Abstract
Acyl-CoA:monoacylglycerol acyltransferase (MGAT) plays a predominant role in the resynthesis of triacylglycerol in the small intestine, but its contribution to triacylglycerol synthesis in other tissues, such as the liver, is not clear. In this study, we identified a novel MGAT gene, which is identical with lysophosphatidylglycerol acyltransferase1 (LPGAT1). Mouse LPGAT1 is expressed in a number of tissues and most highly expressed in the liver. Hepatic LPGAT1 expression in diabetic db/db mice is higher than that in the control db/m mouse, which is consistent with increased hepatic MGAT activity in db/db mouse. To elucidate the role of LPGAT1 gene in lipid metabolism in db/db mice, we constructed an adenovirus of short hairpin RNA (shRNA) targeting LPGAT1 to selectively knockdown LPGAT1 gene expression in the liver. Hepatic MGAT activity and LPGAT1 expression in db/db mice infected with LPGAT1 shRNA adenovirus were significantly lower than those in mice infected with the control virus. Notably, treatment with LPGAT1 shRNA adenovirus caused a marked reduction in serum triacylglycerol and cholesterol levels and a significant increase in hepatic cholesterol level. These findings indicate that LPGAT1, a newly identified MGAT enzyme, plays a significant role in hepatic triacylglycerol synthesis and secretion in db/db mice.
Keywords: MGAT, LPGAT, liver, lipid
Triacylglycerol (TAG), a neutral lipid normally found in lipid droplets, is an important molecule for energy storage and lipid metabolism in eukaryotes (1). TAG is synthesized by two major pathways, i.e., the glycerol 3-phosphate pathway and the monoacylglycerol pathway (2). Both pathways generate diacylglycerol (DAG) that can be used as a substrate for TAG synthesis by acyl CoA:diacylglycerol acyltransferase (DGAT), which is a rate-limiting enzyme for TAG synthesis. In the glycerol 3-phosphate pathway, DAG is synthesized by dephosphorylation of phosphatidic acid, and in the monoacylglycerol pathway, DAG is formed directly from monoacylglycerol and fatty acyl-CoA by monoacylglycerol acyltransferase (MGAT; EC 2.3.1.22). While the glycerol 3-phosphate pathway is ubiquitously present in all tissues, the monoacylglycerol pathway is reported to be present in tissues that have high TAG synthesizing activity, such as the small intestine, liver, and adipose tissue.
The monoacylglycerol pathway is best known for its role in fat absorption in the small intestine because of the high influx of monoacylglycerol from breakdown of dietary TAG. In the intestine of animals, TAG contained in dietary fat is digested by pancreatic lipase into a soluble free fatty acid and 2-monoacylglycerol. These are quickly absorbed into intestinal enterocytes and formed into DAG by MGAT enzyme. Sequentially, DAG is further covalently acylated by DGAT enzyme to form TAG molecule. The newly formed TAG is then packed with proteins and other lipids (cholesterol ester and phospholipid) into chylomicron, which is secreted into the lymph and used as a lipid source in peripheral tissues. On the other hand, MGAT is thought to play a significant role in the adipose tissue and liver, where TAG is actively hydrolyzed to provide fatty acids for lipid metabolism. MGAT may also preserve essential polyunsaturated fatty acids by resynthesizing them into TAG in those tissues. However, to what extent MGAT pathway contributes to total TAG synthesis and which MGAT is responsible for TAG synthesis in those tissues remain unclear.
MGAT protein was partially purified from the rat intestine (3), rat liver (4, 5), rat adipose tissue (6), plant (7), insect (8), and bird (9). Its enzymatic properties, such as lipid cofactor requirement, sensitivity to heat and protease, and substrate specificity, were also studied; however, the MGAT gene had for a long time not been identified. The first reported MGAT gene (MGAT1), which was identified based on sequential homology with members of the DGAT2 gene family, is expressed in the stomach, kidney, white and brown adipose tissues, and liver, but not in the small intestine (10). The second MGAT gene (MGAT2) was also identified as a homolog of DGAT2 (11, 12) and MGAT1 (13). In human, MGAT2 is highly expressed in the small intestine, liver, stomach, kidney, colon, and white adipose tissue. However, in mice, MGAT2 is expressed predominantly in the small intestine (11). Recently, it was reported that mice lacking MGAT2 are unsusceptible to developing obesity, glucose intolerance, hypercholesterolemia, or fatty liver even when fed on a high-fat diet (14). These findings suggest that MGAT2 may play an important role in dietary fat absorption in mouse intestine and consequently be a therapeutic target for treating obesity and other metabolic diseases associated with excessive fat intake. The third MGAT gene (MGAT3), which is highly homologous to MGAT1 and MGAT2, is an intestinal specific enzyme implicated in dietary fat absorption in human only (15).
In our search for new MGAT genes, we found that the lysophosphatidylglycerol acyltransferase 1 (LPGAT1) gene (16) displays MGAT activity and is highly expressed in mouse liver. To elucidate the role of LPGAT1 gene in lipid metabolism in db/db mice, we constructed an adenovirus of short hairpin RNA (shRNA) targeting LPGAT1 to selectively knockdown LPGAT1 gene expression in the liver and determined hepatic MGAT activity, LPGAT1 expression, and serum TAG and cholesterol levels in LPGAT1 shRNA adenovirus-treated db/db mice.
EXPERIMENTAL PROCEDURES
Animal care
All animal work was approved by the Animal Care and Use Committee of Dainippon Sumitomo Pharma. Male diabetic db/db mice and control db/m mice (7W) were purchased from CLEA. Animals were conventionally housed at a constant temperature of 22 ± 3°C on a light/dark cycle with light from 8:00 to 20:00 h. Mice were fed a standard chow diet, and food and water were available to the animals ad libitum.
Cloning of the full-length mouse LPGAT1 cDNA
A mouse cDNA clone was identified in the National Center for Biotechnology Information database based on a sequence motif conserved among members of the reported acyltransferase families and proved to be identical to the LPGAT1 gene (16). A PCR primer pair (forward, 5′-CCACTGAGACAGGACCGAC-3′, and reverse, 5′-ATGTAGCAAGTCCAAGTCAA-3′) was used to amplify the full-length coding region of LPGAT1 gene from mouse adipose tissue cDNA library (Biochain). Amplification was performed by PCR using Ex-Taq polymerase (Takara) and a thermal cycling of 50 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 60 s), resulting in a 1.2 kb cDNA product. This fragment was subcloned into pT7-Blue vector (Novagen) and sequenced.
Northern blot analysis
[α-32P]dCTP-labeled LPGAT1 probe was prepared from full-length cDNA of mouse LPGAT1 gene using a random prime kit (Takara). Normal adult mouse tissue poly(A) RNA blot (Biochain) was then hybridized with the LPGAT1 probe. Hybridization was carried out in DIG Easy Hyb (Boeringer) at 50°C overnight. The hybridized membrane was washed twice at room temperature in 2× SSC buffer containing 0.1% SDS and once at 50°C in 0.1× SSC buffer containing 0.1% SDS to remove excess radiolabeled probe. The blot was exposed to a photoimaging plate, and signals were analyzed by a BAS-2500 system (Fujifilm).
Expression of FLAG-tagged LPGAT1 in Chinese hamster ovary cells
Mouse LPGAT1 cDNA was amplified by PCR with Ex-Taq polymerase (Takara) using a primer pair (forward, 5′-CCAGAATTCATGGCCGTGACCGTG-3′, and reverse, 5′-CCAAAGCTTCAATTCCTAAAATAGACAATGG-3′) designed to add FLAG-tag to the N terminus of LPGAT1 gene, and the resulting 1.1-kb product was subcloned into pCMV-Tag2B vector (Stratagene). Chinese hamster ovary (CHO) cells were transfected with the FLAG-tagged LPGAT1 plasmid premixed with the LipofectAMINE PLUS kit (Invitrogen) according to the manufacturer's instructions. To isolate a stable transfectant, CHO cells transfected with FLAG-tagged LPGAT1 plasmid were cultured in a medium containing 400 μg/ml GENETICIN (Gibco) for 7 days, and a clonal cell was obtained using the limited dilution method. Transient transfectants (24 h after transfection) or stable transfectants were harvested in ice-cold PBS, pelleted by centrifugation, lysed in a buffer containing 50 mM Tris-HCl, pH 7.5, 250 mM sucrose, 1 mM EGTA, 1 mM DTT, and a protease inhibitor cocktail (Sigma). The lysates were sonicated on ice and frozen at −80°C.
Preparation of microsomes from mouse liver
Liver fragments were extracted from mice, and microsomal fractions were prepared as described previously (6). Aliquots of the prepared samples were stored at −80°C until use.
MGAT activity assay
In this study, we used a standard assay to measure MGAT activity. The reaction was conducted at 25°C in a final volume of 0.15 ml. The reaction mixture contained cell lysate/microsome, 50 mM N-(2-acetamido) iminodiacetic acid, pH 6.4, 2.5 mM MgCl2, 1.25 mg/ml BSA, 1 mM EDTA, 30 μM [14C]palmitoyl CoA (Amersham), and 1/10 vol of 1 mM monoacylglycerol (sn-2-monoacylglycerol or rac-1-monoacylglycerol; Sigma) premixed with 0.4% CHAPS. The reaction was initiated by adding palmitoyl CoA and terminated by addition of 0.3 ml heptane/isopropanol/water (80:20:2,v/v). After vigorous vortexing, 0.2 ml heptane and 0.1 ml water were added, and aliquots of the organic phase were extracted and dried in a vacuum evaporator. The extracted lipids were separated by TLC on a Silica Gel TLC plate (150 Å; Whatman) using a solvent system consisting of hexane/diethylether/acetic acid (75:25:1, v/v). TLC plates were exposed to a photoimaging plate, and DAG radioactivity was measured using a BAS-2500 system (Fujifilm). MGAT activity was calculated by DAG synthesized.
Analysis of real-time quantitative RT-PCR
RNA was extracted from the harvested CHO cells or from dissected liver fragments using an RNA purification kit (Qiagen) according to the manufacturer's instructions. Total RNA was used for real-time quantitative PCR analysis, which was performed with an enzyme kit (Power SYBR Green PCR Master Mix) and ABI Prism 7900-HT sequence detector (PE Applied Biosciences). Primers and probes for analysis of the LPGAT1 gene were designed using Primer Express or Oligo Software.
Construction of LPGAT1 shRNA-expressing adenovirus
shRNA sequence for knockdown of mouse LPGAT1 gene (5′-GTTTAGAATGTAAGTCAAACGTGTGCTGTCCGTTTGATTTGCATTCTAAGCCTTTTT-3′) was inserted into the cloning site of pcPURU6i cassette (Takara). The plasmid was digested with EcoRI and BamHI, and the fragment was inserted into the SwaI site of cosmid vector pAxcwit2 (Nippongene). LPGAT1 shRNA adenovirus (AxmU6shRmlPGAT1) and mlamin shRNA adenovirus (AxmU6shRmlamin) were constructed according to the manufacturer's instructions. As an empty control virus for in vivo experiments, adenovirus that contains U6 promoter alone was constructed. HEK-293 cells were infected with the resulting adenovirus, and viruses were purified by two rounds of CsCl centrifugation. Viral particle concentration was determined by absorption of 260 nm.
Western blot analysis
CHO cells stably expressing FLAG-tagged mouse LPGAT1 were infected with mouse LPGAT1 shRNA adenovirus or mouse lamin shRNA adenovirus. Membrane fractions were next prepared, and expression of FLAG-tagged proteins was detected by immunoblotting of membrane samples with anti-FLAG M2 antibody.
Animal treatment
Seven-week-old diabetic db/db mice and control db/m mice were purchased from CLEA. After 7 days of acclimation, db/db mice were randomized to two treatment groups based on body weight and plasma TAG level. LPGAT1 shRNA adenovirus or control virus was diluted and injected intravenously to the mice via the tail vein (6 × 1010 particles/body). Body weight was continuously recorded, and food intake was measured twice a week. After 5 or 16 days, animals were sacrificed, and blood samples were collected from the vena cava inferior and tissues were dissected, weighed, and frozen immediately at −80°C. Plasma TAG, cholesterol, FFA, and HDL-cholesterol levels were measured using Wako's kit according to the manufacturer's instructions. To measure hepatic lipid contents, ∼50 mg liver was homogenized in 0.5 ml of hexane/isopropanol (3:2, v/v), and 0.1 ml of the supernatant was dried at 40°C for >1 h. Lipids were dissolved in ethanol, and lipid content (TAG and cholesterol) was measured as mentioned above.
Statistical analysis
For in vitro experiments, values represent the mean or mean ± SEM of at least two independent measurements. For in vivo experiments, values represent the mean ± SEM of at least five independent measurements per treatment. Statistical difference across treatment groups was determined using the Student's t-test. A P value of <0.05 was considered significant.
RESULTS
Identification of mouse LPGAT1 as a novel MGAT gene
As candidate gene for a novel MGAT, a mouse cDNA clone was identified in the National Center for Biotechnology Information database based on a sequence motif conserved among members of the reported acyltransferase families. A 1.2 kb cDNA fragment was cloned by PCR amplification using cDNA library of mouse adipose tissue. Sequence analysis reveals that the gene encodes a 370 amino acid protein of 43 kDa and carries acyltransferase motifs. This MGAT candidate gene was proved to be identical to the LPGAT1 gene (16). LPGAT1 enzyme is reported to recognize various lysophosphatidylglycerols as substrates, but it is unknown whether the enzyme also recognizes neutral lipids, such as DAG and monoacylglycerol.
Tissue distribution of mouse LPGAT1 mRNA
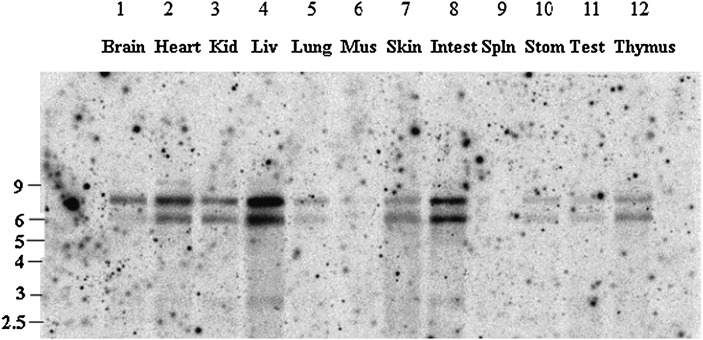
Tissue distribution of mouse LPGAT1 mRNA was analyzed by Northern blot analysis. As previously reported in human (16), two mRNA splice isoforms were detected. Mouse LPGAT1 was expressed in tissues including heart, kidney, liver, skin, intestine, and thymus. Unlike in human, mouse LPGAT1 was most highly expressed in the liver (Fig. 1).
Fig. 1.
Tissue distribution of mouse LPGAT1 mRNA . LPGAT1 expression in mouse tissues was determined by Northern blot analysis. Lane 1, brain; lane 2, heart; lane 3, kidney; lane 4, liver; lane 5, lung; lane 6, skeletal muscle; lane 7, skin; lane 8, small intestine; lane 9, spleen; lane 10, stomach; lane 11, testis; lane 11, thymus.
MGAT activity of recombinant LPGAT1 expressed in CHO cells
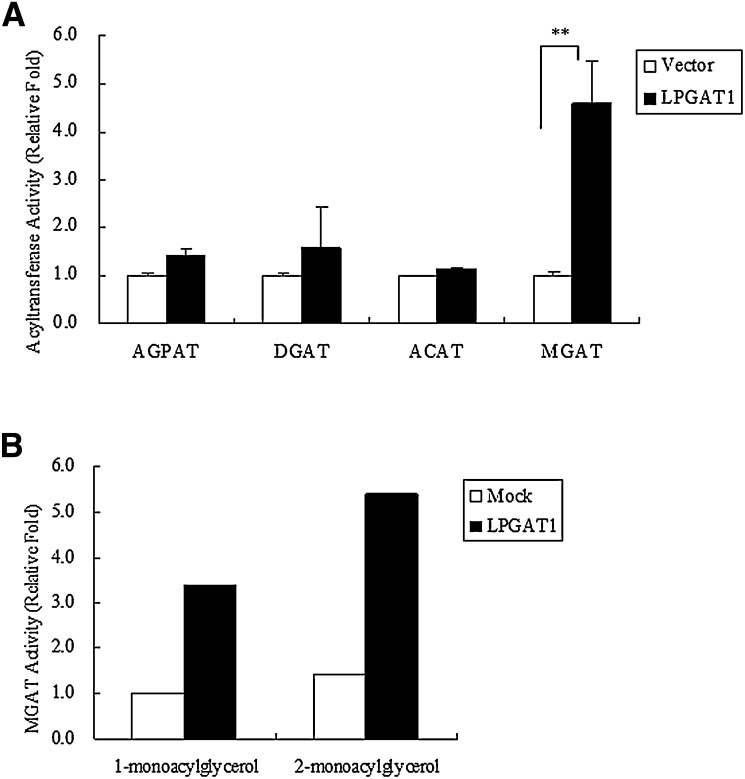
To determine whether the protein encoded by LPGAT1 cDNA possesses acyltransferase activity, LPGAT1 was transiently transfected into CHO cells. Activity of acylglycerophosphate acyltransferase (AGPAT), DGAT, acyl-CoA:cholesterol acyltransferase (ACAT), and MGAT in the cell lysates was measured using lysophosphatidic acid, DAG, cholesterol, and monoacylglycerol as substrates, respectively. Although no significant increase in AGPAT, DGAT, and ACAT activity was detected in the lysates from LPGAT1 transfected cells, MGAT activity was 5-fold higher than that in the lysates of cells transfected with the empty vector (Fig. 2A). To investigate substrate specificity, MGAT assays were performed using rac-1-monoacylglycerol or sn-2-monoacylglycerol as acyl acceptor. Like other MGAT enzymes, LPGAT1 enzyme appears to prefer the sn-2-isomer as acyl acceptor (Fig. 2B).
Fig. 2.
In vitro MGAT activity and substrate specificity of LPGAT1 enzyme. A: CHO cells were transfected with empty vector or mouse LPGAT1, and cell lysates were analyzed for acyltransferase activity. Activity was measured by incorporating [14C]palmitoyl CoA into lysophosphatidic acid, DAG, cholesterol, and monoacylglycerol, resulting in phosphatidic acid (AGPAT), triacylglycerol (DGAT), cholesterol ester (ACAT), and diacylglycerol (MGAT), respectively. Values are given mean ± SEM (n = 3). Significant differences from the control group were determined using Student's t-test. **P < 0.01. B: Preference of LPGAT1 for rac-1-monoacylglycerol and sn-2-monoacylglycerol as acyl acceptor. MGAT activity was determined with 30 μM [14C]palmitoyl CoA and 0.1 mM rac-1-monoacylglycerol or sn-2-monoacylglycerol.
MGAT activity and LPGAT1 expression in the liver of db/db mice mouse
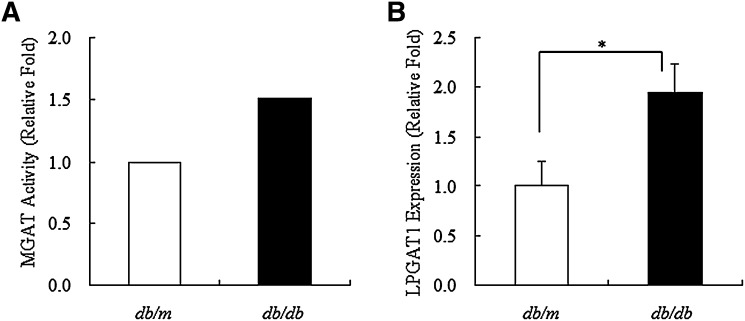
Enzymatic assay showed remarkable difference in hepatic MGAT activity between db/db mice and control db/m mice, i.e., MGAT activity in liver microsomes of db/db mice was 1.5-fold higher than that in liver microsomes of db/m mice (Fig. 3A). Similarly, hepatic LPGAT1 expression in microsomes of db/db mice was 2-fold higher than that in microsomes of db/m mice, which was consistent with increased hepatic MGAT activity in microsomes of db/db mice (Fig. 3B).
Fig. 3.
MGAT activity and LPGAT1 expression in db/m and db/db mice. A: Microsomes were prepared from liver of db/m or db/db mice, and hepatic MGAT activity was measured using 30 μM [14C]palmitoyl CoA and 0.1 mM sn-2-monoacylglycerol. B: Total RNA was prepared from liver of db/m or db/db mice and used for real-time quantitative PCR to evaluate the expression of LPGAT1 gene. Values are given as mean ± SEM (n = 5). Significant differences from the control group were determined using Student's t-test. *P < 0.05.
Construction of LPGAT1 shRNA adenovirus
To elucidate the role of LPGAT1 gene in lipid metabolism in diabetic db/db mice, we used an adenoviral vector to selectively knock down LPGAT1 gene expression. The adenovirus was constructed of shRNA targeting LPGAT1. CHO cells stably expressing mouse LPGAT1 gene were infected with the LPGAT1 shRNA adenovirus or mouse lamin shRNA adenovirus as a negative control. Western blot analysis revealed that LPGAT1 expression was dose-dependently reduced with increasing units of LPGAT1 shRNA adenovirus infection but was not affected by treatment with mouse lamin shRNA, confirming that LPGAT1 shRNA adenovirus specifically and significantly repressed LPGAT1 gene expression (Fig. 4).
Fig. 4.
LPGAT1 shRNA adenovirus-mediated knockdown of LPGAT1 expression in CHO cells. CHO cells stably expressing FLAG-tagged mouse LPGAT1 gene were infected with increasing doses (MOI) mLPGAT1 shRNA adenovirus or control virus, which contained mouse lamin shRNA sequence. Membrane fractions were then prepared, and expression of FLAG-tagged protein was detected by immunoblotting of membrane samples with M2 anti-FLAG antibody.
In vivo knockdown of LPGAT1 expression in the liver of db/db mouse
For in vivo knockdown of LPGAT1 expression, the appropriate dose of LPGAT1 shRNA adenovirus was determined by tail vein injection of different amounts of the empty vector. No effect was observed for a dose up to 6 × 1010 particles per animal, but higher doses led to significant reduction of animal body weight and food intake (data not shown). Therefore, we decided to use LPGAT1 shRNA adenovirus at the dose of 6 × 1010 particles per animal in the in vivo knockdown experiment. Furthermore, we confirmed that infection with LPGAT1 shRNA adenovirus specifically affects hepatic LPGAT1 expression as previously reported (data not shown; 17).
Male db/db mice aged 8 weeks were injected via the tail vein with LPGAT1 shRNA adenovirus. To distinguish virus treatment-related effects from LPGAT1-specific knockdown effects, control animals were injected with an empty virus. Both groups of animals were analyzed on days 5 and 16 after injection.
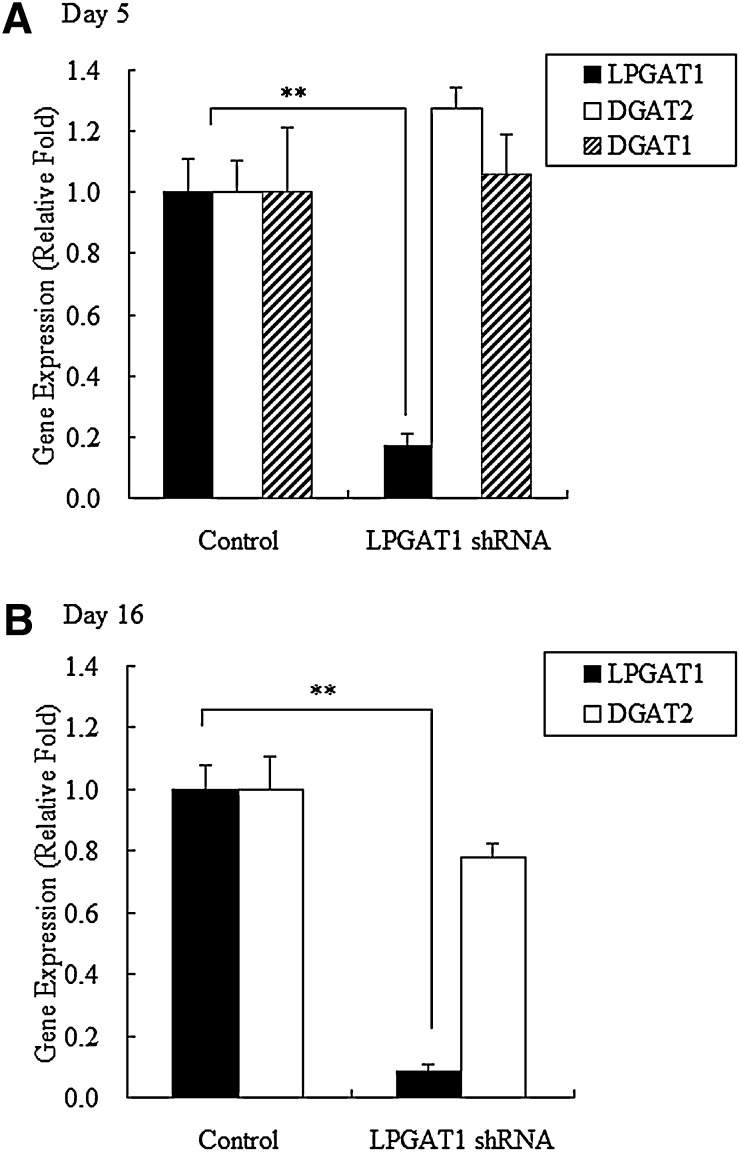
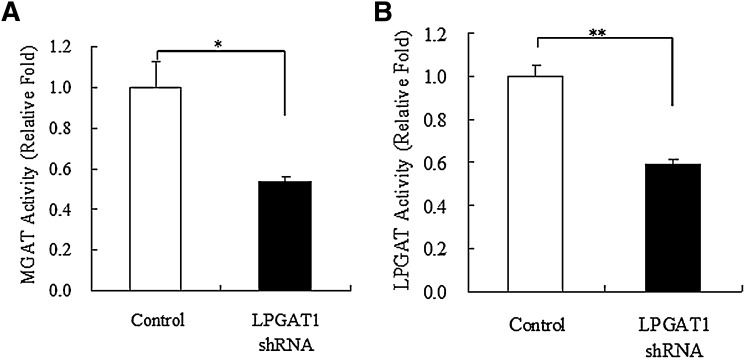
Compared with treatment with the control vector, treatment with LPGAT1 shRNA adenovirus decreased LPGAT1 mRNA levels by 83% on day 5 and by 92% on day 16 after injection. However, no difference in DGAT1 mRNA was observed as compared with control group on day 5, and DGAT2 mRNA levels were unaffected on days 5 and 16 (Fig. 5A, B). These findings indicate that LPGAT1 shRNA adenovirus specifically repressed hepatic LPGAT1 expression for at least 16 days in db/db mice. Enzymatic assay revealed that LPGAT1 shRNA adenovirus-induced decrease in LPGAT1 mRNA levels in db/db mice was associated with reduced hepatic MGAT and LPGAT activities (Fig. 6A, B). However, the degree of reduction of hepatic MGAT and LPGAT activity was less than that predicted from the decrease in LPGAT1 mRNA levels. It is likely that other enzymes besides LPGAT1 contribute to residual MGAT and LPGAT activity.
Fig. 5.
LPGAT1 shRNA adenovirus-mediated knockdown of LPGAT1 expression in liver samples of db/db mice. Male db/db mice were injected intravenously either with the control virus or LPGAT1 shRNA adenovirus. Animals were euthanized 5 days (A) or 16 days (B) after infection, and gene expression levels in liver samples were analyzed by quantitative RT-PCR. Values are given as mean ± SEM (n ≥ 4). Significant differences from the control group were determined using Student's t-test. *P < 0.05.
Fig. 6.
Hepatic MGAT and LPGAT activity in db/db mice treated with LPGAT1 shRNA adenovirus. Hepatic MGAT activity (A) and LPGAT activity (B) in mice infected either with the control virus or LPGAT1 shRNA adenovirus were analyzed. Values are given mean ± SEM (n = 5). Significant differences from the control group were determined using Student's t-test. *P < 0.05.
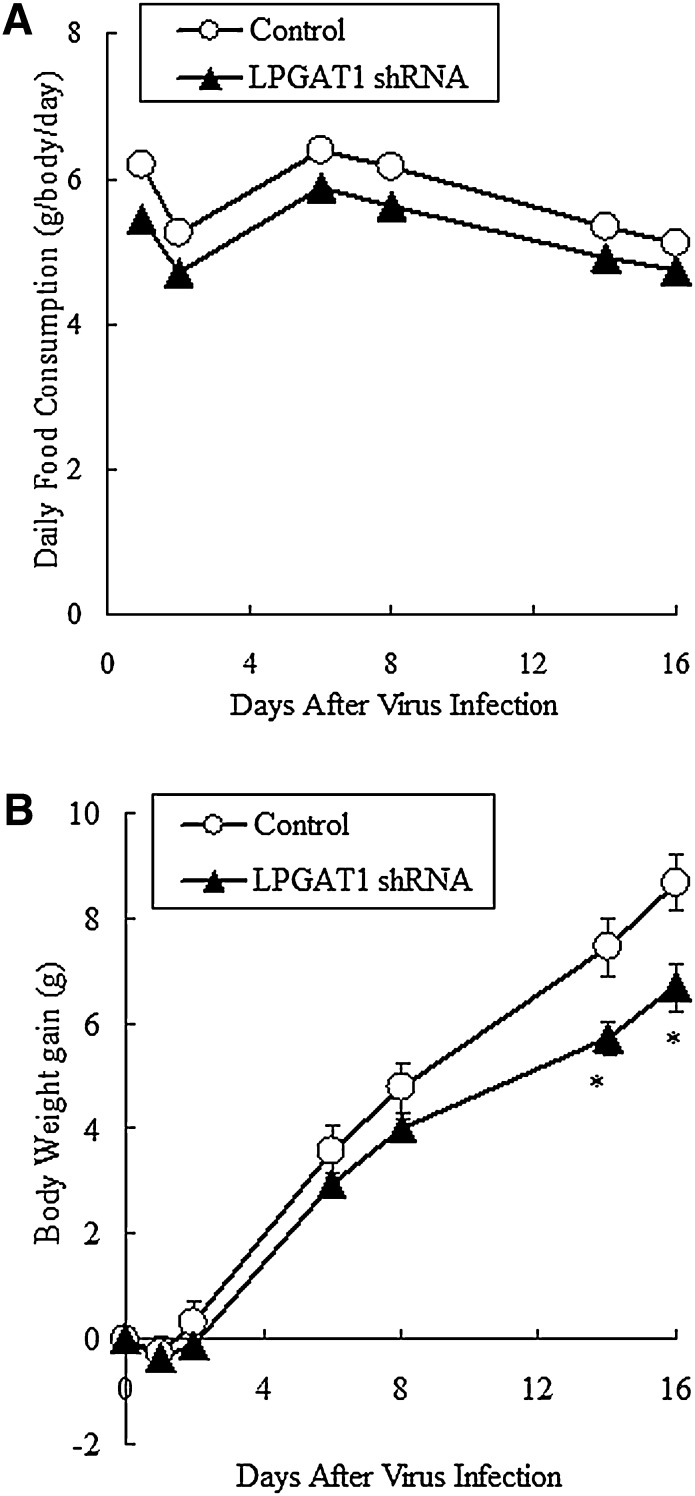
Food consumption and body weight in db/db mice were continuously recorded after virus infection. On day 2 after viral injection, both LPGAT1 shRNA adenovirus-treated mice and control mice showed a slight reduction (15%) in food intake. However after day 2, the animal's food consumption returned to pretreatment level. Throughout the experimental period, LPGAT1 shRNA adenovirus-treated mice consumed the same amount of food as the control mice (Fig. 7A). Body weight was slightly but significantly reduced in LPGAT1 shRNA adenovirus-treated animals compared with the control group (Fig. 7B, Table 1). Liver and epididymal fat weights were comparable between the two groups on day 16 after infection (Table 1).
Fig. 7.
Food consumption and body weight of db/db mice treated with the control virus or LPGAT1 shRNA adenovirus. Animals infected either with the control virus or LPGAT1 shRNA adenovirus were fed ad libitum with standard chow. Food consumption (A) and body weight (B) were recorded for 16 days. Values are given as mean ± SEM (n = 5). Significant differences from the control group were determined using Student's t-test. *P < 0.05.
TABLE 1.
Effects of LPGAT1 shRNA adenovirus infection on body weight and tissue weight after 16 days in db/db mice
| Parameters | Control | LPGAT1 shRNA | P |
|---|---|---|---|
| BW, day 0 (g) | 37.30 ± 0.50 | 36.20 ± 0.50 | 0.158 |
| BW, day 16 (g) | 45.50 ± 0.60 | 43.20 ± 0.40 ** | 0.005 |
| BW gain (g) | 8.20 ± 0.40 | 7.00 ± 0.30 * | 0.026 |
| Liver weight (g) | 3.94 ± 0.23 | 3.83 ± 0.19 | 0.716 |
| Epididymal fat weight (g) | 1.87 ± 0.10 | 1.99 ± 0.08 | 0.384 |
Data are expressed as the mean ± SEM. BW, body weight *P < 0.05, **P < 0.01 by Student's t-test.
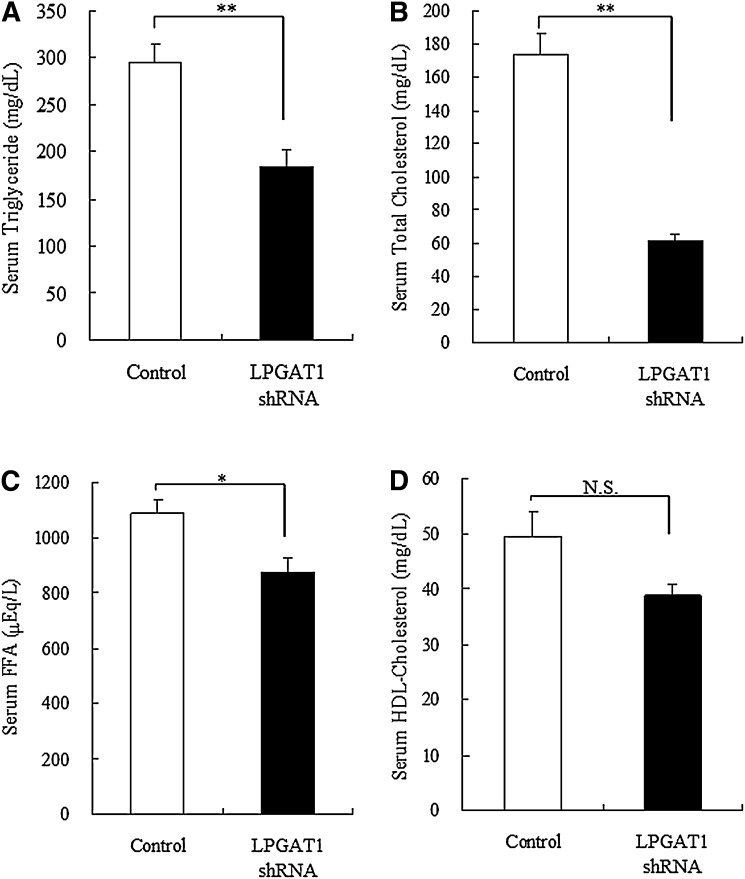
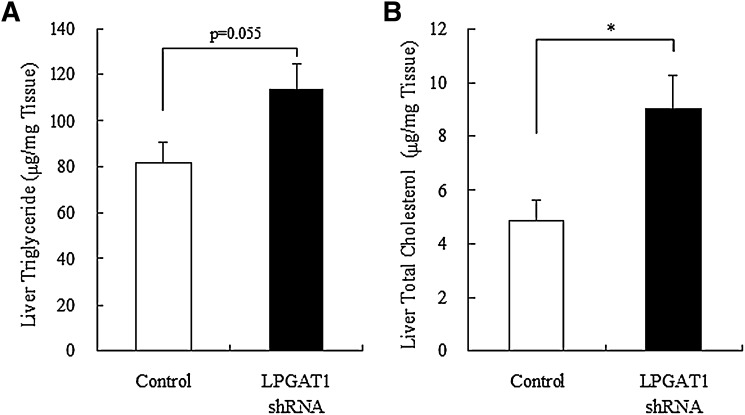
Next, we examined lipids profile in LPGAT1 shRNA adenovirus-treated mice. Notably, treatment with LPGAT1 shRNA adenovirus induced a significant reduction in serum TAG and total cholesterol level and a modest decrease in FFA level (Fig. 8A–C). However, this treatment had no significant effect on HDL-cholesterol level (Fig. 8D). To investigate whether treatment with LPGAT1 shRNA adenovirus caused any change in lipid profile in the liver, hepatic TAG and cholesterol levels were determined. Contrary to serum lipid profile, treatment with LPGAT1 shRNA adenovirus significantly increased total cholesterol level in the liver (Fig. 9B), indicating that the treatment induced cholesterol accumulation in the liver of db/db mice. Hepatic TAG level slightly increased in LPGAT1 shRNA adenovirus-treated animals; however, this increase was not statistically significant (Fig. 9A).
Fig. 8.
Serum lipid concentrations in db/db mice treated with LPGAT1 shRNA adenovirus. Serum TAG (A), total cholesterol (B), FFA (C), and HDL-cholesterol (D) levels of control- or LPGAT1 shRNA-treated db/db mice were determined. Values are given as mean ± SEM (n = 5). Significant differences from the control group were determined using Student's t-test. *P < 0.05; **P < 0.01.
Fig. 9.
Hepatic lipid contents in db/db mice treated with the control virus or LPGAT1 shRNA adenovirus. Hepatic TAG (A) and total cholesterol (B) levels in control- or LPGAT1 shRNA-treated db/db mice were determined. Values are given as mean ± SEM (n = 5). Signifi cant differences from the control group were determined using Student's t-test. *P < 0.05.
DISCUSSION
In our search for a new MGAT gene involved in TAG synthesis in tissues except the small intestine, we isolated a candidate gene from mouse adipose tissue cDNA library based on acyltransferase motifs. The identified gene was identical to LPGAT1. From the results of this study, we conclude that the isolated LPGAT1 enzyme possesses MGAT activity due to the followings reasons: First, LPGAT1 expressing CHO cells showed 5-fold higher MGAT activity than the control vector transfected cells and displayed AGPAT, DGAT, and ACAT activities similar to those in the control cells (Fig. 2A). In addition, like other MGAT enzymes, i.e., MGAT1 (10), MGAT2 (11, 13), and MGAT3 (15), LPGAT1 enzyme appeared to prefer the sn-2-monoacylglycerol to rac-1-monoacylglycerol as acyl acceptor (Fig. 2B). This indicates that LPGAT1 has enzymatic characteristics similar to those of other MGAT enzymes. Second, hepatic LPGAT1 expression in diabetic db/db mice was 2-fold higher than that in the control db/m mice, which was consistent with increased hepatic MGAT activity in db/db mice (Fig. 3). Although expression of MGAT1 in the mouse liver has previously been reported, the level of this expression is very low (10). In addition, we confirmed in this study that MGAT1 expression is not increased in db/db mice compared with the controls (data not shown). It has also been reported that the expression of mouse MGAT2 is localized in the small intestine and not observed in the liver (11, 13). Moreover, MGAT3 is known to be expressed only in the human intestine (15). These findings indicate that increased hepatic MGAT activity in db/db mice may be attributed to upregulation of the LPGAT1 gene in the liver. Third, treatment with LPGAT1 shRNA adenovirus dramatically decreased LPGAT1 mRNA levels on days 5 and 16 after infection in db/db mouse liver. This decrease was associated with reduced hepatic MGAT activity (Fig. 6A), indicating that LPGAT1 contributes to hepatic MGAT activity in db/db mice. From these findings we postulate that LPGAT1 functions as a hepatic MGAT enzyme in mice. On the other hand, the LPGAT1 gene has been demonstrated to also possess LPGAT activity (16) and the decrease of hepatic LPGAT1 expression was associated with reduced hepatic LPGAT activity in this study (Fig. 6B). So, it is probable that LPGAT1 functions either as MGAT or LPGAT enzyme in vivo depending on physiological conditions. For example, under circumstances where tissue monoacylglycerol concentration is high, LPGAT1 acts as an MGAT enzyme, while when lysophosphatidylglycerol level is dominant, it catalyzes LPGAT reaction. This possibility remains to be confirmed. It would also be necessary to examine in details MGAT enzymatic characteristics of LPGAT1, i.e., selectivity toward different acyl CoAs and enzymatic kinetics.
Northern blot analysis indicated that mouse LPGAT1 gene is expressed in a number of tissues, including the heart, kidney, and small intestine, and most highly expressed in the liver (Fig. 1). This is the first MGAT gene to be shown as highly expressed in the mouse liver. As the liver plays a major role in TAG metabolism in vivo, we examined the contribution of MGAT enzyme to this metabolism using RNA interference technology. Obese db/db mice with hyperglycemia and hyperlipidemia were found to have increased hepatic MGAT activity and elevated LPGAT1 expression. These mice were regarded as a good model for examining the effects of MGAT (LPGAT1) knockdown in the liver. Accordingly, we constructed LPGAT1 shRNA adenovirus and confirmed that this adenovirus specifically represses LPGAT1 expression in vitro (Fig. 4) and induced a marked reduction of hepatic LPGAT1 expression and MGAT/LPGAT1 activity in vivo. Treatment with LPGAT1 shRNA adenovirus had no effect on food consumption in db/db mice but slightly decreased body weight (Fig. 7A, B). Liver and epdidymal fat weights in LPGAT1 shRNA adenovirus-treated mice were comparable with those in the control mice (Table 1), indicating that other TAG synthesizing tissues, such as subcutaneous fat, may be responsible for the reduction in body weight.
Notably, treatment with LPGAT1 shRNA adenovirus induced a marked reduction of serum TAG and total cholesterol levels and a modest reduction of free fatty acid in of db/db mice (Fig. 8A–C). Since LPGAT activity is involved neither in TAG nor cholesterol synthesis, we speculate that the detected phenotypes were the results of decreased MGAT activity in the liver. We therefore hypothesize that decreased hepatic MGAT activity led to reduced TAG synthesis in the liver, resulting in reduced VLDL formation. VLDL is the major lipoprotein secreted by the liver and includes TAG and cholesterol as lipid components. Therefore, reduced VLDL formation may lead to decreased VLDL secretion, resulting in reduced serum TAG and cholesterol levels. If this hypothesis is true, hepatic cholesterol, which is independent of TAG synthesis pathway, may accumulate in the liver because of reduced VLDL secretion. This is supported by the observation that hepatic cholesterol level was significantly elevated (Fig. 9B). On the other hand, hepatic TAG level was not significantly affected or slightly increased. One explanation is that MGAT activity is involved only in TAG secretion pathway and not in TAG accumulation pathway, which is mainly via the glycerol-phosphate pathway. If so, LPGAT1 shRNA adenovirus would inhibit TAG secretion pathway but would not affect TAG accumulation. This may lead to decreased VLDL secretion and have no apparent effect on hepatic TAG level. According to a previous study, there seems to be two types of DGAT activity in the rat liver, one is mainly for TAG accumulation and the other is for TAG secretion (18). Thus, it is necessary to examine whether MGAT functions mainly in the TAG secretion pathway.
It has been reported that DGAT1 knockout mice have normal plasma TAG level (19) and that inhibition of DGAT2 expression in obese mice by antisense oligonucleotides results in significant reduction of both plasma TAG level and hepatic TAG content (20). However, in both studies, plasma and hepatic cholesterol levels were not affected in these animals. On the other hand, in this study, LPGAT1 knockdown resulted in drastic reduction in both serum TAG and cholesterol levels, suggesting a unique function of MGAT enzyme.
In conclusion, we found that the LPGAT1 gene encodes MGAT enzyme and is the first MGAT gene highly expressed in the mouse liver. Specific knockdown of hepatic LPGAT1 expression in db/db mice resulted in a marked reduction of serum TAG and cholesterol levels. These findings indicate that LPGAT1 functions as MGAT enzyme and contributes to TAG secretion of the mouse liver.
Acknowledgments
The authors thank Dr. Toshizumi Tanabe (Osaka City University) for advice and discussion of our manuscript. The authors also thank Michio Nakai, Kazuo Komiya, and Haruhisa Iguchi for constructing LPGAT1 shRNA adenovirus and Maki Masuda for her technical assistance.
Footnotes
Abbreviations:
- ACAT
- acyl-CoA:cholesterol acyltransferase
- AGPAT
- acylglycerophosphate acyltransferase
- CHO
- Chinese hamster ovary
- DAG
- diacylglycerol
- DGAT
- diacylglycerol acyltransferase
- LPGAT
- lysophosphatidylglycerol acyltransferase
- MGAT
- monoacylglycerol acyltransferase
- shRNA
- short hairpin RNA
- TAG
- triacylglycerol
REFERENCES
- 1.Coleman R. A., Lee D. P. 2004. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43: 134–176. [DOI] [PubMed] [Google Scholar]
- 2.Bell R. M., Coleman R. A. 1980. Enzymes of glycerolipid synthesis in eukaryotes. Annu. Rev. Biochem. 49: 459–487. [DOI] [PubMed] [Google Scholar]
- 3.Manganaro F., Kuksis A. 1985. Purification and preliminary characterization of 2-monoacylglycerol acyltransferase from rat intestinal villus cells. Can. J. Biochem. Cell Biol. 63: 341–347. [DOI] [PubMed] [Google Scholar]
- 4.Bhat B. G., Bardes E. S-G., Coleman R. A. 1993. Solubilization and partial purification of neonatally expressed rat hepatic microsomal monoacylglycerol acyltransferase. Arch. Biochem. Biophys. 300: 663–669. [DOI] [PubMed] [Google Scholar]
- 5.Bhat B. G., Wang P., Coleman R. A. 1994. Hepatic monoacylglycerol acyltransferase is regulated by sn-1,2-diacylglycerol and by specific lipids in Triton X-100/phospholipid-mixed micelles. J. Biol. Chem. 269: 13172–13178. [PubMed] [Google Scholar]
- 6.Jamdar S. C., Cao W. F. 1992. Properties of monoacylglycerol acyltransferase in rat adipocytes. Arch. Biochem. Biophys. 296: 419–425. [DOI] [PubMed] [Google Scholar]
- 7.Tumaney A. W., Shekar S., Rajasekharan R. 2001. Identification, purification, and characterization of monoacylglycerol acyltransferase from developing peanut cotyledons. J. Biol. Chem. 276: 10847–10852. [DOI] [PubMed] [Google Scholar]
- 8.Arrese E. L., Rojas-Rivas B. I., Wells M. A. 1996. Synthesis of sn-1,2-diacylglycereol by monoacylglycerol acyltransferase from Manduca sexta fat body. Arch. Insect Biochem. Physiol. 31: 325–335. [DOI] [PubMed] [Google Scholar]
- 9.Mostafa N., Bhat B. G., Coleman R. A. 1994. Adipose monoacylglycerol:acyl-coenzyme A acyltransferase activity in the white-throated sparrow (Zonotrichia albicollis): characterization and function in a migratory bird. Lipids. 29: 785–791. [DOI] [PubMed] [Google Scholar]
- 10.Yen C. L., Stone S. J., Cases S., Zhou P., Farese R. V., Jr. 2002. Identification of a gene encoding MGAT1, a monoacylglycerol acyltransferase. Proc. Natl. Acad. Sci. USA. 99: 8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen C. L., Farese R. V., Jr 2003. MGAT2, a monoacylglycerol acyltransferase expressed in the small intestine. J. Biol. Chem. 278: 18532–18537. [DOI] [PubMed] [Google Scholar]
- 12.Lockwood J. F., Cao J., Burn P., Shi Y. 2003. Human intestinal monoacylglycerol acyltransferase: differential features in tissue expression and activity. Am. J. Physiol. Endocrinol. Metab. 285: E927–E937. [DOI] [PubMed] [Google Scholar]
- 13.Cao J., Lockwood J., Burn P., Shi Y. 2003. Cloning and functional characterization of a mouse intestinal acyl-coA: monoacylglycerol acyltransferase, MGAT2. J. Biol. Chem. 278: 13860–13866. [DOI] [PubMed] [Google Scholar]
- 14.Yen C. L., Cheong M., Grueter C., Zhou P., Moriwaki J., Wong J. S., Hubbard B., Marmor S., Farese R. V., Jr 2009. Deficiency of the intestinal enzyme acyl CoA:monoacylglycerol acyltransferase-2 protects mice from metabolic disorders induced by high-fat feeding. Nat. Med. 15: 442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng D., Nelson T. C., Chen J., Walker S. G., Wardwell-Swanson J., Meegalla R., Taub R., Billheimer J. T., Ramaker M., Feder J. N. 2003. Identification of acyl coenzyme A:monoacylglycerol acyltransferase 3, an intestinal specific enzyme implicated in dietary fat absorption. J. Biol. Chem. 278: 13611–13614. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y., Cao J., Shi Y. 2004. Identification and characterization of a gene encoding human LPGAT1, an endoplasmic reticulum-associated lysophosphatidylglycerol acyltransferase. J. Biol. Chem. 279: 55866–55874. [DOI] [PubMed] [Google Scholar]
- 17.Rippmann J. F., Schoelch C., Nolte T., Pavliska H., Marle A., Es H., Prestle J. 2009. Improved lipid profile through liver-specific knockdown of liver X receptor a in KKAy diabetic mouse. J. Lipid Res. 50: 22–31. [DOI] [PubMed] [Google Scholar]
- 18.Owen M. R., Corstorphine C. C., Zammit V. A. 1997. Overt and latent activities of diacylglycerol acyltrasnferase in rat liver microsomes: possible roles in very-low-density lipoprotein triacylglycerol secretion. Biochem. J. 323: 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buhman K. K., Smith S. J., Stone S. J., Repa J. J., Wong J. S., Knapp F. F., Jr., Burri B. J., Hamilton R. L., Abmurad N. A., Farese R. V., Jr 2002. DGAT1 is not essential for intestinal triacylglycerol absorption or chylomicron synthesis. J. Biol. Chem. 277: 25474–25479. [DOI] [PubMed] [Google Scholar]
- 20.Yu X. X., Murray S. F., Pandey S. K., Booten S. L., Bao D., Song X., Kelly S., Chen S., McKay R., Monia B. P., et al. 2005. Antisense oligonucleotide reduction of DGAT2 expression improves hepatic steatosis and hyperlipidemia in obese mice. Hepatology. 42: 362–371. [DOI] [PubMed] [Google Scholar]