Abstract
Isothermal Titration Calorimetry (ITC) provides a sensitive and accurate means by which to study the thermodynamics of RNA folding, RNA binding to small molecules, and RNA–protein interactions. The advent of extremely sensitive instrumentation and the increasing availability of ITC in shared facilities have made it increasingly valuable as a tool for RNA biochemistry. As an isothermal measurement, it allows analysis at a defined temperature, distinguishing it from thermal melting approaches (UV melting and differential scanning calorimetry, for instance) that provide thermodynamic information specific to the melting temperature. Residual structures at low temperature in the unfolded state and heat capacity changes lead to potential differences between thermodynamic values measured by ITC and those derived from melting studies. This article describes how ITC can be put to use in the study of RNA biochemistry.
1. Introduction
Isothermal Titration Calorimetry (ITC) is a methodology that directly measures the heat (q) taken up or given off by a reaction. Since heat is a universal readout for any reaction, no extrinsic labeling of the sample is required. Reactions are followed over a series of small injections such that in the earliest injections, reactions go to completion due to the large molar excess of the titrate (material in the sample cell) relative to the titrant (material in the syringe being added to the cell), but by the end of the titration, no binding occurs because all of the titrate is already in a complex with its binding partner. A single ITC titration allows one to measure an affinity constant for an interaction (Ka), the reaction enthalpy (ΔH) and the stoichiometry (n). This differs from thermal melting studies where for bimolecular interactions, one often collects data over a range of concentrations to obtain comparable thermodynamic data. Since Ka is related to the Gibbs Free Energy change (ΔG), one can solve indirectly the entropy change (ΔS). If reactions are carried out over multiple temperatures, heat capacity changes (ΔCP) of binding can also be determined.
The most common instruments used for the ITC analysis of biological materials are power-compensation calorimeters that measure the power consumption required to keep a sample cell and a control cell at constant temperature during the titration (Wiseman et al., 1989). As energy is released by an exothermic reaction, less power is required to hold the cells at the same temperature and a negative deflection from the baseline is observed. If a reaction is endothermic, a positive deflection occurs. Since practically all biological interactions have at least a small enthalpic component, most binding reactions can be studied in this manner. This chapter will lead the reader through the sample preparation, binding reaction, and data analysis required for the characterization of an RNA binding to another species. Examples of such experiment include two RNAs binding each other to form duplexes or tertiary structures (Mikulecky and Feig, 2006a; Mikulecky et al., 2004; Reymond et al., 2009; Takach et al., 2004; Vander Meulen et al., 2008), RNAs binding to proteins (McKenna et al., 2006; Niedzwiecka et al., 2004; Recht and Williamson, 2004; Recht et al., 2008), or small molecule ligands (Bernacchi et al., 2007; Gilbert and Batey, 2009; Li et al., 2004). Examples of all three classes of experiments have been measured successfully by ITC.
2. Required Materials
VP-ITC or ITC200
Loading syringe
Thermovac station for degassing samples
Titrant solution
Titrate solution
5% solution of Top Job or Mr. Clean (for cell cleaning)
ddH2O
3. Instrumentation
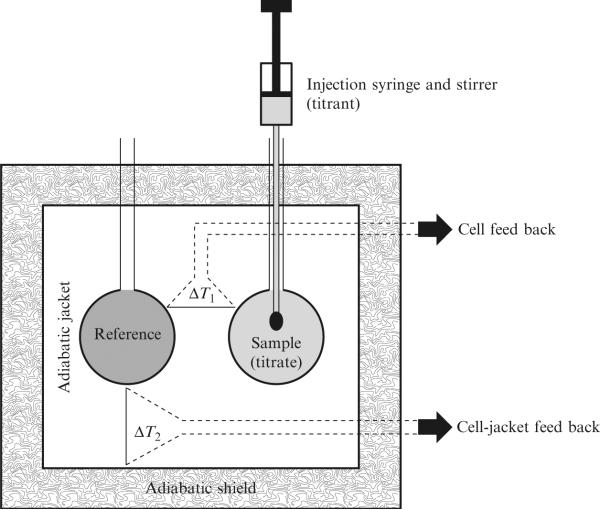
A schematic diagram of an ITC instrument is shown in Fig. 19.1. This schematic is based on the VP-ITC produced by MicroCal (now a subsidiary of GE Healthcare) but is in principle equivalent to the ITC200 (MicroCal), and nano-ITC2G (TA instruments). The auto-ITC200 (MicroCal) has the added attachment of an autoloader to facilitate higher throughput for compound screening. The titrant is loaded into the sample chamber and the sample is sealed with a special injection syringe. The paddle-shaped needle of this syringe doubles as the stirring mechanism within the cell. Injections occur through the action of a screw-driven plunger allowing accurate delivery of as little as 1 μL for the VP-ITC and 0.1 μL for the ITC200. Specific instrument parameters for data collection are described below and include settings for temperature, injection volume, injection speed, and stirring speed.
Figure 19.1.
Schematic diagram of a typical power compensation isothermal titration calorimeter such as the VP-ITC or the ITC200. (Reprinted with permission from Salim and Feig (2009). Copyright, Elsevier, Inc.)
Most casual and first-time ITC users will take advantage of a multiuser facility that has an ITC. This situation poses a significant problem for most RNA experiments. The issue derives from the training and calibration routine popularized by MicroCal involving the binding of CMP to Ribonuclease A (Wiseman et al., 1989). While this is a terrific standard for labs working on proteins and small molecules, it involves loading the sample cell with concentrated nuclease solutions that will readily destroy any RNA sample. For users considering the use of such a facility, it is essential that you speak with the facility manager and inquire about the training and calibration protocols well in advance of running your experiment. Alternative protocols are in use today including titrations involving Ni(II) binding to histidine (Salim and Feig, 2009), Ba(II) binding to 18-Crown-6 (Gilbert and Batey, 2009), or tris base with nitric acid (Baranauskiene et al., 2009). We strongly urge facilities to consider switching to one of these more benign reactions as a training standard. Whether or not RNase A has been in the sample cell recently, it is prudent to test the cell for nuclease contamination in advance of any experiment. Typically, this involves loading an RNA sample in the cell, incubating it for several hours or overnight, followed by PAGE analysis. If sample integrity is compromised, one should perform the stringent decontamination protocols described below.
4. Sample Considerations and Preparation
One of the major drawbacks of ITC is the sample sizes required for analysis. While this has been coming down in recent years, it is still quite significant and limits certain applications, especially with naturally isolated RNAs or those containing synthetic modifications. On the VP-ITC instrument with a 1.44 mL sample cell, a typical titration requires approximately 1.8–2.0 mL of titrate at a concentration of 1–2 μM. The titrant will typically be at a concentration 10–20 times that of the titrate. Even though the injection syringe only holds 240 μL, one realistically needs about 400 μL to fill it in a bubble-free manner. The newer ITC200 has a 200 μL sample cell and 40 μL injection syringe. Sample requirement is thus about sevenfold smaller in this instrument.
While this concentration range is often a good place to start, one may adjust this up or down depending on the specific interaction being studied. The limitations are dependent on the experimental enthalpy of the interaction and the binding constant. When binding enthalpy is low, the experiment becomes heat limited. This problem can be resolved by increasing the sample concentration or the injection volume as either will increase the heat evolved per injection. Ideally, one uses a parameter called the c-value (defined in Eq. (19.1)) to set the concentration of titrate where n is the stoichiometry of the interaction, KB is the affinity constant and MT is the concentration of the titrate in the cell. The c-value must lie between 1 and 1000 for ITC data to be meaningful and optimally between 10 and 100 (Tellinghuisen, 2005). Theoretical studies have shown that the optimal concentration ratio (Rm) of titrant (BT) to titrate (MT) for ITC is defined in Eq. (19.2) and is between 15 and 30 for simple binding systems with 1:1 stoichiometry (Tellinghuisen, 2005).
| (19.1) |
| (19.2) |
The process of making RNA goes beyond the scope of this discussion. However, one does need to be sure that large amounts of high-quality material are available, be it T7 transcribed or synthetic. It should be clean of impurities (typically HPLC, FPLC, or gel purified depending on size) and properly folded. Specific artifacts to watch out for and avoid include the formation of dimeric species due to the high concentrations required in the titration syringe and alternative folds. Such species are particularly prevalent in the analysis of hairpins when annealed at high concentrations. It is strongly recommending that native gel analysis be performed on the material prior to ITC to ensure that material is free from these types of defects as the energy associated with unfolding an alternative conformer will adversely affect the thermodynamic values measured and the presence of inactive conformers can impact the measured stoichiometry of the reaction.
The size of the RNAs to be used also dictates to a great extent what must happen just prior to the experiment. A critical issue for collecting high-quality ITC data is the buffer-match between titrate and titrant solutions. The best way to ensure that both samples are in identical buffer conditions is to dialyze them against the same reservoir overnight prior to the experiment. This process ensures rigorously identical buffer conditions and works well if the RNAs are long, but becomes problematic for short RNAs, like the 6–10 nt species often used to study of duplex formation. In the latter case, the final steps in the purification are often either HPLC purification and ethanol or LiClO4/Acetone precipitation. In these cases, special care should be taken to remove as much salt as possible during workup as mismatch between the ionic strength of the samples leads to significant heats of dilution. This problem is mitigated somewhat by using moderate to high salt (100 mM or greater) binding buffers but can still be a significant source of problems.
Knowledge of the exact concentration of the RNAs is also very important, as any errors will result in a nonintegral stoichiometry. Readers should be aware that approximations based on base composition are useful for very short RNAs or for the determination of ballpark concentrations, but the errors get significant for longer and more structured nucleic acids. It is best to measure exact extinction coefficients for folded RNAs and use those for determining the sample concentrations.
5. Cleaning the Sample Cell and Titration Syringe
Maintaining a clean sample cell is essential for the proper and effective operation of the ITC. While RNAs are typically quite soluble and not particularly prone to fowling of the cell, one is often measuring the binding of the RNA to a protein or small molecule that may have less solubility and greater potential for deposition onto cell walls. On a multiuser instrument, we always recommend performing stringent cleaning prior to beginning a multiday experiment as it is sometimes unclear how careful the previous user has been. This precaution can prevent accidental degradation of precious RNA samples. Three separate cleaning protocols are described below.
5.1. Gentle cleaning before and after runs
Standard cleaning involves rinsing the cell with degassed ddH2O, washing it with 100 mL 5% Mr. Clean or Top Job and then flushing the cell exhaustively with up to 1 L of ddH2O. This procedure can be run overnight as well, soaking the instrument in 5% Mr. Clean followed by flushing the instrument with water the next day. It is essential to remember to also flush and rinse the titration syringe and loading syringes.
5.2. Stringent cleaning and nuclease contamination issues
The preliminary test for all RNA experiments on a new instrument should always be a stability control experiment. If degradation occurs within the instrument, stringent cleaning and decontamination are essential. Sample degradation will dramatically erode data quality and the degradation process itself is exothermic leading to heat evolution over time that affects baseline stability and changes the sample concentration during the course of the experiment. Stringent cleaning of the ITC cell involves filling the sample cell with a 5% (v/v) solution of Contrad-70 and letting it incubate for several hours, often at elevated temperature (up to 65 °C). This step is reasonably effective at removing minor nuclease contamination (such as left over from a protein titration of a previous user) although repeated treatment is sometimes necessary for more significant contamination (experiments involving nuclease, for instance). Stringent cleaning is also required periodically to remove buildup from the cell walls, especially after the use of protein samples.
Protein buildup in the cell can be removed by trypsin or proteinase K treatment of the cell. Our protocol involves filling the cell with a degassed solution of 1 mg/mL solution of proteinase K in 50 mM Tris buffer, pH 7.5, and incubating it in the cell for at least 1 h (but can be left in the cell overnight). The enzyme solution is then removed from the cell and the cell is rinsed with 500 mL ddH2O followed by the gentle cleaning cycle described above.
Serious nuclease contamination occurs if the RNase A calibration routine has been used in the instrument. In these cases, as stated above, it can be extremely difficult to fully decontaminate the instrument. After proteinase K treatment, the cell and titration syringe should be filled with neat RNaseZap (Ambion) and allowed to incubate overnight. After rinsing the cell with deionized water, follow the gentle cleaning protocol above and retest the cell for contamination. This procedure may have to be repeated two or three times before sample integrity is maintained.
6. Collecting Titration Data
Data collection in an ITC titration is a two-step process involving collection of information on the background heat and the heat of a reaction itself. Sources of background heat include buffer mismatch, heats of dilution, heats of mixing, etc. and accounting for this energy is an important part of the data analysis. One of the two protocols for the background measurement can be used, the selection of which dictates the way in which the primary data will be collected. Our preferred method is a procedure involving just a single titration where we collect additional data at the end of a titration after all binding is complete. The second method (which is probably more common in the field of biocalorimetry) involves performing a second independent titration where the titrant is added to a buffer solution in the absence of titrate. We prefer the single titration method because it saves time and material relative to performing an independent series of background injections, but it cannot be used in all cases. In particular, if there is a significant nonspecific binding above and beyond a specific binding event, one must collect the background titration separately (two titration method). Standard data collection parameters for a typical RNA experiment are listed in Table 19.1.
Table 19.1.
Typical experimental parameters for an ITC experimenta
| Experimental parameter | Setting | Comments |
|---|---|---|
| Cell temperature | 2–80 °C | See specific concerns regarding high-temperature experiments in text |
| No of injections | 25–35b | Based on single titration method; fewer injections can be used for the two-titration method |
| Injection volume | First Inj.: 1–2 μl; Rest of Inj.: 7–12 μl | First small injection is used to account for syringe backlash; data from this injection should be omitted from data analysis |
| Injection spacing | 250–500 s | Typical spacing is about 300 s; should be lengthened if titration fails to adequately return to baseline prior to commencing the next injection. Spacing can be shortened if long flat baselines are present between each injection |
| Titrate/titrant concentration | Cell: 1.8 μM; Syringe: 42 μM | Can be optimized depending on reaction heat; this example comes from an RNA-RNA experiment with ΔH of ~ 40 kcal/mol and a 1:1 stoichiometry |
| ITC equilibration options | Automatic | |
| Reference power | 25–30 μCal/s | Setting a much higher value than is required may affect the sensitivity of the instrument |
| Initial delay | 60–100 s | Ensure stable baseline prior to first injection |
| Stirring speed | 270–310 rpm | Faster stirring speeds may lead to high-frequency noise in the data |
| Feedback mode | Fast | |
| Filter period | 2 s | For slow reactions, the filter period can be increased to improve data quality |
Based on use with a VP-ITC from MicroCal. Specific parameter settings might need to be adjusted for use with other instruments.
A terminal ratio of about 3 is obtained for a 1:1 interaction (i.e., the final excess ratio of the titrant over the titrate).
6.1. Choosing who should be titrated into whom
Concern about who should be titrated into whom is pretty common. The simple answer is that for well-behaved systems, it should not matter and one should get equivalent data from either direction. In practice, however, it sometimes does make a big difference. One sample must be 15–20 times more concentrated than the other. For protein or small molecule binding, this fact sometimes leads to solubility problems. For those dealing with RNA-RNA titrations, problems can be manifested in terms of alternative folding problems like dimerization equilibria. In general, if one binding partner has lower solubility, consider using this material as the titrate initially. The other consideration is one of nonspecific binding. This manifests itself in terms of nonlinear baselines at the end of the titration and noninteger stoichiometries.
6.2. Data collection procedure using the single titration method
Degas 2 mL of 1–2 μM solution of RNA 1 (titrate), 300 μL of a 15–30 m–μ solution of RNA 2 (titrate) and some excess buffer using the thermovac aparatus. Rinse the cell with titration buffer and empty completely. Fill the sample cell with titrate being careful not to bend the cell-loading syringe or to introduce bubbles into the cell. Then load the syringe with the titrate solution. Save the excess solutions from loading the cell and use this material to determine the exact concentration of the reagents in the cell and the syringe. Carefully affix the titration syringe into the top of the sample cell and prepare the instrument by setting the data collection parameters. The small initial injection of 1–2 μL will not be used in the data analysis. This step prevents an artifact associated with the mechanical backlash of the screw mechanism in the drive syringe from which under delivers titrant in this first injection (Mizoue and Tellinghuisen, 2004). Plan the experiment to ensure that 5–10 data points are collected after complete saturation is achieved, as these will be used to define the background heat for the reaction. For a 1:1 stoichiometry experiment, this will often require that you titrate out to a molar ratio of ~2.5. The integrated injection heats from the last few injections will be fit to a linear model and extrapolated through the entire dataset as the background correction.
6.3. Data collection procedure using the two-titration method
This procedure is similar to the single titration method except that less titrant is used in the initial titration with a terminal molar ratio of approximate 1.5–2 instead of 2.5–3. After the initial data collection is complete, empty and rinse the cell and refill the instrument with buffer. Then, using the exact same instrument protocol as for the first titration, titrate the titrant into buffer. This will serve as the background titration and integrated heats of injection from this run are subtracted from the experimental run. Note that in this case, the titrant is clean but dilute at the end of the background titration and can be recovered so long as no degradation occurred.
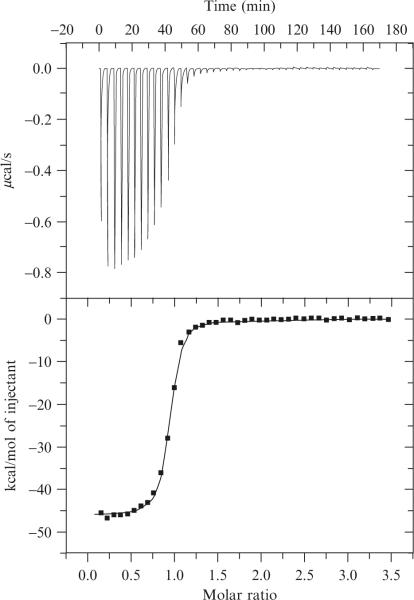
If the experiment has worked well, data should look like that shown in Fig. 19.2. Specific things to look for in the raw data are: (i) low noise in the heat versus time curve; (ii) clean, well-shaped peaks; (iii) level baseline after each injection (injection i should not tail into injection i+ 1); (iv) clear evidence of saturation behavior; (v) two or more points with maximal heat evolution at the beginning of the titration; (vi) several points within the transition region; (vii) small integrated heat from injections after saturation was achieved. Problems with (i-iv) typically derive from data collection parameter issues such as stirring speeds, injection volumes, and injection rates. Data quality problems with (v-vii) usually derived from issues related to titrate or titrant concentrations. In general, when working with small and precious samples with low heat output (small molecule binding to RNAs), the best data often results from using fewer injections. The limit of this is actual a titration involving just a single injection which gives very accurate ΔH parameters but provides no information on binding affinity. If the reaction being studied has a large ΔH (such as most RNA duplex formation experiments), then a larger number of small volume injections often works well in our hands.
Figure 19.2.
Example ITC data from an experiment measuring the association of two single-stranded 7-mer RNAs. Data collected at 15 °C in 50mM HEPES, pH 7.5, and 1 M NaCl. Top panel. Power versus time curve. Bottom panel. Integrated injection enthalpy plotted versus the mole ratio of the reactants. ΔH = −46.1 ± 0.2 kcal/mol, Ka = 4.3 × 10−7 M and n = 0.91 ± 0.1 (Reprinted with permission from Takach et al. (2004). Copyright, American Chemical Society.)
7. Data Processing and Analysis
Data analysis is typically performed using the Origin software supplied by the manufacturer. The automated routines allow for numerical integration of the peaks and subtraction of background heat from either the onetitration or two-titration methods. Note that the heat from injection i (qi) is given by the formula in Eq. (19.3) where n is the reaction stoichiometry, F is the fractional saturation of the reaction, MT is the titrate concentration, ΔH is the reaction enthalpy and V is the cell volume. For a simple binding equilibrium when the concentration of titrant (BT) is known, one can solve for the fractional saturation at any point along the titration based on Eqs. (19.4) and (19.5). Equation (19.5) is then fit by nonlinear least squares methods to determine n, KB and ΔH (Wiseman et al., 1989).
| (19.3) |
| (19.4) |
| (19.5) |
where
More complicated binding models involving sequential parallel binding equilibria are available if necessary, but knowledge of the system involved should be used to determine if their use is warranted.
8. Special Considerations
8.1. The importance of pH and proton association/dissociation reactions
The experimental pH of a titration is of special interest in some ITC studies. Due to the polyanionic nature of RNA and the typical positive charge of RNA binding proteins, there is the potential for changes in protonation state to occur during binding. While this occasionally occurs in simple RNA folding interactions where intrinsic pKas are altered, it is common in RNA–protein interactions. One must pay attention to it in the case of ITC because it leads to an enthalpic artifact that one must correct for if it occurs. If a proton is lost or taken up during the binding event, the reaction is accompanied by (de)protonation of the buffer. This results in a very specific artifact in which the measured enthalpies become buffer dependent. This problem can be solved by either shifting the pH of the experiment to make the proton inventory zero or remeasuring the thermodynamic values in a second buffer at the same pH. In this case, if one knows the ionization enthalpies of both buffers, one can calculate the corrected enthalpy change of the reaction (ΔHcorr) by accounting for the ΔHion and the number of protons released or taken up (Δn)using Eqs. (19.6) and (19.7). Tables of the protonation enthalpies (ΔHion) for common biological buffers can be found in the literature (Feig, 2007; Fukada and Takahashi, 1998).
| (19.6) |
| (19.7) |
8.2. Temperature-dependent phenomena
Measurement temperature is a major concern with the evaluation of thermodynamic parameters of RNA interactions by ITC. While the ability to set the experimental temperature is one of the major strengths of ITC relative to thermal melting analysis, it is also one of the greatest weaknesses if sufficient care is not taken to recognize and account for potential artifacts.
There are two main issues at play. The first has to do with residual structure. When RNA folding thermodynamics is studied by thermal melting, the unfolded or unbound form is a high-temperature state whereas the structured form is the low-temperature state. ITC, on the other hand, is an isothermal measurement as the name implies. Both the bound and free states are at modest temperature and that means that the uncomplexed form of the RNA may exhibit significant residual structure (Feig, 2007; Holbrook et al., 1999; Mikulecky and Feig, 2006a,b). Thus, the heats that are measured in this experiment are really the sum of the typically unfavorable unfolding of whatever residual structure exists in the initial state followed by the typically favorable free energy associated with folding into the final bound state. Depending on the RNAs being studied and the extent of the folding of the starting materials, this can lead to very complex temperature dependencies yielding highly nonlinear heat capacity changes (Feig, 2007; Mikulecky and Feig, 2006b).
A second effect occurs at high temperature. As we know from thermal melting analysis, as one approaches the TM for a given RNA folding transition, the amount of product that forms at equilibrium is temperature dependent. In ITC, one relies on the fact that all of the titrant can and will bind the titrate. However, if a fraction of the material is thermally unfolded at a given temperature, this material may not bind, affecting the fractional saturation F at equilibrium, Eq. (19.4). Under these conditions, if one studies an RNA by ITC at the high temperature (a good working definition of high temperature for this discussion is within 20 °C of the melting transition), the data might need to be corrected for the percentage of unfolded material. Note, there are many systems that might be of interest for study with TMs in the range of 50–60 °C. For these systems, this effect already leads to measureable deviations at 37 °C, so care must be taken to know the TM behavior of any systems being studying.
9. Conclusions
ITC can be put to excellent use in the analysis of RNA folding and RNA-binding interactions. Care must be taken, however, to design the experiments to respect the idiosyncrasies of this biopolymer and understand how these thermodynamic values differ from those obtained by thermal melting. With due care and increasingly available instruments, ITC is likely to be of exceptional value to RNA scientists interested in fundamental folding and binding phenomena.
REFERENCES
- Baranauskiene L, et al. Titration calorimetry standards and the precision of isothermal titration calorimetry data. Int. J. Mol. Sci. 2009;10:2752–2762. doi: 10.3390/ijms10062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S, et al. Aminoglycoside binding to the HIV-1 RNA dimerization initiation site: Thermodynamics and effect on the kissing-loop to duplex conversion. Nucleic Acids Res. 2007;35:7128–7139. doi: 10.1093/nar/gkm856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig AL. Applications of isothermal titration calorimetry in RNA biochemistry and biophysics. Biopolymers. 2007;87:293–301. doi: 10.1002/bip.20816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada H, Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
- Gilbert SD, Batey RT. Monitoring RNA-ligand interactions using isothermal titration calorimetry. Methods Mol. Biol. 2009;540:97–114. doi: 10.1007/978-1-59745-558-9_8. [DOI] [PubMed] [Google Scholar]
- Holbrook JA, et al. Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: Interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry. 1999;38:8409–8422. doi: 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- Li TK, et al. Drug targeting of HIV-1 RNA. DNA hybrid structures: Thermodynamics of recognition and impact on reverse transcriptase-mediated ribonuclease H activity and viral replication. Biochemistry. 2004;43:9732–9742. doi: 10.1021/bi0497345. [DOI] [PubMed] [Google Scholar]
- McKenna SA, et al. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J. Mol. Biol. 2006;358:1270–1285. doi: 10.1016/j.jmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Mikulecky PJ, Feig AL. Heat capacity changes associated with DNA duplex formation: Salt- and sequence-dependent effects. Biochemistry. 2006a;45:604–616. doi: 10.1021/bi0517178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Feig AL. Heat capacity changes associated with nucleic acid folding. Biopolymers. 2006b;82:38–58. doi: 10.1002/bip.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, et al. Entropy-driven folding of an RNA helical junction: An isothermal titration calorimetric analysis of the hammerhead ribozyme. Biochemistry. 2004;43:5870–5881. doi: 10.1021/bi0360657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoue LS, Tellinghuisen J. The role of backlash in the “First injection anomaly” in isothermal titration calorimetry. Anal. Biochem. 2004;326:125–127. doi: 10.1016/j.ab.2003.10.048. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka A, et al. Thermodynamics of mRNA 5′ cap binding by eukaryotic translation initiation factor eIF4e. Biochemistry. 2004;43:13305–13317. doi: 10.1021/bi0491651. [DOI] [PubMed] [Google Scholar]
- Recht MI, Williamson JR. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J. Mol. Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Recht MI, et al. Monitoring assembly of ribonucleoprotein complexes by isothermal titration calorimetry. Methods Mol. Biol. 2008;488:117–127. doi: 10.1007/978-1-60327-475-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond C, et al. Monitoring of an RNA multistep folding pathway by isothermal titration calorimetry. Biophys. J. 2009;96:132–140. doi: 10.1016/j.bpj.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim NN, Feig AL. Isothermal titration calorimetry of RNA. Methods. 2009;47:198–205. doi: 10.1016/j.ymeth.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takach JC, et al. Salt-dependent heat capacity changes for RNA duplex formation. J. Am. Chem. Soc. 2004;126:6530–6531. doi: 10.1021/ja0316263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen J. Optimizing experimental parameters in isothermal titration calorimetry. J. Phys. Chem. B Condens Matter Mater. Surf. Interfaces Biophys. 2005;109:20027–20035. doi: 10.1021/jp053550y. [DOI] [PubMed] [Google Scholar]
- Vander Meulen KA, et al. Thermodynamics and folding pathway of tetraloop receptor-mediated RNA helical packing. J. Mol. Biol. 2008;384:702–717. doi: 10.1016/j.jmb.2008.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman T, et al. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–135. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]