and Mu trnS-17 promoters with base substitutions at either −10, −35 or both elements (indicated by −10, −35 and −10/−35) used for competition assay. The sequences of conserved −10, −35 elements and TG motif of the trnS-17 promoter are underlined, and base substitutions in each Mu promoter are italicized. Lower panel: the fractional retention of the preformed σA-trnS promoter DNA complex as a function of the molar ratio of competitor DNA to 32P-labeled trnS spacing variant ([Competitor]/[Labeled DNA]). The trnS spacing variant is indicated by trnS-X (X is the length of spacer DNA ranging from 14 to 21 bp) above each chart. In this assay, 6 µM σA and 0.5 nM trnS-X promoter DNA were used to generate the σA-trnS-X promoter DNA complex. The molar ratios of competitor DNA to the labeled trnS promoter DNA were 0:1, 1:1, 2:1, 5:1, 10:1, 25:1, 50:1 and 100:1. The band density of each preformed binary complex in the absence of competitor DNA was referred to as 100%.
Construction of the Mu G3b and trnS-17 promoters used for competition assay
The mutation at the −10, −35 element or TG motif of G3b or trnS-17 promoter was created using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene). The templates and primer pairs used for the construction were shown in Supplementary Table S1. All of the promoter mutations were confirmed by DNA sequencing.
To prepare competitor DNA, the Wt and Mu G3b promoter DNA fragments (182 bp in length, spanning from +67 to −115 of the G3b promoter) were synthesized by polymerase chain reaction (PCR). The Wt G3b promoter DNA-containing plasmid, pCT-G3b and the pCT-G3b-derived plasmids containing mutation at −10, −35 element, TG motif or either two elements of the G3b promoter were used as templates and BC1041-BamHI (5′-GAGCTCGGATCCAGAGAACGTAGACAACAA-3′) and BC1046-HindIII (5′-CTGCAGAAGCTTGCCATTTCTTCGTCCCACTTTC-3′) as primers. Similarly, the Wt and Mu trnS-17 promoter DNA fragments (95 bp in length, spanning from +15 to −80 of the trnS-17 promoter) were synthesized by PCR. The Wt trnS promoter DNA-containing plasmid, pOtrnS and the pOtrnS-derived plasmids containing mutation at either −10, −35 or both elements of the trnS-17 promoter were used as templates, and trnSF5 (5′-CATTATAAAAGTCGTCACG-3′) and trnSR3 (5′-TGTGAATAATGAGACAAGAC-3′) were used as primers. The promoter DNA fragments thus synthesized were further run on a 2% agarose gel, cut out from the gel, and purified using the Micro-Elute DNA Clean/Extraction Kit (GeneMark). The purified DNA was diluted with DNA-binding buffer [50 mM Tris–HCl, pH 7.9, 10 mM MgCl2, 160 mM KCl, 0.1 mM EDTA, 0.1 mM DTT and 5% (v/v) glycerol] before being used.
Synthesis and labeling of the promoter DNA fragments
The Wt and Mu G3b promoter DNA were synthesized by PCR using the pCT-G3b or the pCT-G3b-derived plasmids containing mutation at −10, −35 or both elements of the G3b promoter as DNA templates and BC1041-BamHI and BC1046-HindIII as primers. The promoter DNA fragments of trnS-X were synthesized by PCR using the pOtrnS (containing trnS-17) and pOtrnS-derived plasmids containing trnS-X (X is from 14 to 21 except 17) as templates and trnSF5 and trnSR3 as primers. To label the 5′-end of promoter DNA fragments, either reverse or forward primer was labeled by [γ-32P]-adenosine triphosphate (ATP) prior to PCR. For labeling template strand DNA, reverse primer (BC1046-HindIII or trnSR3) was 32P–labeled; for labeling non-template strand DNA, forward primer (BC1041-BamHI or trnSF5) was 32P–labeled. To label primer, 20 pmol of the primer DNA was mixed with 20 pmol of [γ-32P]-ATP and 10 U of T4 polynucleotide kinase (Roche) in 20 µl of phosphorylation buffer (50 mM Tris–HCl, pH 8.2, 10 mM MgCl2, 0.1 mM EDTA, 5 mM DTT and 0.1 mM spermidine), then the mixture was incubated at 37°C for 2 h. The labeled promoter DNA fragments were run on a 2% agarose gel, cut out from the gel and purified using the Micro-Elute DNA Clean /Extraction Kit (GeneMark). The purified DNA was diluted with DNA-binding buffer before being used.
Electrophoretic mobility shift assay
For electrophoretic mobility shift assay (EMSA), the σA or σA-RNAP was incubated with 32P-labeled G3b (1 nM) or trnS (0.5 nM) promoter DNA at 37°C for 10 min. The final volume of the binding mixture was adjusted to 10 µl by adding the DNA-binding buffer. The binding of σA to promoter DNA was performed in the presence of 0.1 µM heparin. The complex of σA and promoter DNA was then run on a 5% non-denaturing polyacrylamide gel in 1× tris–acetate–EDTA (TAE) buffer (40 mM tris–acetate, pH 8.5 and 2 mM EDTA) at 4°C. The complex of σA-RNAP and promoter DNA was run at room temperature. Finally, the gel was dried and analyzed with a Fuji BAS 2500-phosphoimager. For reconstitution of holo RNAP, core RNAP and 10-fold molar excess of σA were incubated at 37°C for 10 min.
In EMSA-based competition assay, the preformed σA-G3b promoter DNA complex obtained by incubating 32P-labeled G3b promoter DNA (1 nM) with an amount of σA in the presence of 0.1 µM heparin at 37°C for 10 min was challenged with the cold Wt, Mu G3b promoter DNA or non-promoter DNA (tgbp2 cDNA of Bamboo mosaic virus) for another 10 min. Then, the samples were run on a 5% non-denaturing polyacrylamide gel in a 4°C-cold chamber. Similarly, the σA-trnS-X promoter DNA complex was preformed by mixing 32P-labeled trnS-X promoter (0.5 nM) with σA in the presence of 0.1 µM heparin. Then, the cold Wt or Mu trnS-17 promoter DNA was added. The mixture was further incubated at 37°C for 10 min before being electrophoresed at 4°C. The fractional retention of the preformed binary complex was measured as a function of the molar ratio of competitor DNA to labeled promoter DNA.
DNase I footprinting assay
The σA or σA-RNAP was incubated with 0.5 nM 32P-labeled G3b or trnS promoter DNA for 30 min at various temperatures (20, 25 and 37°C). The final volume of the binding mixture was 40 µl and the final concentrations of buffer components in the binding mixture were 41.6 mM Tris–HCl, pH 7.9, 10 mM MgCl2, 0.1 mM DTT, 0.35 mM EDTA, 60 mM KCl, 100 µg/ml bovine serum albumin (BSA) and 17.6% glycerol. After incubation, an appropriate amount of DNase I (Roche) was added to digest the DNA. For binding at 20°C, 0.6 (for G3b promoter) or 0.25 U (for trnS promoter) of DNase I was added; for binding at 25°C, 0.4 U (for G3b promoter) of DNase I was added; for binding at 37°C, 0.25 (for G3b promoter) or 0.1 U (for trnS promoter) of DNase I was added. After a proper period of time, 40 µl of stop buffer (50 mM Tris–HCl, pH 7.9, 200 mM NaCl, 20 mM EDTA and 1 mg/ml glycogen) was added to stop the digestion reaction. The digested DNA was extracted with phenol–chloroform. The DNA fragments in the upper aqueous layer were collected and precipitated with 0.3 M NaOAc and 2.5-fold volumes of 100% ethanol, washed twice with 70% ethanol, heat dried (60°C), dissolved in 10 µl of loading buffer (98% deionized formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol) and electrophoresed with an 8% (w/v) polyacrylamide/8 M urea sequencing gel in 1× tris–borate–EDTA (TBE) buffer (90 mM tris–boric acid, pH 8.5 and 2 mM EDTA).
RESULTS
Purification of the B. subtilis primary σ, σA, with a core-independent promoter DNA-binding activity
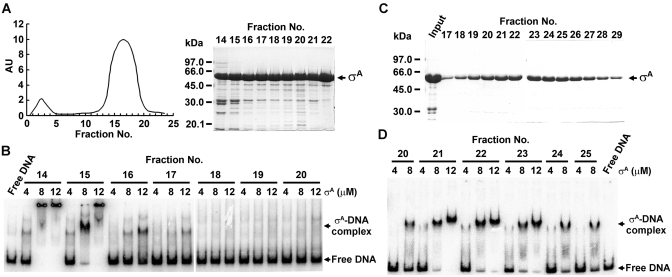
The B. subtilis σA is a primary σ factor. To obtain the σA active in promoter DNA binding, we modified the methods used to purify the overexpressed σA from E. coli (16,47,48). Basically, the σA-containing inclusion bodies were denatured with 7 M Gn–HCl and diluted slowly with refolding buffer drop by drop, rather than stepwise. Then, the refolded σA was subjected to purification with two consecutive molecular sieving columns, Superdex HR-200 and SD-75, rather than with an anion exchange (DEAE cellulose) and a molecular sieving (Sephadex-G75) column. With these modifications, the σA with promoter DNA-binding activity was achieved. Shown in Figure 1A and B are the results of σA purification with Superdex HR-200 and the promoter DNA-binding activities of the fractionated σA as examined by EMSA. Significant binding of the tested ϕ29 phage G3b promoter DNA was observed for the σA eluted earlier from the Superdex HR-200 column (fraction nos 14–16), suggesting that the σA active in promoter DNA binding has a relatively looser conformation. Because the σA sample was still low in homogeneity at this step, we further purified it with an SD-75 molecular sieving column. As shown in Figure 1C and D, the σA eluted from the SD-75 column (fraction nos 21–23) had a much improved homogeneity (∼95%) and was also active in promoter DNA binding with a relatively high apparent dissociation constant (Kd) of about 5 µM, which is similar to that reported for the complex of B. subtilis σD and its cognate promoter (17). Because the active σA sample is free from the β-subunit of E. coli core RNAP and lack of transcription activity (Supplementary Figure S1), we assumed that the σA, by itself, is able to bind the G3b promoter DNA.
Figure 1.
Purification of the B. subtilis σA with a core-independent promoter DNA-binding activity. (A and C) Purification of the in vitro refolded B. substilis σA by molecular sieving columns, Superdex HR-200 (A) and SD-75 (C). AU as shown in the left panel of (A) indicates the absorption unit of the fractionated samples. The fraction no. shown on top of the SDS–polyacrylamide gels indicates the fraction of the σA-containing sample collected from the two columns. (B and D) The G3b promoter DNA-binding activity of σA fractionated by Superdex HR-200 (B) and SD-75 (D) columns. The numbers above the horizontal lines are the fractions of σA-containing sample collected from the two columns. The numbers shown below the horizontal lines are the concentrations (µM) of σA used.
To rule out the possibility that the promoter DNA-binding activity of the in vitro refolded σA is just an artifact due to protein refolding, we further purified the σA overproduced in E. coli in a soluble form at low temperature (16°C) and analyzed its promoter DNA-binding activity. Similar to the refolded σA, the soluble σA was able to bind the G3b promoter DNA independent of core RNAP (Supplementary Figure S2A and B), supporting the hypothesis that the B. subtilis σA is an intrinsic DNA-binding protein.
σA binds preferentially to the promoter −10 element in vitro
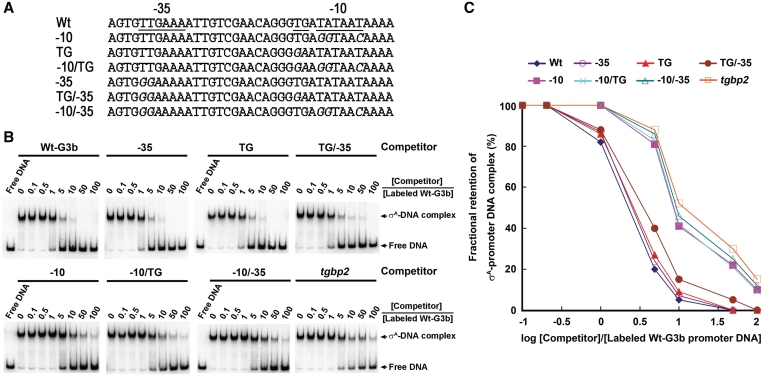
To investigate whether σA has preference for a specific promoter element, six Mu G3b promoter DNA with single or pairwise substitutions at −10 element, −35 element and TG motif (Figure 2A) were adopted for competition assay by EMSA. In this analysis, the binary complex preformed by σA and labeled Wt G3b promoter DNA was challenged with various concentrations of the unlabeled Wt, Mu G3b promoter DNA and non-promoter DNA (the full-length tgbp2 cDNA of Bamboo mosaic virus). As shown in Figure 2B, the preformed binary complex was relatively more sensitive to challenge with the Wt or the Mu G3b promoter DNA having base substitutions at either −35, TG or −35/TG element than to challenge with those having substitutions at −10 element (including −10, −10/TG and −10/-35) or with the non-promoter DNA. The requirement of the promoter −10 element for efficient competition was also visualized when the correlation between the fractional retention of the preformed σA-promoter DNA complex and the log [Competitor]/[Labeled G3b promoter DNA] was examined (Figure 2C). Clearly, σA binds preferentially to the −10 element of G3b promoter in vitro. Moreover, the high efficiency of the non-promoter DNA, albeit lower than that of the Wt G3b promoter DNA, to challenge the preformed σA-Wt G3b promoter DNA complex suggested that the σA also binds the G3b promoter DNA with low sequence selectivity.
Figure 2.
σA binds preferentially to the promoter −10 element. (A) Mutations of the G3b promoter DNA. The sequences of the −10 element, TG motif and −35 element of the Wt G3b promoter are underlined. −10, TG, −10/TG, −35, TG/−35 and −10/−35 indicate the six promoter mutations used for competition assay. Base substitutions in each Mu promoter are italicized. (B and C) Fractional retention of the σA-promoter DNA complex as a function of increasing molar ratio of competitor DNA to 32P-labeled G3b promoter DNA. In the EMSA-based competition assay (B), 8 µM σA and 1 nM 32P-labeled G3b promoter DNA were used to generate the σA-G3b promoter DNA complex. The competitor DNA used to challenge the σA-G3b promoter DNA complex was shown above the horizontal lines. The molar ratios of competitor DNA to the labeled G3b promoter DNA were 0:1, 0.1:1, 0.5:1, 1:1, 5:1, 10:1, 50:1 and 100:1 as indicated under the horizontal lines. In analysis of the correlation between the fractional retention of the preformed binary complex and the log [Competitor]/[Labeled G3b promoter DNA] (C), the band density of each preformed binary complex in the absence of competitor DNA was referred to as 100%. The experiment was repeated twice and the same tendency was obtained.
The low sequence selectivity of σA in vitro
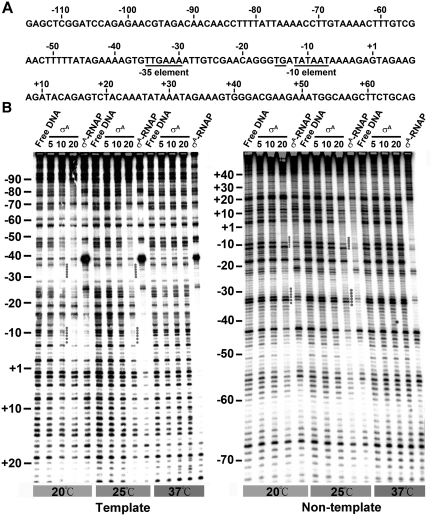
The low sequence selectivity of σA was confirmed by DNase I footprinting which probes the protein-DNA interaction in a time- and population-averaged manner. This assay was performed under various temperatures (20, 25 and 37°C) and σA concentrations (5, 10 and 20 µM), where both partial and complete titrations of the labeled G3b promoter DNA (spanning from −115 to +67 of the promoter) occurred. The specific interaction of σA-RNAP with G3b promoter DNA was also examined for reference. The specific footprint of σA-RNAP on the template strand DNA spanned from about −70 to +1 at 20°C, and extended downstream into +17 at 25 and 37°C (left panel of Figure 3B). Downstream extension of the specific σA-RNAP footprint on the non-template strand DNA was also observed as binding temperature was increased from 20°C (−42 to +10) to 37°C (−42 to +20) (right panel of Figure 3B). Similar footprint extension was reported for E. coli RNAP on various promoters (49,50). Different from the above observations, the footprints of σA on both strands of the G3b promoter DNA were only detected at lower temperatures (20 and 25°C) with σA concentration higher than 10 µM (Figure 3B). The presence of complete or nearly complete protection of the −10 region as well as partial protection near the −35 region supported the idea that σA binds preferentially at least to the −10 element of G3b promoter (Figure 2C). Moreover, the DNA protected by σA was much wider than that protected by σA-RNAP in both the upstream and downstream regions of the core promoter. These results indicated that, despite the preferential binding for −10 element, the σA also binds DNA with low sequence selectivity. The low sequence selectivity of σA could be substantiated by the facts that the σA has a similar affinity for the G3b promoter DNA and the tgbp3 non-promoter DNA which is relatively A-T rich (Supplementary Figure S3B) and that the full-length tgbp2 cDNA is able to compete with the Wt G3b promoter DNA for σA, albeit less efficiently (Figure 2B and C). Similar observations were reported for the B. subtilis σA fragment lacking Region 1 (51) and the E. coli GSTσ(360) (52).
Figure 3.
The low sequence selectivity of σA. (A) The nucleotide sequence of non-template strand DNA of the G3b promoter from −115 to +67. The bases of the −35 element, TG motif and −10 element are underlined. (B) The footprinting patterns of σA and σA-RNAP on the template and non-template strand DNA at the designed temperature (20, 25 or 37°C). The concentrations (µM) of σA used for the footprinting assay are shown under the horizontal lines and the concentration of σA-RNAP used is 100 nM. The numbers shown on the left of each panel are positions relative to the transcription start site of the G3b promoter. The dotted lines indicate the –10 and –35 regions of the G3b promoter DNA protected by σA.
The promoter-specific interaction of σA is −10 specific in vitro
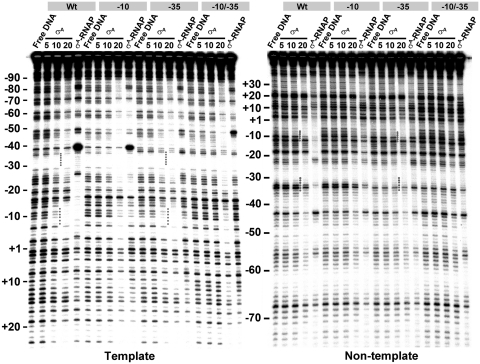
To confirm that σA binds preferentially to the promoter −10 element or that the σA-promoter interaction is −10 specific in vitro, the footprinting patterns of σA on the Wt and the three G3b promoter DNA with base substitutions at either −10, −35 or −10/−35 elements were compared. The footprints of σA at both −10 and −35 elements were observed on both strands of each promoter DNA in the presence of a higher concentration (20 µM) of σA, no matter whether there is a promoter mutation or not (Figure 4). However, the footprints of σA at both −10 and −35 elements (as indicated by dotted line) were only observed for the Wt and the Mu promoter DNA with base substitutions at −35 element as the σA concentration was decreased to 10 µM. No significant footprint of σA at −10 element was detected for the G3b promoter DNA with −10 or −10/−35 substitutions under the same condition. This result supports the idea that the in vitro σA-promoter interaction is −10 specific.
Figure 4.
The effect of base substitutions on the footprinting patterns of σA on the G3b promoter DNA. The effect of base substitutions on the footprinting patterns of σA and σA-RNAP on the template (left panel) and non-template strand DNA (right panel) at 20°C. Wt, −10, −35 and −10/−35 indicate the Wt G3b promoter and the Mu G3b promoter containing base substitution at −10, −35 or both elements, respectively. Other details for experimental conditions and labeling of the figures are the same as those described in the legend to Figure 3.
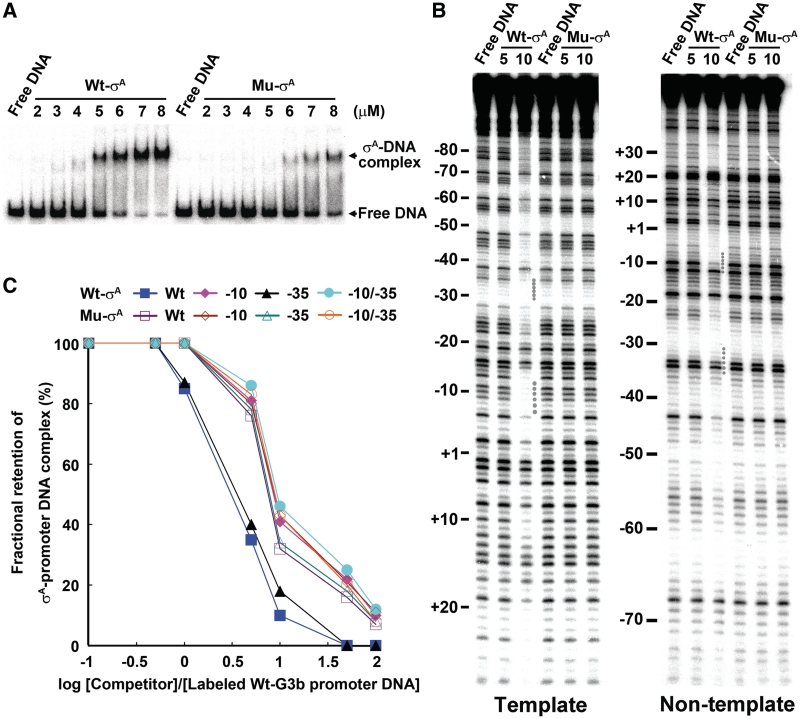
Furthermore, we compared the promoter DNA-binding activities of the Wt- and the Mu-σA of which Gln-196 and Thr-199, which are assumed to be critical for the sequence-selective interaction of σA-RNAP with −12T of promoter DNA (51,53), are replaced with Ala. To achieve accurate comparison, we purified the Wt- and Mu-σA overproduced in E. coli in a soluble form and analyzed their promoter DNA-binding activities. A relatively weaker affinity of the Mu-σA, comparing the Wt, for promoter DNA was observed in EMSA (Figure 5A). Moreover, the footprints of the Mu-σA at −10 and −35 elements on both strands of the G3b promoter DNA were undetectable, whereas those of the Wt-σA were easily detected under the same condition (Figure 5B). More importantly, the −10 preference or −10 specificity became insignificant for the Mu-σA as revealed from competition assay using the Wt G3b promoter DNA or the G3b promoter DNA containing base substitutions at −35 element as a competitor (Figure 5C). The requirement of the conserved amino acid residues in Region 2.4 of σA (Figure 5) and conserved bases in promoter −10 element for efficient σA-promoter DNA interaction (Figure 4) indicated that the promoter interaction of σA in vitro is indeed −10 specific.
Figure 5.
The effect of Gln-196-Ala and Thr-199-Ala substitutions in Region 2.4 of σA on the promoter DNA-binding activity of σA. The σA with Gln-196-Ala and Thr-199-Ala substitutions was designated as Mu-σA. (A) The effect of Gln-196-Ala and Thr-199-Ala substitutions in Region 2.4 of σA on the promoter DNA-binding activity of σA as examined by EMSA. (B) The effect of Gln-196-Ala and Thr-199-Ala substitutions in Region 2.4 of σA on the promoter DNA-binding activity of σA as examined by DNase I footprinting. In both (A) and (B), the numbers shown under Wt-σA or Mu-σA are the concentrations (µM) of σA. Other details for labeling of the figures are the same as those described in the legend to Figure 3. (C) The effect of Gln-196-Ala and Thr-199-Ala substitutions in Region 2.4 of σA on the preferential promoter −10 binding of σA. In the analysis of fractional retention of the Wt- or Mu-σA-promoter DNA complex as a function of increasing molar ratio of competitor to 32P-labeled G3b promoter DNA, 8 µM Wt-σA or 15 µM Mu-σA was mixed with 1 nM 32P-labeled G3b promoter DNA to generate the σA-promoter DNA complex. For competition, the molar ratios of the competitor DNA to the labeled G3b promoter DNA were 0:1, 0.1:1, 0.5:1, 1:1, 5:1, 10:1, 50:1 and 100:1. The band density of each preformed binary complex in the absence of competitor DNA was referred to as 100%. The experiment was repeated twice and the same tendency was obtained.
The −10 specific promoter interaction of σA is unable to allow the σA to discern the optimal promoter spacing in vitro
To see whether σA, by itself, is able to discern the optimal promoter spacing as reported previously (30), the spacing variants of another B. subtilis promoter, trnS, of which the −10, −35 elements, TG motif, and spacing (17 bp) are highly conserved, were constructed. The resultant spacing variants were named as trnS-X, in which X is the length of spacer DNA ranging from 14 to 21 bp (Figure 6A). Moreover, to diminish the potential non-specific binding of σA to the spacing variants in both upstream and downstream regions of the promoter, a shorter DNA fragment of each spacing variant, corresponding to the region from +15 to −80 of the trnS-17 promoter, was adopted to perform EMSA.
The results of EMSA are shown in Figure 6B. σA bound to the spacing variants with about equal efficiency; however, three distinct binding strengths of σA-RNAP were observed among the spacing variants. Optimal binding was seen for trnS-17. Moderate binding was detected for trnS-16 and trnS-18. Lowest binding was observed for the rest of trnS spacing variants with a shorter (14- or 15-bp) or a longer (19-, 20- or 21-bp) spacer. Since the footprint of σA on the trnS promoter was detectable at 20 but not 37°C, we thus also carried out the binding analysis at 20°C throughout the EMSA. Similar EMSA results were obtained for both σA and σA-RNAP (Supplementary Figure S4). Therefore, σA, by itself, is unable to discern the optimal promoter spacing (17 ± 1 bp). To accomplish this function, the assistance from core RNAP is required.
To see whether the −10 specific interaction of σA also occurs with the trnS spacing variants, three Mu trnS-17 promoters with base substitutions at −10, −35 or −10/−35 elements were constructed (top panel of Figure 6C) and then used for EMSA-based competition analyses. As shown in the bottom panel of Figure 6C, the preformed binary complex of σA and each spacing variant was relatively more sensitive to challenge with the Wt or the Mu trnS-17 promoter containing base substitutions at −35 element than with those bearing base substitutions at −10 element (including −10 and −10/−35 mutations). Clearly, similar to the −10 specific G3b promoter interaction of σA, the σA also has a preference for binding to the −10 element of the trnS spacing variants. These results also indicated that the promoter −10 specific interaction of σA is unable to allow the σA to discern the optimal promoter spacing in vitro.
σA contacts the trnS spacing variants in a flexible manner, in addition to the −10 specific interaction
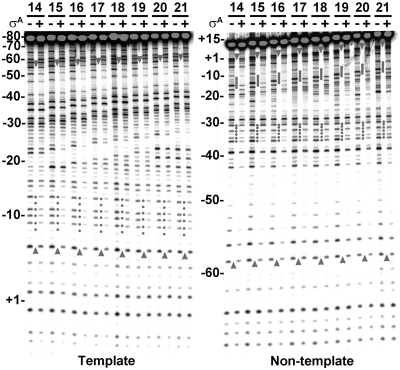
To confirm that σA binds the trnS spacing variants through preferential interaction with their promoter −10 elements, DNase I footprinting assay was performed. To start with, we screened the optimal σA concentration for specific interaction between σA and trnS-17 promoter at 20°C. Specific footprints of σA on both DNA strands of the trnS-17 promoter, spanning from about −58 to −6 on the template and from about −59 to +3 on the non-template strand DNA (as encompassed by the upward and downward arrowheads), were detected when 20 µM of σA was used for binding analysis (Supplementary Figure S5). Thus, the same condition was adopted for DNase I footprinting analyses of σA on other trnS spacing variants. As shown in Figure 7, similar protection patterns, except those in the spacers, were observed for σA on the trnS spacing variants. Moreover, clear footprint was observed at −10 element on the template strand (left panel), but at both −10 and −35 elements on the non-template strand DNA (right panel). Furthermore, there was a gradual upstream extension of the σA footprint on both DNA strands (indicated by the arrowhead) as the spacing was increased from 14 to 21 bp. Since σA only binds specifically to the promoter −10 element of the trnS spacing variants (Figure 6C) and since there is a gradual upstream extension of the σA footprint on the trnS promoter DNA in response to the increase of spacer length, we favored that, in addition to the −10 specific promoter interaction, σA also contacts the −35 element and its upstream sequence of each trnS spacing variant in a flexible manner.
Figure 7.
σA contacts the trnS spacing variants in a flexible manner in addition to the −10 specific interaction. The footprinting patterns of σA on the template (left panel) and non-template strand DNA (right panel) of the trnS spacing variants at 20°C. In this assay, 20 µM σA was used for footprinting assay. The numbers above the horizontal lines are the spacer lengths of the trnS spacing variants. The plus and minus signs denote the addition and omission of σA, respectively. The numbers shown on the left of each panel are positions relative to the transcription start site of the trnS-17 promoter. The DNA protected by σA on both the template and non-template strand DNA of each spacing variant are encompassed by the upward and downward arrowheads. The promoter −10 and −35 elements protected by σA on each trnS spacing variant are indicated by dotted lines.
σA-RNAP, but not σA alone, can recognize optimal promoter spacing
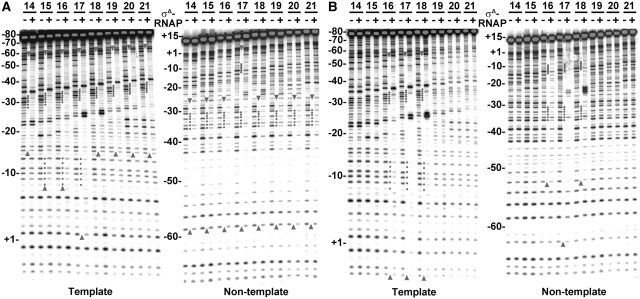
To examine the effect of core RNAP on binding of σA to the trnS spacing variants, the footprints of σA-RNAP on the spacing variants were analyzed at both 20°C and 37°C. Shown in Figure 8A are the footprints of σA-RNAP on the trnS spacing variants at 20°C. Significant footprints of σA-RNAP at the −10 and −35 elements of both DNA strands were detected only for the trnS variants with an optimal spacing (16 and 17 bp). Weak or no footprint of σA-RNAP at the −10 element was observed for the variants with a shorter (14- or 15-bp) or a longer (18-, 19-, 20- or 21-bp) spacer; the footprint on this type of promoter was restricted to the region spanning from the upstream of the −10 element to the position corresponding to −60 of trnS-17 on the template strand DNA and from the −35 element to the position corresponding to −59 of trnS-17 on the non-template strand DNA. Clearly, the σA-RNAP mainly binds the −35 element and its upstream sequence at 20°C if the promoter spacing is non-optimal.
Figure 8.
The footprinting patterns of trnS spacing variants in response to σA-RNAP binding. (A and B) The footprinting patterns of σA-RNAP on the trnS spacing variants at 20 (A) or 37°C (B). The concentration of σA-RNAP used for footprinting assay was 15 nM. Details for labeling the figures are the same as those described in the legend to Figure 7.
Shown in Figure 8B are the footprinting patterns of σA-RNAP on both DNA strands of the trnS spacing variants at 37°C. Again, significant footprints of σA-RNAP at both −10 and −35 elements, consistent with the existence of specific interaction between σA-RNAP and the two promoter elements (Supplementary Figure S6), were observed only for the trnS variants with an optimal spacing, especially that with a 17-bp spacer. No or very weak footprint of σA-RNAP was detected on both DNA strands if the spacer is shorter (14- or 15-bp) or longer (19-, 20- or 21-bp). Taken together, these results demonstrated that core RNAP not only enables σA to discern the optimal promoter spacing but also to simultaneously and specifically recognize both promoter elements as previously reported (27,29).
DISCUSSION
We have documented the in vitro core-independent promoter-specific interaction of the B. subtilis primary σ factor, σA. This interaction is −10 specific; however, it is unable to allow the σA to discern the optimal promoter spacing. To fulfill this goal, the σA requires assistance from core RNAP.
The detection of core-independent −10 specific promoter interaction of σA in vitro is contrary to previous findings with the E. coli σ70 (22,23) and T. maritima σA (24,25), for which it has been reported that Region 1.1 of σ acts as an auto-inhibitor for σ-promoter interaction, and that the promoter interaction in the absence of core RNAP would require deletion of Region 1.1 of σ, just as seen for the σD of B. subtilis (17). Reasons responsible for these contradictory results are complicated. We thought that our ability to detect the core-independent −10 specific promoter interaction of σA is ascribed to the isolation of σA with a proper promoter DNA-binding conformation through the use of a molecular sieving column, Superdex HR-200 (Figure 1) and the use of a relatively low temperature and low ionic strength for DNase I footprinting (Figure 3). The early elution of the σA active in promoter-DNA binding from the Superdex HR-200 column suggests that the σA has a relatively looser conformation. This idea can be supported by our preliminary observation that a σA preparation more active in promoter DNA binding possesses a higher intensity of tryptophan fluorescence (the two tryptophan residues of the B. subtilis σA are located just at the N-terminal end of the promoter −10-binding helix of σA). Possibly, the close proximity of the negatively charged Region 1.1 to the positively charged promoter recognition domains of σA (25) is at least to a certain extent alleviated during the slow refolding of the denatured σA. Moreover, the need of a low temperature and low ionic strength for the detection of an efficient interaction between σA and promoter DNA by DNase I footprinting suggests that the σA-promoter DNA interaction is an exothermic reaction, in which electrostatic and/or hydrogen bonding occur.
The preferential binding of σA, by itself, to the promoter −10 (TATAAT) but not −35 element in vitro (Figures 2B, C and 6C) is consistent with the findings that the consensus sequences of GTA(C/T)AATGGGA and TGTAGAAT, both of which contain the TATAAT-like sequence, are required for single stranded deoxyribonucleic acid (ssDNA) aptamer binding to the Thermus aquaticus free σA (54,55). It is also consistent with our observation that the −10 specific promoter interaction of σA is unable to allow the σA to discern the optimal promoter spacing (Figure 6). The lack of binding specificity of the promoter −35 element by σA can be attributed to the presence of Region 1.1 of σA, which makes σ Region 4.2 inaccessible and thus prevents the interaction between σ Region 4.2 and the promoter −35 element (22–24). The ability of σA to contact the promoter −35 element and its upstream sequence of each trnS spacing variant in a flexible manner (Figure 7) indicates that after the promoter −10 specific interaction of σA, an extensive σA-promoter contact in the −35 and its upstream region occurs. It also suggests that the structure between the two DNA-binding domains of σA is flexible enough for σA to contact the two promoter regions separated by various spacings. The structural flexibility of E. coli σ70 in recognition of the −10- and −35-like sequences has also been reported in the λQ-engaged transcription elongation complex (56). The structural flexibility between the two DNA-binding domains of σA can be attributed to the existence of a relatively unstructured linker, Region 3.2, in σA according to the modeled structure of thermophilic eubacteria σA (57,58). In fact, our analysis of the structural property of the B. subtilis σA by PONDR® also revealed the existence of a disordered flexible Region 3 in σA (Supplementary Figure S7). Thus, the ability of σA in the context of RNAP to discern the optimal promoter spacing must be due to the change of the linker domain from an extended unfolded conformation to a hairpin loop structure fixed in the binary complex of RNAP and promoter DNA (58). This change makes the distance (∼50 Å) between the two DNA-binding domains of σ only more compatible with the ∼17-bp (57.8 Å) spacing (27,59).
The low sequence selectivity of σA (Figure 3 and Supplementary Figure S3) together with the in vitro −10 specific promoter interaction of σA (Figures 2B, C and 6C) suggests σ can either directly recognize the promoter −10 element or indirectly through non-specific interaction with the A-T rich or promoter −10-like sequences ubiquitous on the chromosome DNA, especially in the upstream region of a promoter, and through sliding to the promoter site in a way similar to that proposed for the sliding of E. coli lac repressor to its operator (60,61). Investigation of the existence of in vivo σA-promoter interaction, which is critical to σ function, is currently undergoing in the lab.
FUNDING
National Science Council of Taiwan, Republic of China (NSC 92-2311-B-005-021, NSC 94-2311-B-005-006, and NSC 94-2311-2752-B-005-001-PAE). Funding for open access charge: National Science Council of Taiwan, Republic of China.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the National Science Council of Taiwan, Republic of China, for supporting the research.
REFERENCES
- 1.Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 2.Juang YL, Helmann JD. A promoter melting region in the primary sigma factor of Bacillus subtilis. Identification of functionally important aromatic amino acids. J. Mol. Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- 3.Panaghie G, Aiyar SE, Bobb KL, Hayward RS, de Haseth PL. Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex. J. Mol. Biol. 2000;299:1217–1230. doi: 10.1006/jmbi.2000.3808. [DOI] [PubMed] [Google Scholar]
- 4.Rong JC, Helmann JD. Genetic and physiological studies of Bacillus subtilis sigma A mutants defective in promoter melting. J. Bacteriol. 1994;176:5218–5224. doi: 10.1128/jb.176.17.5218-5224.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomsic M, Tsujikawa L, Panaghie G, Wang Y, Azok J, deHaseth PL. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli sigma(70) in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase-promoter complexes. J. Biol. Chem. 2001;276:31891–31896. doi: 10.1074/jbc.M105027200. [DOI] [PubMed] [Google Scholar]
- 6.Fenton MS, Lee SJ, Gralla JD. Escherichia coli promoter opening and −10 recognition: mutational analysis of sigma 70. EMBO J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 8.Helmann JD, Chamberlin MJ. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 9.Lonetto MA, Brown KL, Rudd KE, Buttner MJ. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc. Natl Acad. Sci. USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardella T, Moyle H, Susskind MM. A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J. Mol. Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- 11.Keener J, Nomura M. Dominant lethal phenotype of a mutation in the −35 recognition region of Escherichia coli sigma 70. Proc. Natl Acad. Sci. USA. 1993;90:1751–1755. doi: 10.1073/pnas.90.5.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 13.Marr MT, Roberts JW. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- 14.Siegele DA, Hu JC, Walter WA, Gross CA. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 15.Zuber P, Healy J, Carter HL, 3rd, Cutting S, Moran CP, Jr, Losick R. Mutation changing the specificity of an RNA polymerase sigma factor. J. Mol. Biol. 1989;206:605–614. doi: 10.1016/0022-2836(89)90569-x. [DOI] [PubMed] [Google Scholar]
- 16.Chang BY, Shyu YT, Doi RH. The interaction between Bacillus subtilis sigma-A (sigma A) factor and RNA polymerase with promoters. Biochimie. 1992;74:601–612. doi: 10.1016/0300-9084(92)90131-w. [DOI] [PubMed] [Google Scholar]
- 17.Chen YF, Helmann JD. The Bacillus subtilis flagellar regulatory protein sigma D: overproduction, domain analysis and DNA-binding properties. J. Mol. Biol. 1995;249:743–753. doi: 10.1006/jmbi.1995.0333. [DOI] [PubMed] [Google Scholar]
- 18.Kudo T, Doi RH. Free sigma factor of Escherichia coli RNA polymerase can bind to DNA. J. Biol. Chem. 1981;256:9778–9781. [PubMed] [Google Scholar]
- 19.Kudo T, Jaffe D, Doi RH. Free sigma subunit of Bacillus subtilis RNA polymerase binds to DNA. Mol. Gen. Genet. 1981;181:63–68. doi: 10.1007/BF00339006. [DOI] [PubMed] [Google Scholar]
- 20.Ramesh U, Meares CF. Footprint of the sigma protein. Biochem. Biophys. Res. Commun. 1989;160:121–125. doi: 10.1016/0006-291x(89)91629-x. [DOI] [PubMed] [Google Scholar]
- 21.Wellman A, Meares CF. Footprint of the sigma protein: a re-examination. Biochem. Biophys. Res. Commun. 1991;177:140–144. doi: 10.1016/0006-291x(91)91959-g. [DOI] [PubMed] [Google Scholar]
- 22.Dombroski AJ, Walter WA, Gross CA. Amino-terminal amino acids modulate sigma-factor DNA-binding activity. Genes Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- 23.Dombroski AJ, Walter WA, Record MT, Jr, Siegele DA, Gross CA. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 24.Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW. Autoregulation of a bacterial sigma factor explored by using segmental isotopic labeling and NMR. Proc. Natl Acad. Sci. USA. 2002;99:8536–8541. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz EC, Shekhtman A, Dutta K, Pratt MR, Cowburn D, Darst S, Muir TW. A full-length group 1 bacterial sigma factor adopts a compact structure incompatible with DNA binding. Chem. Biol. 2008;15:1091–1103. doi: 10.1016/j.chembiol.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borukhov S, Severinov K. Role of the RNA polymerase sigma subunit in transcription initiation. Res. Microbiol. 2002;153:557–562. doi: 10.1016/s0923-2508(02)01368-2. [DOI] [PubMed] [Google Scholar]
- 27.Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 28.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 29.Young BA, Anthony LC, Gruber TM, Arthur TM, Heyduk E, Lu CZ, Sharp MM, Heyduk T, Burgess RR, Gross CA. A coiled-coil from the RNA polymerase beta' subunit allosterically induces selective nontemplate strand binding by sigma(70) Cell. 2001;105:935–944. doi: 10.1016/s0092-8674(01)00398-1. [DOI] [PubMed] [Google Scholar]
- 30.Dombroski AJ, Johnson BD, Lonetto M, Gross CA. The sigma subunit of Escherichia coli RNA polymerase senses promoter spacing. Proc. Natl Acad. Sci. USA. 1996;93:8858–8862. doi: 10.1073/pnas.93.17.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helmann JD. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoyama T, Takanami M, Ohtsuka E, Taniyama Y, Marumoto R, Sato H, Ikehara M. Essential structure of E. coli promoter: effect of spacer length between the two consensus sequences on promoter function. Nucleic Acids Res. 1983;11:5855–5864. doi: 10.1093/nar/11.17.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaurin B, Grundstrom T, Edlund T, Normark S. The E. coli beta-lactamase attenuator mediates growth rate-dependent regulation. Nature. 1981;290:221–225. doi: 10.1038/290221a0. [DOI] [PubMed] [Google Scholar]
- 36.Russell DR, Bennett GN. Construction and analysis of in vivo activity of E. coli promoter hybrids and promoter mutants that alter the −35 to −10 spacing. Gene. 1982;20:231–243. doi: 10.1016/0378-1119(82)90042-7. [DOI] [PubMed] [Google Scholar]
- 37.Hidalgo E, Demple B. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 1997;16:1056–1065. doi: 10.1093/emboj/16.5.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandecki W, Reznikoff WS. A lac promoter with a changed distance between −10 and −35 regions. Nucleic Acids Res. 1982;10:903–912. doi: 10.1093/nar/10.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkhill J, Brown NL. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 1990;18:5157–5162. doi: 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auble DT, deHaseth PL. Promoter recognition by Escherichia coli RNA polymerase. Influence of DNA structure in the spacer separating the −10 and −35 regions. J. Mol. Biol. 1988;202:471–482. doi: 10.1016/0022-2836(88)90279-3. [DOI] [PubMed] [Google Scholar]
- 41.Ayers DG, Auble DT, deHaseth PL. Promoter recognition by Escherichia coli RNA polymerase. Role of the spacer DNA in functional complex formation. J. Mol. Biol. 1989;207:749–756. doi: 10.1016/0022-2836(89)90241-6. [DOI] [PubMed] [Google Scholar]
- 42.McKane M, Gussin GN. Changes in the 17 bp spacer in the PR promoter of bacteriophage lambda affect steps in open complex formation that precede DNA strand separation. J. Mol. Biol. 2000;299:337–349. doi: 10.1006/jmbi.2000.3757. [DOI] [PubMed] [Google Scholar]
- 43.Mulligan ME, Brosius J, McClure WR. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J. Biol. Chem. 1985;260:3529–3538. [PubMed] [Google Scholar]
- 44.Stefano JE, Gralla JD. Spacer mutations in the lac ps promoter. Proc. Natl Acad. Sci. USA. 1982;79:1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Travers AA. Structure and function of E. coli promoter DNA. CRC Crit. Rev. Biochem. 1987;22:181–219. doi: 10.3109/10409238709101483. [DOI] [PubMed] [Google Scholar]
- 46.Warne SE, deHaseth PL. Promoter recognition by Escherichia coli RNA polymerase. Effects of single base pair deletions and insertions in the spacer DNA separating the −10 and −35 regions are dependent on spacer DNA sequence. Biochemistry. 1993;32:6134–6140. doi: 10.1021/bi00075a003. [DOI] [PubMed] [Google Scholar]
- 47.Chang BY, Doi RH. Overproduction, purification, and characterization of Bacillus subtilis RNA polymerase sigma A factor. J. Bacteriol. 1990;172:3257–3263. doi: 10.1128/jb.172.6.3257-3263.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang BY, Doi RH. Conformational properties of Bacillus subtilis RNA polymerase sigma A factor during transcription initiation. Biochem. J. 1993;294:43–47. doi: 10.1042/bj2940043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofer B, Muller D, Koster H. The pathway of E. coli RNA polymerase–promoter complex formation as visualized by footprinting. Nucleic Acids Res. 1985;13:5995–6013. doi: 10.1093/nar/13.16.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schickor P, Metzger W, Werel W, Lederer H, Heumann H. Topography of intermediates in transcription initiation of E. coli. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang X, Lopez de Saro FJ, Helmann JD. Sigma factor mutations affecting the sequence-selective interaction of RNA polymerase with −10 region single-stranded DNA. Nucleic Acids Res. 1997;25:2603–2609. doi: 10.1093/nar/25.13.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dombroski AJ. Recognition of the −10 promoter sequence by a partial polypeptide of sigma 70 in vitro. J. Biol. Chem. 1997;272:3487–3494. [PubMed] [Google Scholar]
- 53.Kenney TJ, Moran CP., Jr Genetic evidence for interaction of sigma A with two promoters in Bacillus subtilis. J. Bacteriol. 1991;173:3282–3290. doi: 10.1128/jb.173.11.3282-3290.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, Kuznedelov K, Merkiene E, Stavrovskaya E, Klimasauskas S, et al. A basal promoter element recognized by free RNA polymerase sigma subunit determines promoter recognition by RNA polymerase holoenzyme. Mol. Cell. 2006;23:97–107. doi: 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 55.Sevostyanova A, Feklistov A, Barinova N, Heyduk E, Bass I, Klimasauskas S, Heyduk T, Kulbachinskiy A. Specific recognition of the −10 promoter element by the free RNA polymerase sigma subunit. J. Biol. Chem. 2007;282:22033–22039. doi: 10.1074/jbc.M702495200. [DOI] [PubMed] [Google Scholar]
- 56.Devi PG, Campbell EA, Darst SA, Nickels BE. Utilization of variably spaced promoter-like elements by the bacterial RNA polymerase holoenzyme during early elongation. Mol. Microbiol. 2010;75:607–622. doi: 10.1111/j.1365-2958.2009.07021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 58.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 59.Callaci S, Heyduk E, Heyduk T. Core RNA polymerase from E. coli induces a major change in the domain arrangement of the sigma 70 subunit. Mol. Cell. 1999;3:229–238. doi: 10.1016/s1097-2765(00)80313-5. [DOI] [PubMed] [Google Scholar]
- 60.Barkley MD. Salt dependence of the kinetics of the lac repressor–operator interaction: role of nonoperator deoxyribonucleic acid in the association reaction. Biochemistry. 1981;20:3833–3842. doi: 10.1021/bi00516a026. [DOI] [PubMed] [Google Scholar]
- 61.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.