Abstract
Plasma HDL cholesterol levels (HDL-C) are an independent predictor of coronary artery disease (CAD). We have completed a genome-wide linkage scan for HDL-C in a US cohort consisting of 388 multiplex families with premature CAD (GeneQuest). The heritability of HDL-C in GeneQuest was 0.37 with gender and age as covariates (P = 5.1 × 10−4). Two major quantitative trait loci (QTL) for log-transformed HDL-C adjusted for age and gender were identified onto chromosomes 7p22 and 15q25 with maximum multipoint logarithm of odds (LOD) scores of 3.76 and 6.69, respectively. Fine mapping decreased the 7p22 LOD score to a nonsignificant level of 3.09 and split the 15q25 QTL into two loci, one minor QTL on 15q22 (LOD = 2.73) that spanned the LIPC gene, and the other at 15q25 (LOD = 5.63). A family-based quantitative transmission disequilibrium test (QTDT) revealed significant association between variant rs1800588 in LIPC and HDL-C in the GeneQuest population (P = 0.0067), which may account for the minor QTL on 15q22. The 15q25 QTL is the most significant locus identified for HDL-C to date, and these results provide a framework for the ultimate identification of the underlying HDL-C variant and gene on chromosomes 15q25, which will provide insights into novel regulatory mechanisms of HDL-C metabolism.
Keywords: genetics, linkage, single nucleotide polymorphism, high density lipoprotein cholesterol, coronary artery disease, myocardial infarction, genome-wide scan, quantitative trait locus, LIPC gene
Coronary artery disease (CAD) and its principal clinical complication of acute myocardial infarction (MI) represent the most important causes of death and disability in the developed world. According to the American Heart Association's 2009 statistical update, nearly 16.8 million Americans are affected with CAD, and 7.9 million have had a myocardial infarction. On average, an American will suffer from a coronary event every 26 s, and about every minute somebody will die from one (1). The lifetime risk of developing CAD after age 40 is 49% for men and 32% for women (1).
A decreased concentration of plasma HDL-cholesterol (HDL-C) is a major risk factor for CAD, and epidemiological evidence from several longitudinal studies, including the Framingham Heart Study, indicate that HDL-C is an independent predictor of atherosclerosis in both men and women (2, 3). Families that have early onset, heritable CAD more frequently have low HDL-C than the general population. In men with CAD, low HDL-C is the most common lipid abnormality observed, affecting half the patients (4). Due to its anti-atherogenic properties, even small variations in HDL cholesterol levels are physiologically important. For each 1 mg/dl increase in HDL-C levels, there is a decrement of 2–3% in CAD risk (5). In addition to its prevalence in CAD patients, reduced HDL-C levels are a cornerstone of the metabolic syndrome because low HDL-C is associated with insulin resistance and abdominal obesity in humans (6). With the problem of obesity continuing to escalate in the United States, metabolic syndrome poses a major public health threat affecting 22% of the adult population (7). Because low HDL-C is prevalent in patients with both metabolic syndrome and CAD, the challenge of elucidating the causes of variation in HDL-C levels and discovering new drug treatments for the condition will continue to be critical.
Plasma HDL-C levels have a strong genetic component; approximately 50% of variation in human populations is due to genetic factors (8, 9). While differences in plasma HDL-C have long been recognized to be controlled by genetic factors, our current understanding of the genetics of variation in HDL-C levels is largely based on studies of extreme monogenic HDL-C conditions. Although variation in genes caused by rare mutations may make some contribution to HDL-C levels (10, 11), the majority of genetic variation in genes that control HDL-C levels in the general population has yet to be identified (12). Identification of the genes and genetic variants that control HDL-C concentrations are critical for preventative cardiology in reducing the public health burden of CAD and the metabolic syndrome.
Genome-wide linkage and association scans provide comprehensive and unbiased approaches to identify HDL genes and may lead to the elucidation of unrecognized genetic pathways in HDL metabolism. Genome-wide association is more powerful than genome-wide linkage analysis to detect common alleles at a locus, but it is less powerful if the extreme phenotypes of interest are due to the segregation of many relatively rare alleles at that locus. Furthermore, whereas allelic associations can be due to spurious causes, especially heterogeneity/population stratification, linkage analysis is not subject to such type 1 errors. To date, multiple quantitative trait loci (QTLs) have been identified that show strong evidence of linkage for HDL-C levels (summarized in Table 1). Recent genome-wide association studies (GWAS) also identified multiple loci represented by various single nucleotide polymorphisms (SNP) associated with HDL-C (13–18). The previous work demonstrates the complex inheritance of genetic factors that influence this major CAD risk factor.
TABLE 1.
Summary of QTLs identified for HDL-C by genome-wide linkage analysis
| QTL for HDL-Ca | Number of Families | LOD Score | Study |
|---|---|---|---|
| 2q21.3 | 330 families (Framingham Heart Study) | 3.5 | Arya et al. (44) |
| 4q21.21 | 13 French-Canadian families | 4.6 | Yu et al. (29) |
| 5p13.3 | 101 Caucasian families (NHLBI Study) | 3.6 | Peacock et al. (31) |
| 6q23.1 (HDL3-C) | 330 families (Framingham Heart Study) | 4.0 | Yang et al. (45) |
| 7p15-22 | 388 Caucasian families (GeneQuest) | 3.76 | Present study |
| 7q31.32 | 295 African-American diabetic sib pairs | 4.3 | Adeyemo et al. (46) |
| 8q23.1-24.22 | 25 Finnish families | 4.7 | Soro et al. (33) |
| 8q23.1-24.22 | 10 Mexican-American families (San Antonio Heart Study) | 4.9 | Almasy et al. (30) |
| 9p21.3 | 27 Mexican-American families | 3.4 | Arya et al. (47) |
| 11q23.3 | 105 families from Utah | 3.5 | Kort et al. (48) |
| 12q14.1 | 292 pedigrees (Quebec Family Study) | 4.1 | Bosse et al. (49) |
| 15q22.31b | 10 Mexican-American families (San Antonio Heart Study) | 3.3 | Almasy et al. (30) |
| 15q21-26 | 388 Caucasian families (GeneQuest) | 6.69 | Present study |
| 16q22.3-23.1 | 10 Mexican-American (San Antonio Heart Study) | 4.3 | Mahaney et al. (50) |
| 16q22.3-23.1 | 48 Dutch and Finnish families | 3.4 | Pajukanta et al. (51) |
Abbreviations: HDL-C, high density lipoprotein cholesterol; LOD, logarithm of odds; NHLBI, National Heart, Lung, and Blood Institute; QTL, quantitative trait locus.
Chromosome location.
Unesterified HDL2a-C.
In the present study, we describe a whole genome linkage scan to identify chromosomal regions influencing HDL-C levels in families with premature CAD and MI. Five candidate QTLs were localized to chromosomes 3p25, 7p22, 13q12, 13q32, and 15q25 with maximum multipoint logarithm of odds (LOD) scores of 4.10, 4.21, 4.66, 3.95, and 7.57, respectively. After log-transformation and adjustment of age and gender, 15q25 and 7p22 QTLs remained significant with maximum multipoint LOD scores of 6.69 and 3.76, respectively. Further fine mapping resulted in the drop of the LOD score of 7p22 QTL to 3.09 and of 15q25 to 5.63. Interestingly, the promoter SNP of LIPC rs1800588 showed significant association with HDL in the family-based quantitative transmission disequilibrium test (QTDT) analysis. To the best of our knowledge, the 15q25 QTL is the most significant locus identified for HDL-C to date. Our study provides a framework for the ultimate cloning and identification of genes that regulate plasma HDL-C levels.
MATERIAL AND METHODS
Clinical data
The study population consists of 714 Caucasian individuals from 388 families with familial premature CAD and MI as described previously (19). Patients were recruited by cardiologists and data coordinators at the Cleveland Clinic Foundation over an approximately five-year period. Institutional review boards approved protocols, and informed consent was obtained from every study participant. For recruitment, each proband in a family was required to have a living sibling meeting the same criteria. Participants answered a health questionnaire, had anthropomorphic measures taken, and had fasted blood drawn for measurement of serum markers and DNA extraction. HDL-C was measured by standard laboratory procedures.
Genotyping
DNA was extracted from whole blood using Puregene Kits (Gentra). Genome-wide genotyping of microsatellite markers was performed by the National Heart, Lung, and Blood Institute (NHLBI) Mammalian Genotyping Services directed by Dr. James L. Weber at Center for Medical Genetics, Marshfield Clinic, using Screening Set 11 with 408 markers that span the human genome at approximately every 10 cM (http://research.marshfieldclinic.org/genetics/geneticResearch/screeningSets.asp). For fine mapping, additional markers were selected from the Marshfield database, synthesized, tagged with 6′-FAM (Sigma), and genotyped using an ABI 3100 genetic analyzer (Applied Biosystems) as previously described (20). The quality of genotyping for markers used for fine mapping was high.
For genotyping SNPs, the TaqMan PreDesigned SNP Genotyping Assays were performed on an ABI PRISM 7900HT Sequence Detection System as previously described (21, 22).
Genetic statistical analyses
Before linkage scanning, obvious pedigree errors, data errors, genotyping errors, and locus-order errors that commonly occur with a large-scale linkage analysis were corrected. Allele frequencies for all markers genotyped for the cohort were estimated by maximum likelihood methods using the S.A.G.E. program FREQ (23). Pedigree relationships were checked using RELTEST, which uses a Markov process model of allele sharing along the chromosome and classifies pairs of pedigree members according to their true relationship by use of genome-scan data (24). Twenty-seven of 428 pedigrees with uncorrectable errors were excluded from further linkage analysis. The S.A.G.E. program MARKERINFO was used to detect any Mendelian inheritance inconsistencies. Three families with inconsistent Mendelian inheritance were eliminated from the study. Three pairs of monozygotic twins were identified in GeneQuest and excluded from further statistical analysis. Only 388 Caucasian families with HDL-C data were analyzed for linkage.
A genome-wide linkage analysis was performed using the program GENEHUNTER (GENEHUNTER 2.1 package, Whitehead Institute, Cambridge, MA) using the sibs quantitative trait mapping function, maximum likelihood QTL variance estimation. All sib pairs were used for analysis. Positions of markers were from Center for Medical Genetics (see http://research.marshfieldclinic.org/genetics/ for Marshfield Genetic Database marker information). Age and sex were determined to be important covariates for HDL-C levels and were modeled in the linkage analysis using general linear regression with SAS Version 9.00. To evaluate the significance of the linkage results, we followed the criteria proposed by Lander and Kruglyak (25) specifically for sib pair linkage analysis in humans. LOD scores for suggestive, significant, and highly significant evidence of linkage are 2.2, 3.6, and 5.4, respectively.
A family-based association study of SNPs with HDL-C was carried out using quantitative transmission disequilibrium tests (QTDT2.5.1, http://www.sph.umich.edu/csg/abecasis/QTDT/) (26).
RESULTS
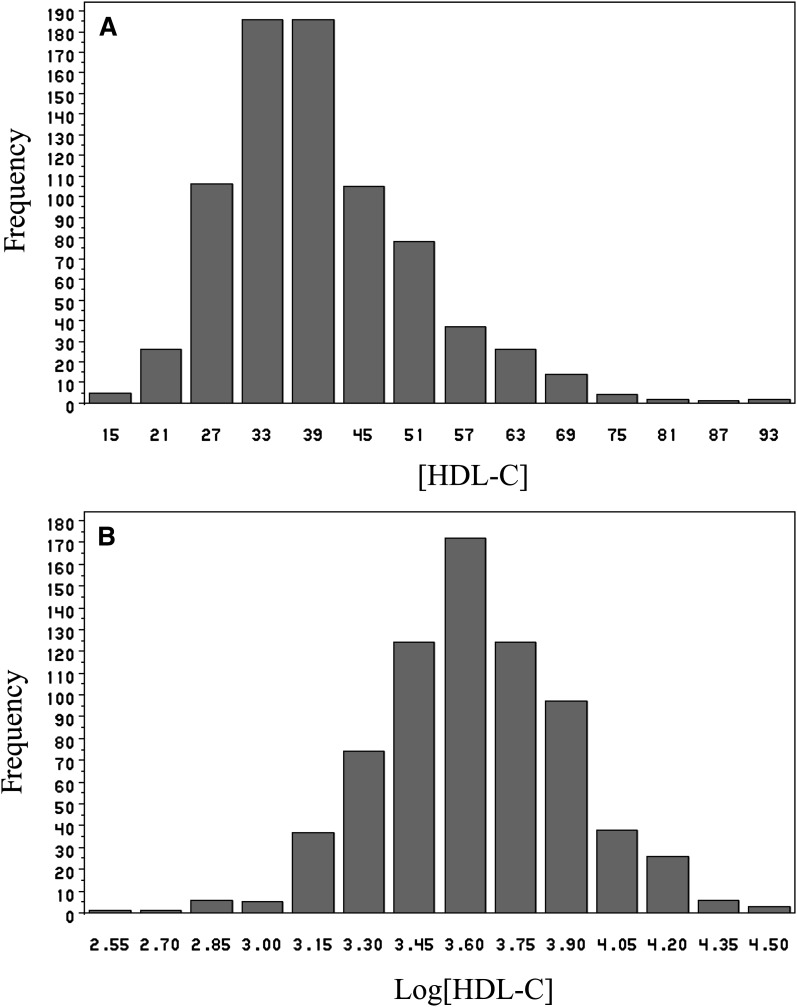
We have completed a genome-wide linkage analysis to identify QTLs for plasma HDL-C levels in a well-characterized US cohort consisting of multiplex families (GeneQuest). HDL-C values were available for 67% of the GeneQuest study population. A total of 714 Caucasian persons in 388 families with HDL-C data available were analyzed. The clinical and demographic features of the study population are shown in Table 2. The mean value of HDL-C in the population was low (39.2 mg/dl). As the HDL-C levels did not present with a normal distribution (Fig. 1A), the values were also log-transformed prior to analysis. The transformed HDL-C values are shown in Fig. 1B. Age and sex were determined to be significantly correlated with HDL-C values (P = 0.0024 and P < 0.0001, respectively). Therefore, HDL-C values were also adjusted for age and gender using general linear regression analysis.
TABLE 2.
Clinical and demographic features of the study population
| Feature | Value |
|---|---|
| Number of pedigrees | 388 |
| Number of family members (N) | 714 |
| Gender (M/F) | 480/234 |
| Age (years) | 49.6 ± 7.8a |
| Ethnicity | Caucasian |
| Smoking (%) | 81.5 |
| BMI (kg/m ) | 29.4 ± 5.7a |
| Hypertension | 333/714 (46.7%) |
| CAD | 694/714 (97.2%) |
| MI | 384/714 (53.8%) |
| Non-CAD | 20/714 (2.8%) |
| Diabetes (NIDDM) | 78/714 (10.9%) |
| Diabetes (IDDM) | 31/714 (4.3%) |
| Total cholesterol (mg/dl) | 220.2 ± 55.3a |
| HDL cholesterol (mg/dl) | 39.2 ± 11.4a |
| LDL cholesterol (mg/dl) | 133.8 ± 43.4a |
Abbreviations: BMI, body mass index; CAD, coronary artery disease; IDDM, insulin-dependent diabetes mellitus; MI, myocardial infarction; NIDDM, noninsulin-dependent diabetes mellitus.
Data are mean ± SD.
Fig. 1.
Distribution of the HDL-C concentrations [HDL-C] (A) and log transformed HDL-C concentrations (B) in the study population. HDL-C, high density lipoprotein cholesterol.
A residual heritability estimate of HDL-C in the study population was calculated as 0.37 with gender and age as covariates (P = 5.1 × 10−4; SOLAR, http://solar.sfbrgenetics.org/). Genome-wide genotyping was carried out with 408 polymorphic markers that span the entire human genome at approximately every 10 cM. We performed both single-point and multipoint linkage analyses using the GeneHunter sibs quantitative trait mapping function and maximum likelihood QTL variance estimation, the results of which are shown in Table 3 and Figs. 2–5.
TABLE 3.
Summary of chromosomal regions linked to plasma HDL-C levels in a Caucasian premature CAD and MI population (GeneQuest)
| Uncorrected [HDL-C] |
[HDL-C]a |
Log[HDL-C]a |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome and Marker | Location | Map Position (cM) | Position (Mb) | Single-Point LOD | Multipoint LOD | Single-Point LOD | Multipoint LOD | Single-Point LOD | Multipoint LOD |
| Chr 3p | |||||||||
| MFD433 | 3p26 | 11.0 | 3.6 | 5.26 | 3.89 | 3.76 | 3.16 | 1.54 | 1.71 |
| GATA131D09 | 3p26 | 19.3 | 5.5 | 2.95 | 3.47 | 2.42 | 3.57 | 0.62 | 1.49 |
| D3S4545 | 3p25 | 26.0 | 10.8 | 2.60 | 3.11 | 3.40 | 4.10 | 1.76 | 2.27 |
| Chr 7p | |||||||||
| D7S2477 | 7p22 | 0 | 2.6 | 1.81 | 3.10 | 1.99 | 3.37 | 0.05 | 2.73 |
| D7S3056 | 7p22 | 7.0 | 4.5 | 1.26 | 4.58 | 1.28 | 4.21 | 1.83 | 3.76 |
| D7S3047 | 7p21 | 17.0 | 8.5 | 4.26 | 3.51 | 4.12 | 2.55 | 3.62 | 2.07 |
| D7S1802 | 7p21 | 33.0 | 20.2 | 4.24 | 4.46 | 4.41 | 3.21 | 2.26 | 1.74 |
| D7S1808 | 7p15 | 42.0 | 27.9 | 3.69 | 1.79 | 3.02 | 1.76 | 1.19 | 1.56 |
| Chr 13q | |||||||||
| ATA5A09N | 13q12 | 20.0 | 28.7 | 6.42 | 4.79 | 6.17 | 4.66 | 4.24 | 3.10 |
| D13S1493 | 13q13 | 26.0 | 32.0 | 2.98 | 2.87 | 2.84 | 2.36 | 1.43 | 1.38 |
| Chr 13q31-32 | |||||||||
| D13S317 | 13q31 | 64.0 | 81.6 | 2.14 | 3.27 | 2.11 | 3.95 | 1.96 | 1.57 |
| D13S793 | 13q32 | 76.0 | 96.6 | 2.93 | 3.60 | 2.75 | 3.47 | 1.45 | 2.63 |
| Chr 15q | |||||||||
| D15S643 | 15q21 | 52.0 | 55.3 | 2.87 | 2.10 | 3.12 | 1.72 | 2.42 | 2.20 |
| D15S1507 | 15q22 | 60.0 | 60.9 | 4.40 | 2.94 | 4.37 | 4.75 | 3.19 | 3.09 |
| D15S818 | 15q24 | 72.0 | 70.7 | 2.23 | 4.37 | 2.83 | 4.91 | 2.06 | 4.23 |
| D15S655 | 15q25 | 83.0 | 84.2 | 6.28 | 5.91 | 6.76 | 7.57 | 5.79 | 6.69 |
| D15S652 | 15q26 | 90.0 | 88.7 | 4.77 | 5.03 | 5.72 | 6.72 | 4.44 | 6.18 |
Abbreviations: CAD, coronary artery disease; HDL-C, high density lipoprotein cholesterol; LOD, logarithm of odds; MI, myocardial infarction.
Adjusted for age and gender.
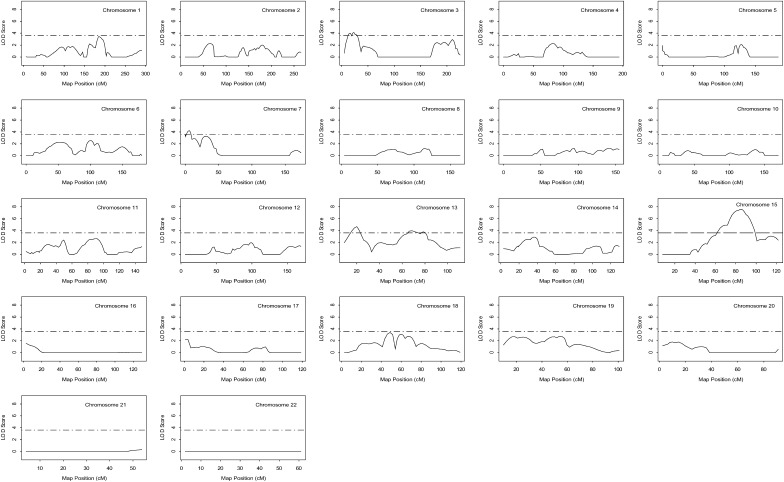
Fig. 2.
Likelihood plots for QTLs for HDL-C adjusted for age and gender. The Y-axis of each plot is the LOD score; the X-axis is the marker map position. Solid lines represent the multipoint linkage analysis; horizontal dashed lines indicate the significance threshold which is equal to a LOD score value of 3.6. Significant linkages to chromosomes 3p25, 7p22, 13q12, 13q32, and 15q25 were detected with multipoint allele sharing LOD scores of 4.10, 4.21, 4.66, 3.95, and 7.57, respectively. HDL-C, high density lipoprotein cholesterol; LOD, logarithm of odds; QTL, quantitative trait locus.
Fig. 3.
Detailed likelihood plots for significant QTLs for HDL-C on chromosomes 3p25, 7p21, 13q12, 13q32, and 15q25. The Y-axis of each plot is the LOD score; the X-axis is the marker map position. Solid lines represent the multipoint linkage analysis; horizontal dashed lines indicate the significance threshold which is equal to a LOD score value of 3.6. HDL-C, high density lipoprotein cholesterol; LOD, logarithm of odds; QTL, quantitative trait locus.
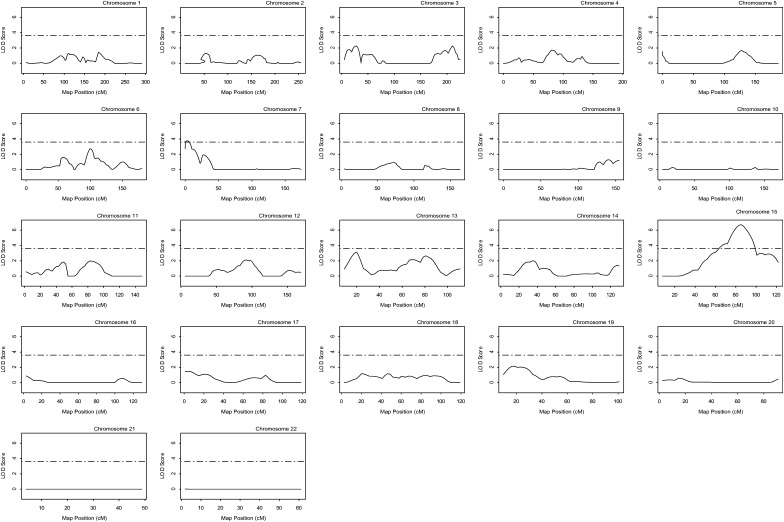
Fig. 4.
Likelihood plots for QTLs for log-transformed HDL-C adjusted for age and gender. Horizontal dashed lines indicate the significance threshold which is equal to a LOD score value of 3.6. HDL-C, high density lipoprotein cholesterol; LOD, logarithm of odds; QTL, quantitative trait locus.
Fig. 5.
Detailed likelihood plots for significant QTLs for log transformed HDL-C on chromosomes 7p21 and 15q25 adjusted for age and gender. Horizontal dashed lines indicate the significance threshold which is equal to a LOD score value of 3.6. HDL-C, high density lipoprotein cholesterol; LOD, logarithm of odds; QTL, quantitative trait locus.
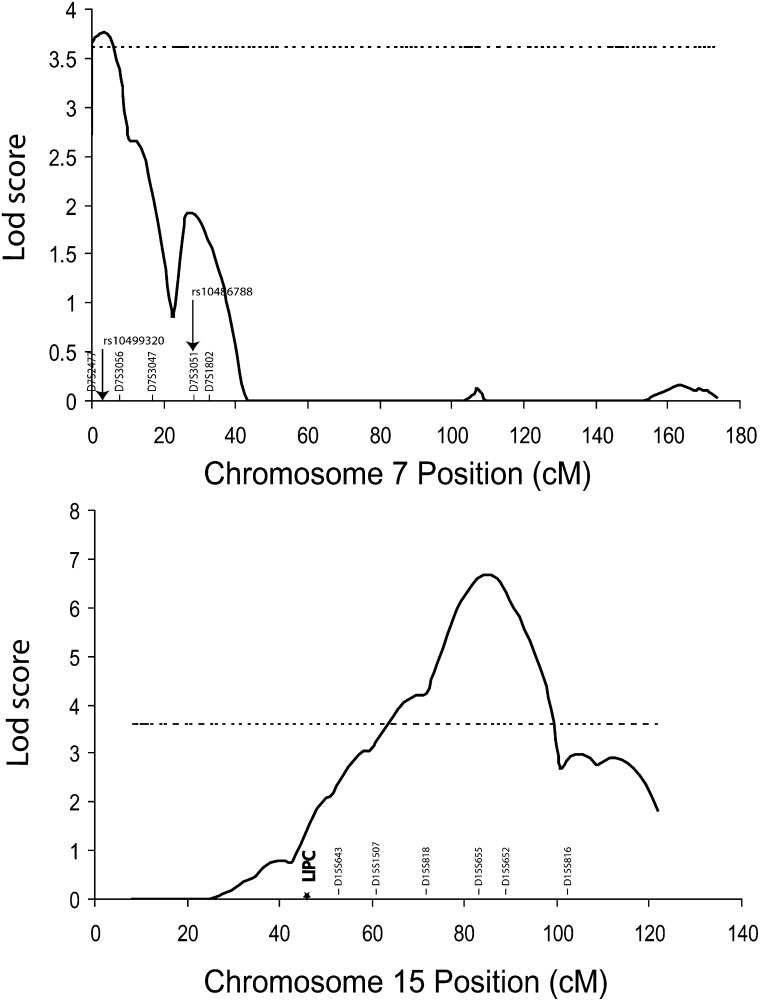
Of the loci identified for HDL-C adjusted for age and gender, the 15q25 region displayed the strongest evidence for linkage to HDL-C. Model-free multipoint linkage analysis revealed high significance at 86 cM in a region between markers D15S655 (83 cM) and D15S652 (90cM) as shown in Fig. 3. Additionally, single-point linkage analysis confirmed that high significance was reached at both markers: D15S655 (LOD 6.76) and D15S652 (LOD 5.72) (Table 3). Therefore, for the chromosome 15 locus, the maximum multipoint and single-point LOD scores were 7.57 and 6.76, respectively.
Four additional loci were detected with significant evidence of multipoint linkage: 3p25, 7p22, 13q12, and 13q31-32. The second strongest locus for HDL-C was at 13q12 (20 cM) with maximum multipoint evidence of linkage equal to a LOD of 4.66 (Fig. 3). This locus is in close proximity to marker ATA5A09N (20 cM), and single-point analysis confirmed significant evidence of linkage at ATA5A09N with a LOD of 6.17 (Table 3). Maximum multipoint linkage for chromosome 3p25 (LOD = 4.10) was reached at 23 cM between markers GATA131D09 (19.3 cM) and D3S4545 (26 cM) (Table 3 and Fig. 3). The chromosome 7p locus followed with maximum multipoint evidence of linkage equal to a LOD of 4.21 at 6.1 cM (Fig. 3). Single-point analysis yielded a significant result at the closest marker D7S3056 (LOD = 4.11; 7.0 cM) (Table 3). Finally, multipoint analysis for the 13q31-32 locus reached an LOD score of 3.95 at 68.8 cM between markers D13S317 (64 cM) and D13S793 (76.0 cM) (Fig. 2).
For HDL-C levels without adjustment of age and gender, we also obtained evidence of linkage for the five putative QTLs on chromosome 15q25, 3p25, 7p22, 13q12, and 13q31-32. The maximum multipoint/single-point LOD scores were 5.91/6.28, 3.47/2.95, 4.58/4.24, 4.79/6.42, and 3.60/2.93, respectively (Table 3). For HDL-C after log-transformation and adjustment for age and gender, the maximum multipoint/single-point LOD scores were 6.69/5.79, 2.27/1.76, 3.76/3.63, 3.10/4.24, and 2.63/1.9, respectively (Figs. 4 and Table 3).
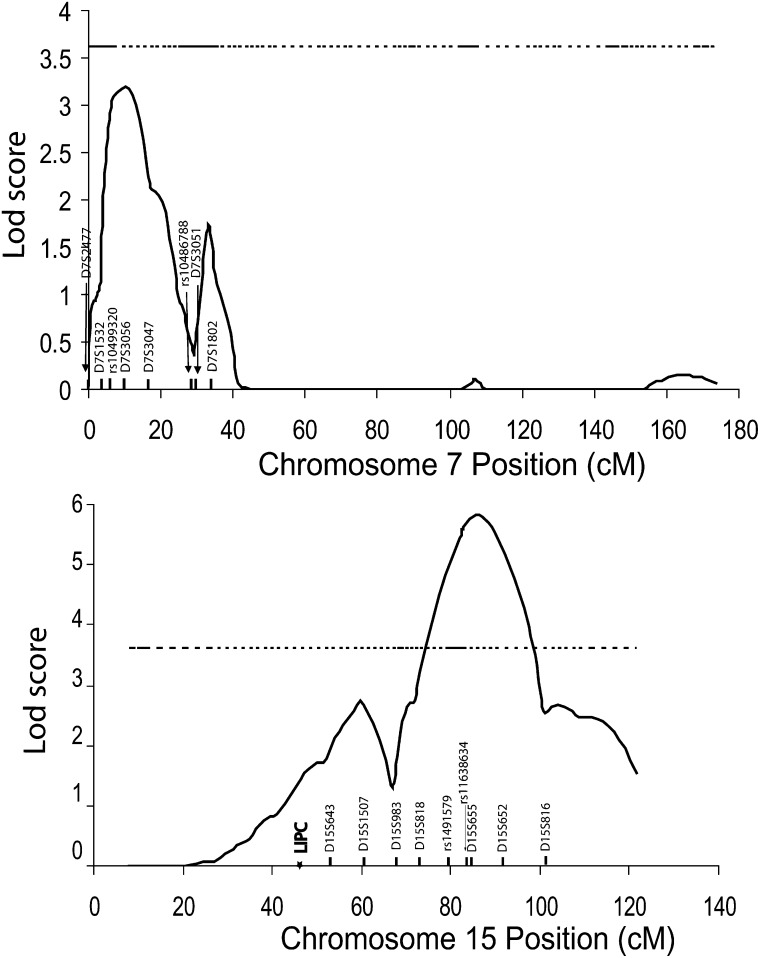
For the two strongest linkage loci on 7p22 and 15q25, fine mapping was carried out with additional microsatellite markers and di-allelic SNP markers. For the 7p22 QTL, we genotyped the GeneQuest families with D7S1532 and two candidate SNPs, rs10499320 and rs10486788, which in the Framingham Heart Study showed potential association with HDL-C (13). Linkage analysis for HDL-C after log-transformation and adjustment for age and gender showed a major, complete linkage peak (half peak before fine mapping). However, the maximum LOD score of 7p22 dropped to 3.09, which did not exceed the significance threshold of a LOD score of 3.6 (Fig. 6 and Table 4). The size of the one-LOD drop interval was not changed by fine mapping. A QTDT did not identify any association between rs10499320 and rs10486788 and HDL-C (P > 0.05) (Table 5). Haplotypes formed by these two SNPs were predicted by PHASE software (http://stat.washington.edu/stephens/software.html), and none of the haplotypes showed any association with HDL-C levels (data not shown).
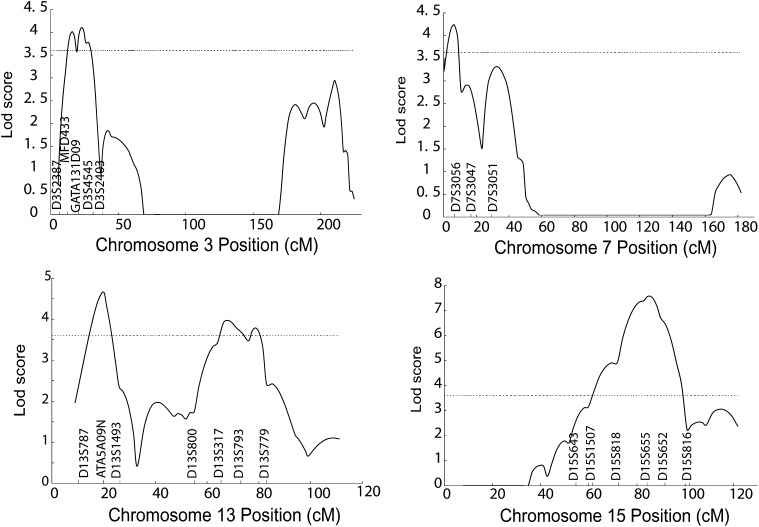
Fig. 6.
Fine mapping of the 7p22 and 15q25 QTLs for log transformed HDL-C adjusted for age and gender. Horizontal dashed lines indicate the significance threshold which is equal to a LOD score value of 3.6. HDL-C, high density lipoprotein cholesterol; LOD, logarithm of odds; QTL, quantitative trait locus.
TABLE 4.
Fine mapping of the 7p22 and 15q25 QTLs for log[HDL-C] after adjustment for age and gender
| Chromosome and Marker | Location | Map Position (cM) | Position (Mb) | Single-Point LOD | Multipoint LOD |
|---|---|---|---|---|---|
| Chr 7p | |||||
| D7S1532 | 7p22 | 3.13 | 2.4 | 0.19 | 1.04 |
| rs10499320 | 7p22 | 4.15 | 3.1 | 1.07 | 2.15 |
| D7S3056 | 7p22 | 7.0 | 4.5 | 1.60 | 3.09 |
| D7S3047 | 7p21 | 17.0 | 8.5 | 3.37 | 2.12 |
| Chr 15q | |||||
| D15S643 | 15q21 | 51.8 | 55.3 | 1.98 | 1.76 |
| D15S1507 | 15q22 | 59.8 | 60.9 | 2.54 | 2.73 |
| D15S983 | 15q23 | 66.9 | 66.4 | 0.79 | 1.30 |
| D15S818 | 15q24 | 71.8 | 70.7 | 2.35 | 2.75 |
| rs1491579 | 15q25 | 78.9 | 82.0 | 2.67 | 4.83 |
| rs11638634 | 15q25 | 82.8 | 85.7 | 2.80 | 5.47 |
| D15S655 | 15q25 | 82.8 | 85.8 | 4.48 | 5.63 |
| D15S652 | 15q26 | 89.8 | 88.7 | 3.42 | 5.51 |
Abbreviations: HDL-C, high density lipoprotein cholesterol; LOD, logarithm of odds; QTL, quantitative trait locus.
TABLE 5.
Assessment of two SNPs at chromosome 7p22 for association with HDL-C using a QTDT in GeneQuest
| Chromosome 7p22 | QTDT Analysis | ||
|---|---|---|---|
| SNP | Gene/Position | F | P |
| rs10499320 | N/A | 0.42 | 0.6578 |
| rs10486788 | N/A | 0.73 | 0.4801 |
Abbreviations: HDL-C, high density lipoprotein cholesterol; QTDT, quantitative transmission disequilibrium test; SNP, single nucleotide polymorphisms.
For fine mapping of the 15q25 QTL, we studied D15S983 and two SNPs, rs1491579 and rs1638634, adjacent to marker D15S655 with the highest LOD score. The fine mapping study splits the QTL into two linkage peaks, one with a maximum multipoint LOD score of 2.73 at chromosome 15q22 covering the LIPC gene (encoding hepatic lipase) and the other with a maximum multipoint LOD score of 5.63 remaining at chromosome 15q25 (Fig. 6 and Table 4). The fine mapping sharpened the major linkage peak by narrowing the one-LOD drop interval from 14.8 cM to 9.1 cM. A QTDT did not identify any association between rs1491579 and rs1638634 and HDL-C (P > 0.05) (Table 5). None of the haplotypes formed by these two SNPs showed any association with HDL-C levels (data not shown).
Recent genome-wide SNP association studies and earlier candidate gene analysis revealed association of SNPs in the LIPC gene with HDL-C levels (14, 27). Because LIPC is located within the small QTL for HDL-C on 15q22 (Fig. 6), we assessed its association with HDL-C in the GeneQuest families using a family-based QTDT, focusing on the promoter and exonic SNPs. For the LIPC promoter, we selected SNP rs1800588 because this promoter SNP has been reported to be associated with plasma HDL-C levels (14). Furthermore, two exonic SNPs, rs690 and rs6083, were selected among tagging SNPs for LIPC identified by the Tagger program and Haploview 4.1 using the threshold minor allele frequency of 0.3 and R2 of 0.6. As shown in Table 6, significant association with HDL-C levels was identified for SNP rs1800588 (P = 0.0067), but not with rs690 or rs6083.
TABLE 6.
Assessment of SNPs at chromosome 15q22-25 for association with HDL-C using a QTDT in GeneQuest
| Chromosome 15q22-25 | QTDT Analysis | ||
|---|---|---|---|
| SNP | Gene/Position | F | P |
| rs1800588 | LIPC/promoter | 5.05 | 0.0067 |
| rs690 | LIPC/exon 4 | 1.72 | 0.1791 |
| rs6083 | LIPC/exon 5 | 0.21 | 0.8128 |
| rs1491579 | SH3GL3/intron 4 | 1.56 | 0.2117 |
| rs11638634 | N/A | 0.58 | 0.5576 |
Abbreviations: HDL-C, high density lipoprotein cholesterol; QTDT, quantitative transmission disequilibrium test; SNP, single nucleotide polymorphisms.
DISCUSSION
The present study reports evidence from genome-wide linkage analysis that multiple QTLs influence HDL-C levels in a cohort of premature CAD and MI families (GeneQuest). In particular, we identified one locus on chromosome 15q25 with highly significant evidence of linkage to date, displaying a LOD score that is greater than the cutoff LOD score of 5.40 for highly significant evidence of linkage at 86 cM. Four regions were also detected with evidence of linkage to HDL-C levels on chromosomes 3p25, 7p22, 13q12, and 13q32.
Supporting the current findings, several independent studies provide evidence for the candidate QTLs on chromosomes 15q25, 13q12, 3p25, and 13q32. Linkage for HDL-C was observed on chromosome 15 in Turkish families with dyslipidemia (LOD = 3.05 at 15q23 at 66.5 Mb) (28); French Canadian families (LOD = 1.6 at 15q25.1 at 78.1 Mb) (29); and for unesterified HDL2b-C in Mexican-American families (LOD = 2.54 at 15q25.3 at 83.2 Mb) (30). Our data, together with these results, suggests that the HDL-C gene at this locus may play a role in HDL-C metabolism in several ethnic populations. On chromosome 13, suggestive evidence of linkage was detected in families from the NHLBI family heart study (LOD = 2.36 at 13q13.2 at 32.9 Mb) (31) and families with type 2 diabetes (LOD = 2.01 at 13q13-14 at 32.2-41.0 Mb) (32). For chromosome 3, suggestive evidence for an HDL-C locus was identified in 25 Finnish families (LOD = 2.1 at 3p26.3-24.3 at 0-17 Mb) (33). The chromosome 13q32 locus was previously reported using a Bayesian Markov Chain–Monte Carlo (MCMC) approach to map HDL-C QTLs in FCHL families (Intensity Ratio = 13 at 13p32 at 96.6-100.3 Mb) (34). The chromosome 13q32 region has also been identified for linkage to total cholesterol, LDL cholesterol, and various lipid-related traits in two studies involving families with familial hypercholesterolemia (35, 36). In total, these independent reports provide some support that the genetic regions identified in the current study harbor genetic variants that regulate HDL-C levels.
Our genome-wide linkage scan and heritability analysis of a US Caucasian population clearly indicate that HDL-C and CAD/MI are complex traits with mixed contributions from multiple genetic and environmental factors. Various studies using transgenic and gene-targeted mice have revealed >100 genes influencing the development of atherosclerotic lesions (37, 38), and a larger number of genes influencing HDL metabolism is expected to be identified. It is interesting to note that a recent study (39) using bivariate linkage analysis of coronary artery calcification (CAC), a measure of atherosclerosis determined by electron beam–computed tomography, provided evidence of two regions with pleiotropic effects on CAC and HDL-C on chromosomes 4p16 (MLS = 3.03, P = 0.00084) and 9p12 (MLS = 3.21, P = 0.00056), which may suggest an underlying genomic mechanism for pleiotropism. In future studies we can explore gene-gene interactions for HDL-C (including related plasma parameters) and clinical cardiovascular phenotypes as both data are available, and significant genetic loci were identified for both phenotypes.
A recent genome-wide SNP association study involving 1,087 Framingham Heart Study offspring cohort participants identified two SNPs (rs10499320, rs10486788) within the 1-LOD- and 2-LOD-drop interval of our 7p15-22 candidate HDL-C QTL that showed positive association with HDL-C levels (P = 8.5 × 10−4, 8.8 × 10−4, respectively) (13). Our QTDT did not detect any association between HDL-C and SNP rs10499320 or between HDL-C and SNP rs10486788 in the GeneQuest families.
For chromosome 15q25 QTL for HDL-C, fine mapping studies split the QTL into two separate QTLs, one major QTL on 15q25 with a maximum LOD score of 5.63 and the other minor QTL on 15q22 that showed a maximum LOD score of 2.73 and covered the LPIC gene. Two SNPs, including rs1491579 and rs11638634 close to marker D15S655 showing the maximum LOD score, were analyzed for association with HDL-C, but no significant association was detected. For the minor QTL at 15q22, we analyzed the LIPC gene for its association with HDL-C using a family-based QTDT in the GeneQuest families. Interestingly, a LIPC promoter SNP, rs1800588, showed significant association with HDL-C, but two exonic SNPs, rs690 and rs6083, were not associated with HDL-C in the GeneQuest population (Table 6). The rs1800588 association may account for the minor QTL on 15q22 identified by fine mapping. These results are identical to those generated by a population-based association study reported in a Turkish population (27). To the best of our knowledge, this is the first family-based QTDT study to demonstrate the association between LIPC and HDL-C. On the other hand, the specific gene responsible for the major 15q25 HDL-C QTL remains to be identified.
Due to its highly significant linkage to HDL-C, the chromosome 15 locus provides the most promise for gene identification. Several interesting candidate genes reside within this genetic interval. The most promising candidate genes for HDL-C at this locus include START domain containing 5 (STARD5), PL1N1, ADANTSL3, and endonuclease VIII-like 1 (NEIL1). STARD5 is a member of the steroidogenic acute regulatory lipid transfer (START) domain superfamily of proteins involved in several pathways of intracellular trafficking and metabolism of cholesterol (40). PLIN1 is a cAMP-dependent protein kinase substrate in adipocytes that plays a role in lipolysis and has been shown to be associated with metabolic variables in Caucasian women (41). ADAMTSL3 belongs to the ADAMTS metalloprotease family (42). NEIL1 knockout mice developed metabolic syndrome with severe obesity, dyslipidemia, and fatty liver disease (43). It would be interesting to determine whether SNPs in these genes may account for the major 15q25 HDL-C QTL.
One limitation of the current study is that empirical P values for each QTL based on trait or marker resimulation data could not be estimated because such a program was not implemented in the GeneHunter package. The other limitation is that the HDL-C QTLs identified in this study were mostly derived from patients and families with CAD and MI (97.2% of the GeneQuest population) (Table 2), which may be enriched for low HDL-C levels. These QTLs may be different from those modulating HDL-C in general healthy populations.
CONCLUSION
The genome-wide linkage analysis for HDL-C QTLs described here provides highly significant evidence for the presence of a locus on chromosome 15q25 controlling HDL-C values. The 15q25 QTL is the most significant locus identified for HDL-C and represents a novel candidate genetic region that influences plasma HDL-C levels. These results will lay the foundation for the successful identification of genetic variants that influence HDL-C at this QTL.
Acknowledgments
The authors thank J. Weber and the NHLBI Mammalian Genotyping Service for genotyping of microsatellite markers.
Footnotes
Abbreviations:
- CAD
- coronary artery disease
- GWAS
- genome-wide association study
- HDL-C
- high density lipoprotein cholesterol
- LOD
- logarithm of odds
- MI
- myocardial infarction
- MCMC
- Bayesian Markov Chain Monte Carlo
- QTDT
- quantitative transmission disequilibrium test
- QTL
- quantitative trait locus
- SNP
- single nucleotide polymorphisms
This study was supported by a grant from the National Heart, Lung and Blood Institute Genotyping Service (Q.K.W.); National Institutes of Health Grants P50 HL-077107 (Q.K.W., E.J.T., R.C.E.) and P50 HL-081011 (Q.K.W.); Research Grant GM-28356 from the National Institute of General Medical Sciences (R.C.E.); China National High Technology (863 Scientific Program 2006AA02Z476); an AHA grant 09BGIA2460022 (G.Q.S.); and the National Basic Research Program of China (973 Program 2007CB512002). Some of the results in this article were obtained by using the program package S.A.G.E., which is supported by a US Public Health Service Resource Grant (RR03655) from the National Center for Research Resources. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Lloyd-Jones D., Adams R., Carnethon M., De S. G., Ferguson T. B., Flegal K., Ford E., Furie K., Go A., Greenlund K., et al. 2009. Heart disease and stroke statistics–2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 119: 480–486. [DOI] [PubMed] [Google Scholar]
- 2.Boden W. E. 2000. High-density lipoprotein cholesterol as an independent risk factor in cardiovascular disease: assessing the data from Framingham to the Veterans Affairs High–Density Lipoprotein Intervention Trial. Am. J. Cardiol. 86: 19L–22L. [DOI] [PubMed] [Google Scholar]
- 3.Wilson P. W., Abbott R. D., Castelli W. P. 1988. High density lipoprotein cholesterol and mortality. The Framingham Heart Study. Arteriosclerosis. 8: 737–741. [DOI] [PubMed] [Google Scholar]
- 4.Genest J. J., McNamara J. R., Salem D. N., Schaefer E. J. 1991. Prevalence of risk factors in men with premature coronary artery disease. Am. J. Cardiol. 67: 1185–1189. [DOI] [PubMed] [Google Scholar]
- 5.Gordon D. J., Probstfield J. L., Garrison R. J., Neaton J. D., Castelli W. P., Knoke J. D., Jacobs D. R., Jr., Bangdiwala S., Tyroler H. A. 1989. High-density lipoprotein cholesterol and cardiovascular disease. Four prospective American studies. Circulation. 79: 8–15. [DOI] [PubMed] [Google Scholar]
- 6.Despres J. P., Lemieux I., Dagenais G. R., Cantin B., Lamarche B. 2000. HDL-cholesterol as a marker of coronary heart disease risk: the Quebec cardiovascular study. Atherosclerosis. 153: 263–272. [DOI] [PubMed] [Google Scholar]
- 7.Ford E. S., Giles W. H., Dietz W. H. 2002. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. JAMA. 287: 356–359. [DOI] [PubMed] [Google Scholar]
- 8.Wang X., Paigen B. 2002. Quantitative trait loci and candidate genes regulating HDL cholesterol: a murine chromosome map. Arterioscler. Thromb. Vasc. Biol. 22: 1390–1401. [DOI] [PubMed] [Google Scholar]
- 9.Wang X., Le Roy I., Nicodeme I, E., Li R., Wagner R., Petros C., Churchill G. A., Harris S., Darvasi A., Kirilovsky J., et al. 2003. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 13: 1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen J. C., Kiss R. S., Pertsemlidis A., Marcel Y. L., McPherson R., Hobbs H. H. 2004. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 305: 869–872. [DOI] [PubMed] [Google Scholar]
- 11.Frikke-Schmidt R., Nordestgaard B. G., Jensen G. B., Tybjaerg-Hansen A. 2004. Genetic variation in ABC transporter A1 contributes to HDL cholesterol in the general population. J. Clin. Invest. 114: 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qasim A., Rader D. J. 2006. Human genetics of variation in high-density lipoprotein cholesterol. Curr. Atheroscler. Rep. 8: 198–205. [DOI] [PubMed] [Google Scholar]
- 13.Kathiresan S., Manning A. K., Demissie S., D'Agostino R. B., Surti A., Guiducci C., Gianniny L., Burtt N. P., Melander O., Orho-Melander M., et al. 2007. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med. Genet. 8(Suppl. 1): S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., Mannucci P. M., Anand S., Engert J. C., Samani N. J., Schunkert H., et al. 2009. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kooner J. S., Chambers J. C., Aguilar-Salinas C. A., Hinds D. A., Hyde C. L., Warnes G. R., Gomez Perez F. J., Frazer K. A., Elliott P., Scott J., et al. 2008. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 40: 149–151. [DOI] [PubMed] [Google Scholar]
- 18.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q., Rao S., Shen G. Q., Li L., Moliterno D. J., Newby L. K., Rogers W. J., Cannata R., Zirzow E., Elston R. C., et al. 2004. Premature myocardial infarction novel susceptibility locus on chromosome 1p34–36 identified by genomewide linkage analysis. Am. J. Hum. Genet. 74: 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Chen S., Yoo S., Chakrabarti S., Zhang T., Ke T., Oberti C., Yong S. L., Fang F., Li L., et al. 2008. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 135: 1017–1027. [DOI] [PubMed] [Google Scholar]
- 21.Abdullah K. G., Li L., Shen G. Q., Hu Y., Yang Y., MacKinlay K. G., Topol E. J., Wang Q. K. 2008. Four SNPS on chromosome 9p21 confer risk to premature, familial CAD and MI in an American Caucasian population (GeneQuest). Ann. Hum. Genet. 72: 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen G. Q., Li L., Girelli D., Seidelmann S. B., Rao S., Fan C., Park J. E., Xi Q., Li J., Hu Y., et al. 2007. An LRP8 variant is associated with familial and premature coronary artery disease and myocardial infarction. Am. J. Hum. Genet. 81: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S.A.G.E. Statistical Analysis for Genetic Epidemiology, Release 5.3. Cork, Ireland. [Google Scholar]
- 24.Olson J. M. 1999. Relationship estimation by Markov-process models in a sib-pair linkage study. Am. J. Hum. Genet. 64: 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lander E., Kruglyak L. 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. [comments] Nat. Genet. 11: 241–247. [DOI] [PubMed] [Google Scholar]
- 26.Abecasis G. R., Cardon L. R., Cookson W. O. 2000. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 66: 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodoglugil U., Williamson D. W., Mahley R. W. 2010. Polymorphisms in the hepatic lipase gene affect plasma HDL-C levels in a Turkish population. J. Lipid Res. 51: 422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Y., Wyszynski D. F., Waterworth D. M., Wilton S. D., Barter P. J., Kesaniemi Y. A., Mahley R. W., McPherson R., Waeber G., Bersot T. P., et al. 2005. Multiple QTLs influencing triglyceride and HDL and total cholesterol levels identified in families with atherogenic dyslipidemia. J. Lipid Res. 46: 2202–2213. [DOI] [PubMed] [Google Scholar]
- 29.Dastani Z., Quiogue L., Plaisier C., Engert J. C., Marcil M., Genest J., Pajukanta P. 2006. Evidence for a gene influencing high-density lipoprotein cholesterol on chromosome 4q31.21. Arterioscler. Thromb. Vasc. Biol. 26: 392–397. [DOI] [PubMed] [Google Scholar]
- 30.Almasy L., Hixson J. E., Rainwater D. L., Cole S., Williams J. T., Mahaney M. C., VandeBerg J. L., Stern M. P., MacCluer J. W., Blangero J. 1999. Human pedigree-based quantitative-trait-locus mapping: localization of two genes influencing HDL-cholesterol metabolism. Am. J. Hum. Genet. 64: 1686–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peacock J. M., Arnett D. K., Atwood L. D., Myers R. H., Coon H., Rich S. S., Province M. A., Heiss G. 2001. Genome scan for quantitative trait loci linked to high-density lipoprotein cholesterol: The NHLBI Family Heart Study. Arterioscler. Thromb. Vasc. Biol. 21: 1823–1828. [DOI] [PubMed] [Google Scholar]
- 32.Elbein S. C., Hasstedt S. J. 2002. Quantitative trait linkage analysis of lipid-related traits in familial type 2 diabetes: evidence for linkage of triglyceride levels to chromosome 19q. Diabetes. 51: 528–535. [DOI] [PubMed] [Google Scholar]
- 33.Soro A., Pajukanta P., Lilja H. E., Ylitalo K., Hiekkalinna T., Perola M., Cantor R. M., Viikari J. S., Taskinen M. R., Peltonen L. 2002. Genome scans provide evidence for low-HDL-C loci on chromosomes 8q23, 16q24.1–24.2, and 20q13.11 in Finnish families. Am. J. Hum. Genet. 70: 1333–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gagnon F., Jarvik G. P., Badzioch M. D., Motulsky A. G., Brunzell J. D., Wijsman E. M. 2005. Genome scan for quantitative trait loci influencing HDL levels: evidence for multilocus inheritance in familial combined hyperlipidemia. Hum. Genet. 117: 494–505. [DOI] [PubMed] [Google Scholar]
- 35.Al-Kateb H., Bahring S., Hoffmann K., Strauch K., Busjahn A., Nurnberg G., Jouma M., Bautz E. K., Dresel H. A., Luft F. C. 2002. Mutation in the ARH gene and a chromosome 13q locus influence cholesterol levels in a new form of digenic-recessive familial hypercholesterolemia. Circ. Res. 90: 951–958. [DOI] [PubMed] [Google Scholar]
- 36.Kocher O., Yesilaltay A., Cirovic C., Pal R., Rigotti A., Krieger M. 2003. Targeted disruption of the PDZK1 gene in mice causes tissue-specific depletion of the high density lipoprotein receptor scavenger receptor class B type I and altered lipoprotein metabolism. J. Biol. Chem. 278: 52820–52825. [DOI] [PubMed] [Google Scholar]
- 37.Lusis A. J., Fogelman A. M., Fonarow G. C. 2004. Genetic basis of atherosclerosis: part I: new genes and pathways. Circulation. 110: 1868–1873. [DOI] [PubMed] [Google Scholar]
- 38.Lusis A. J., Fogelman A. M., Fonarow G. C. 2004. Genetic basis of atherosclerosis: part II: clinical implications. Circulation. 110: 2066–2071. [DOI] [PubMed] [Google Scholar]
- 39.Turner S. T., Peyser P. A., Kardia S. L., Bielak L. F., Sheedy P. F., III, Boerwinkle E., de Andrade A. M. 2006. Genomic loci with pleiotropic effects on coronary artery calcification. Atherosclerosis. 185: 340–346. [DOI] [PubMed] [Google Scholar]
- 40.Alpy F., Tomasetto C. 2005. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 118: 2791–2801. [DOI] [PubMed] [Google Scholar]
- 41.Qi L., Shen H., Larson I., Schaefer E. J., Greenberg A. S., Tregouet D. A., Corella D., Ordovas J. M. 2004. Gender-specific association of a perilipin gene haplotype with obesity risk in a white population. Obes. Res. 12: 1758–1765. [DOI] [PubMed] [Google Scholar]
- 42.Hall N. G., Klenotic P., Anand-Apte B., Apte S. S. 2003. ADAMTSL-3/punctin-2, a novel glycoprotein in extracellular matrix related to the ADAMTS family of metalloproteases. Matrix Biol. 22: 501–510. [DOI] [PubMed] [Google Scholar]
- 43.Vartanian V., Lowell B., Minko I. G., Wood T. G., Ceci J. D., George S., Ballinger S. W., Corless C. L., McCullough A. K., Lloyd R. S. 2006. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl. Acad. Sci. USA. 103: 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arya R., Lehman D., Hunt K. J., Schneider J., Almasy L., Blangero J., Stern M. P., Duggirala R. 2003. Evidence for bivariate linkage of obesity and HDL-C levels in the Framingham Heart Study. BMC Genet. 4(Suppl. 1): S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Q., Lai C. Q., Parnell L., Cupples L. A., Adiconis X., Zhu Y., Wilson P. W., Housman D. E., Shearman A. M., D'Agostino R. B., et al. 2005. Genome-wide linkage analyses and candidate gene fine mapping for HDL3 cholesterol: the Framingham Study. J. Lipid Res. 46: 1416–1425. [DOI] [PubMed] [Google Scholar]
- 46.Adeyemo A. A., Johnson T., Acheampong J., Oli J., Okafor G., Amoah A., Owusu S., Agyenim-Boateng K., Eghan B. A., Jr., Abbiyesuku F., et al. 2005. A genome wide quantitative trait linkage analysis for serum lipids in type 2 diabetes in an African population. Atherosclerosis. 181: 389–397. [DOI] [PubMed] [Google Scholar]
- 47.Arya R., Duggirala R., Almasy L., Rainwater D. L., Mahaney M. C., Cole S., Dyer T. D., Williams K., Leach R. J., Hixson J. E., et al. 2002. Linkage of high-density lipoprotein-cholesterol concentrations to a locus on chromosome 9p in Mexican Americans. Nat. Genet. 30: 102–105. [DOI] [PubMed] [Google Scholar]
- 48.Kort E. N., Ballinger D. G., Ding W., Hunt S. C., Bowen B. R., Abkevich V., Bulka K., Campbell B., Capener C., Gutin A., et al. 2000. Evidence of linkage of familial hypoalphalipoproteinemia to a novel locus on chromosome 11q23. Am. J. Hum. Genet. 66: 1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bosse Y., Chagnon Y. C., Despres J. P., Rice T., Rao D. C., Bouchard C., Perusse L., Vohl M. C. 2004. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Quebec Family Study. J. Lipid Res. 45: 419–426. [DOI] [PubMed] [Google Scholar]
- 50.Mahaney M. C., Almasy L., Rainwater D. L., VandeBerg J. L., Cole S. A., Hixson J. E., Blangero J., MacCluer J. W. 2003. A quantitative trait locus on chromosome 16q influences variation in plasma HDL-C levels in Mexican Americans. Arterioscler. Thromb. Vasc. Biol. 23: 339–345. [DOI] [PubMed] [Google Scholar]
- 51.Pajukanta P., Allayee H., Krass K. L., Kuraishy A., Soro A., Lilja H. E., Mar R., Taskinen M. R., Nuotio I., Laakso M., et al. 2003. Combined analysis of genome scans of dutch and finnish families reveals a susceptibility locus for high-density lipoprotein cholesterol on chromosome 16q. Am. J. Hum. Genet. 72: 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]