Abstract
We investigated the role of proprotein convertase subtilisin/kexin type 9 (PCSK9) in the resistance of dyslipidemic hamsters to statin-induced LDL-cholesterol (LDL-C) reduction and the molecular mechanism by which statins modulated PCSK9 gene expression in vivo. We utilized the fructose diet-induced dyslipidemic hamsters as an in vivo model and rosuvastatin to examine its effects on liver PCSK9 and LDL receptor (LDLR) expression and serum lipid levels. We showed that rosuvastatin induced PCSK9 mRNA to a greater extent than LDLR mRNA in the hamster liver. The net result was that hepatic LDLR protein level was reduced. This correlated closely with an increase in serum LDL-C with statin treatment. More importantly, we demonstrated that in addition to an increase in sterol response element binding protein 2 (SREBP2) expression, rosuvastatin treatment increased the liver expression of hepatocyte nuclear factor 1 α (HNF1α), the newly identified key transactivator for PCSK9 gene expression. Our study suggests that the inducing effect of rosuvastatin on HNF1α is likely a underlying mechanism accounting for the higher induction of PCSK9 than LDLR because of the utilization of two transactivators (HNF1α and SREBP2) in PCSK9 transcription versus one (SREBP2) in LDLR transcription. Thus, the net balance is in favor of PCSK9-induced degradation of LDLR in the hamster liver, abrogating the effect of rosuvastatin on LDL-C lowering.
Keywords: LDL receptor, rosuvastatin, berberine
The number of LDL receptors (LDLR) expressed on the surface of hepatocytes is a primary determinant for plasma LDL-cholesterol (LDL-C) levels (1). Hepatic LDLR mediates the uptake of LDL particles from the circulation and delivers the receptor-bound LDL to the endosomal system for degradation while the LDLR returns to the cell surface. Statins are competitive inhibitors of HMG-CoA reductase, the rate-limiting enzyme in the cellular cholesterol biosynthetic pathway. The inhibition of cholesterol de novo synthesis leads to increased numbers of cell surface LDLR by activation of LDLR gene transcription. Thus, statins are the most widely prescribed drugs to treat hypercholesterolemia and combined hyperlipidemia.
Contrary to human studies in which statins effectively lower plasma total cholesterol (TC) and LDL-C, in animal studies, the LDL reducing effects of statins exhibit great species differences. Statins reduce plasma TC and LDL-C as the major drug action in rabbits, miniature pigs, dogs, monkeys, and guinea pigs (2–4) but not in rodents such as rats and hamsters (5–7). In rats and hamsters fed either a normal chow or an atherogenic diet, statins only effectively lower plasma triglyceride (TG) but not plasma TC or LDL-C. For example, in dyslipidemic hamsters fed a high-fructose diet, rosuvastatin treatment for 10 days at a daily dose of 10 mg/kg lowered plasma TG from 2 mM to 0.95 mM, a 52% reduction, but the plasma TC was unchanged in these animals (8). In another study of aged male rats fed a normal rodent diet, atorvastatin treatment of 21 days at a therapeutic dose of 10 mg/kg reduced plasma TG by 36% compared with the control group; again, the plasma TC was not reduced by atorvastatin treatment (9). Although it has been well established that the inhibition of hepatic TG secretion is responsible for the TG lowering effect of statins in rodents (5, 10), the mechanisms underlying the high resistance to statin-induced cholesterol reduction observed in these animal species are not fully understood.
Recent studies have identified proprotein convertase subtilisin/kexin type 9 (PCSK9) as a critical new player in LDL metabolism (11–13). PCSK9, a member of the subtilisin family of serine proteases, is highly expressed in adult liver hepatocytes and in small intestinal enterocytes (14). It is synthesized as a 72 kDa zymogen that undergoes autocatalytic cleavage in the endoplasmic reticulum into a heterodimer of a prosegment (122 amino acids) and a 60 kDa active form, bound together noncovalently. The processed PCSK9 is rapidly and efficiently secreted from liver-derived cells in culture and is abundant in human plasma where it modulates LDL-C levels by downregulation of hepatic LDLR. PCSK9 binds to the EGF-A extracellular domain of LDLR and the PCSK9:LDLR protein complex is endocytosed and degraded in the lysosome compartment (15, 16). Thus, PCSK9 plasma levels directly influence the level of circulating LDL-C (17, 18).
Previous studies have demonstrated that PCSK9 gene transcription is under the control of sterol response element binding proteins (SREBPs) (19, 20). Statin treatment depletes the intracellular cholesterol pool, which mobilizes the intracellular proteolytic processing machinery to release ER-bound SREBP2. The processed mature form of SREBP2 enters the nucleus where it binds to the SRE-1 element of the PCSK9 promoter and activates transcription. Because of the coexistence of an SRE-1 motif in the LDLR and PCSK9 promoters, statin treatment leads to increased transcription of both LDLR and its natural inhibitor, PCSK9 (21, 22), the protein product of which in turn tends to reduce LDLR protein levels (23, 24). This intrinsic regulatory loop has been recognized as a potentially undesirable limitation to statin therapeutic efficacy in further lowering plasma LDL-C (13, 25).
Recently, we have identified a highly conserved hepatocyte nuclear factor 1 (HNF1) binding site residing 28 bp upstream from SRE-1 as a critical sequence motif for PCSK9 transcription (26). Inactivation of this regulatory binding sequence either by nucleotide mutation or alternatively by depletion of its cognate binding protein HNF1α in liver-derived HepG2 cells severely reduced the expression of PCSK9 mRNA and protein to almost undetectable levels, thus confirming the importance of this motif and transcription factor HNF1α to the gene expression of PCSK9. Furthermore, we demonstrated that berberine (BBR), a natural hypocholesterolemic compound, inhibits PCSK9 transcription through the HNF1 motif and SRE-1 site. BBR treatment lowered the cellular abundance of HNF1α protein and the active form of SREBP2 in HepG2 cells, which led to a strong inhibition of PCSK9 expression. We further demonstrated that BBR can counteract the PCSK9 transcription stimulating effects of lovastatin, simvastatin, and fluvastatin in HepG2 cells.
In light of these new findings in PCSK9-mediated LDL-C metabolism and its regulation by statins and BBR, we decided to investigate whether PCSK9 plays a critical role in the resistance of dyslipidemic hamsters to statin treatment in LDL-C reduction and if BBR could enhance statin efficacy by mitigating the PCSK9-mediated LDLR degradation under in vivo conditions. In the present study, we utilized the high fructose diet-induced dyslipidemic hamsters as our in vivo model to examine the effects of rosuvastatin on liver PCSK9 and LDLR expression and serum lipid levels. We also examined the effects of BBR alone and in combination with rosuvastatin in these biological events. Our studies yielded compelling results that rosuvastatin induced a dose-dependent elevation of PCSK9 mRNA to an extent that far exceeded the increase in LDLR mRNA expression in the hamster liver. As the net result, hepatic LDLR protein expression was not increased but rather reduced. This correlated closely with an increase in serum LDL-C levels by the statin treatment. More importantly, we demonstrated, for the first time, that statin treatment not only increased the expression of SREBP2 but also stimulated the mRNA and protein expression of hepatic HNF1α, another key activator for PCSK9 transcription. Altogether, these new findings provide a molecular mechanism accounting for, at least in part, the high resistance of hamsters and perhaps rats to statin treatment with regard to LDL-C reduction. This mechanism might also have implications for the clinical findings of relatively reduced efficacy of statins at higher doses in human studies.
MATERIALS AND METHODS
Animal diet and drug treatment
Male Syrian Golden hamsters with body weights of 100–120 g were purchased from Harlan Sprague Dawley. Rosuvastatin was obtained from AstraZeneca, UK and BBR chloride was purchased from Sigma. Hamsters were housed (3 animals/cage) under controlled temperature (72°F) and lighting (12 h light/dark cycle). Animals had free access to autoclaved water and food. After an acclimatization period of 7 days, hamsters were switched to a high-fructose diet (60% fructose; Dyets, Inc., Bethlehem, PA) to induce dyslipidemia (7, 8, 10). After 21 days, while continuously on the fructose diet, hamsters were randomly divided into 5 groups (n = 9 per group) and were given a daily dose of rosuvastatin at 10 mg/kg, rosuvastatin 20 mg/kg, BBR 100 mg/kg, or rosuvastatin 10 mg/kg plus BBR 100 mg/kg once a day by oral gavage. The control group received equal volume of vehicle (0.5 ml of 10% 2-hydroxypropyl-β-cyclodextrin in autoclaved water) by oral gavage. The drug treatment lasted 7 days. Twenty-four hours after the last drug treatment, all animals were euthanized. At the time of dissection, body weight, liver weight, and the gross morphology of the liver were recorded. Livers were immediately removed, cut into small pieces, and stored at –80°C for RNA isolation and protein isolation. Animal use and experimental procedures were approved by the Institutional Animal Care and Use Committee of the VA Palo Alto Health Care System.
Serum isolation and cholesterol determination
Blood samples (0.2 ml) were collected from the retro-orbital plexus using heparinized capillary tubes under anesthesia (2–3% isoflurane and 1–2 L/min oxygen) after a 16 h fast (5 PM to 9 AM) before and after the drug treatments. Serum was isolated at room temperature and stored at −80°C. Standard enzymatic methods were used to determine TC, TG, LDL-C, and HDL-C with commercially available kits purchased from Stanbio Laboratory (Texas). Insulin level was determined by a commercial kit obtained from LINCO Research (cat. no. EZRMI-13K). Each sample was assayed in duplicate.
HPLC analysis of lipoprotein profiles
A total of 20 μl of each serum sample from the same treatment group (n = 9 per group) was pooled. Cholesterol and TG levels of each of the major lipoprotein classes, including chylomicron, VLDL, LDL, and HDL in the pool sera, were analyzed with a dual detection HPLC system consisting of two tandem connected TSKgel Lipopropak XL columns (300 × 7.8-mm; Tosoh, Japan) at Skylight Biotech, Inc. (Tokyo, Japan) (27). Cholesterol and TG contents in each fraction were determined.
RNA isolation and real-time RT-PCR
Total RNA was isolated from flash-frozen hamster tissues using an RNeasy kit (Qiagen, CA) and from HepG2 cells. RNA integrity was confirmed by agarose gel electrophoresis and ethidium bromide staining. Two micrograms of total RNA was reverse-transcribed with a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) using random primers according to the manufacturer's instructions. Real-time PCR was performed on the ABI PRISM® 7900HT Sequence Detection System (Foster City, CA) with SYBR PCR master mix (Applied Biosystems). Each cDNA sample was run in duplicate. Primer sequences used in real-time PCR were:
Human LDLR: forward, 5′-GACGTGGCGTGAACATCTG-3′; reverse, 5′-CTGGCAGGCAATGCTTTGG-3′; human PCSK9: forward, 5′-AGGGGAGGACATCATTGGTG-3′; reverse, 5-CAGGTTGGGGGTCAGTACC-3; human HNF1α: forward, 5′-GTGGCGAAGATGGTCAAGTCC-3′, reverse, 5′-CCCTTGTTGAGGTGTTGGG-3′; human SREBP2: forward, 5′-CCCTGGGAGACATCGACGA-3′, reverse, 5′-CGTTGCACTGAAGGGTCCA-3′; human GAPDH: forward, 5′-ATGGGGAAGGTGAAGGTCG-3′, reverse, 5′-GGGGTCATTGATGGCAACAATA-3′; hamster LDLR: forward, 5′-CTCCACTCTATCTCCAGCATTG-3′, reverse, 5′-TTTCAGCCACCAAATTAACATC-3′; Hamster SREBP1: forward, 5′-GCACTTTTTGACACGTTTCTTC-3′, reverse: 5′-CTGTACAGGCTCTCCTGTGG-3′; hamster SREBP2 forward, 5′-GAGAGCTGTGAATTTTCCAGTG-3′, reverse: 5′-CTACAGATGATATCCGGACCAA-3′; hamster GAPDH: forward, 5′-ACCCAGAAGACTGTGGATGG-3′, reverse, 5′-CGACATGTGAGATCCACGAC-3′.
Because gene sequences for hamster PCSK9 and HNF1α are not available yet, we employed the following strategy to obtain real-time RT-PCR primers to measure hamster PCSK9 and HNF1α mRNA levels. For hamster HNF1α gene, we first used available human, mouse, and rat HNF1α cDNA sequences to design multiple sets of primers with the assistance of an online version of primer 3 software (http://frodo.wi.mit.edu/primer3/). PCR was carried out with each of the primer sets and a hamster cDNA pool. The amplified products were separated on agarose gel and the primer pair resulting in a single PCR product of the expected size (204 bp) was identified. This procedure was followed by another round of new primer selection to amplify a segment of hamster HNF1α gene that encompasses the region amplified by the first pair of correct primers. The resulting larger PCR product of 552 bp was gel purified and sequenced to unveil the precise sequence of the first primer pair for hamster HNF1α gene which was subsequently used in the real-time PCR reaction. A similar approach was used to obtain real-time PCR primers for hamster PCSK9 (real-time PCR product size: 216 bp; sequencing PCR products size: 249 bp and 360 bp).
The primer sequences used in real-time PCR were: hamster PCSK9: forward, 5′- ATCCTCACAGGCCTGGAGTT-3′, reverse, 5′-CTGTGATGACCTCTGGAGCA-3′; hamster HNF1α: forward, 5′-ATTTGCAGCAGCACAATATC-3′, reverse 5′-GTGGGCTCTTCGATCAGTCC-3′.
Western blot analysis of LDLR in liver tissues and in HepG2 cells
Approximately 90–100 mg of hamster's frozen liver tissue from each animal was homogenized in 1 ml RIPA buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, pH 7.4) containing 1 mM PMSF and protease inhibitor cocktail (Roche). After protein quantitation using BCATM protein assay reagent (PIERCE), an equal amount of homogenate proteins from three liver samples of the same treatment group was pooled together and a total of 15 pooled samples from all five treatment groups were resolved by SDS-PAGE and LDLR protein was detected by immunoblotting using a rabbit anti-LDLR antibody obtained from Biovision (Mountain View, CA). The membranes were reprobed with an anti-γ-tubulin antibody (Santa Cruz Biotechnology). Immunoreactive bands of predicted molecular mass were visualized using an ECL plus kit (GE Healthcare Life Sciences, Piscataway, NJ) and quantified with the KODAK Molecular Imaging Software (Kodak, New Haven, CT). The amount of LDLR protein in individual hamster livers was also determined by the above procedure and normalized with tubulin signals. Utilizing the same procedure, HNF1α expression in liver tissues of hamsters was detected by anti-HNF1α antibodies (sc-6547) obtained from Santa Cruz Biotechnology, Inc. Rabbit anti-PCSK9 antibody and anti-SREBP2 antibody were previously described (20). Monoclonal anti-β-actin antibody (Clone AC-15, Sigma) was used to probe β-actin in HepG2 cells as a normalization control for protein loading.
Hamster liver nuclear extract preparation
Individual hamster liver nuclear extracts from control and rosuvastatin 20 mg/kg-treated groups were prepared as described (28). Briefly, ∼100 mg frozen hamster liver tissue were dounce-homogenized 15 times in buffer A [10 mM KCl, 1.5 mM MgCl2, 10 mM HEPES, pH 7.9, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail (Roche), and phosphatase inhibitor cocktail (Sigma)]. After centrifugation at 2,000 g for 10 min at 4°C, the pellet was resuspended in the same buffer and incubated on ice for 10 min, followed by dounce-homogenization of 10 times and centrifugation at 2,000 g for 10 min at 4°C. The nuclei-containing pellet was resuspended in buffer B (420 mM NaCl, 10 mM KCl, 20 mM HEPES, pH 7.9, 20% glycerol, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail, and phosphatase inhibitor cocktail) and extracted for 30 min at 4°C on a shaking rotor. After centrifugation at 16,000 g for 15 min at 4°C, the supernatant was collected and stored in −80°C. The protein concentration was determined using BCA protein assay kit (Thermo Scientific).
Electrophoretic mobility shift assays
Wild-type and mutated oligonucleotide probes were annealed and end-labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP. Each binding reaction comprised 10 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM DTT, 50 mM KCl, 2.5% glycerol, 1 μg of poly (dI-dC), 0.05% NP-40, and 10 μg of liver nuclear extracts in a final volume of 20 μl. The nuclear extracts were incubated with 0.4–0.5 ng of 32P-labeled probe (1× 105 cpm) for 10 min at room temperature. The reaction mixtures were loaded onto a 5% polyacrylamide gel and run in 0.5× TBE buffer at 30 mA for 2 h at 4°C. Gels were dried and visualized on a PhosphoImager. The sense sequences of electrophoretic mobility shift assay probes were as follows: HNF1-wt: 5′- AGTCCGGGGGTTCCGTTAATGTTTAAT CAGATAGGATC-3′; HNF1-mu: 5′-GTCCGGGGGTTCCGTTcgTGTTgccTCAGATAGGATC-3′. The HNF1 binding site is underlined.
Identification of HNF1 binding site of hamster PCSK9 promoter
To identify the putative HNF1 binding site on hamster PCSK9 promoter, we first compared the sequences encompassing the HNF1 site from human, mouse, and rat PCSK9 promoters and selected regions that are highly homologous among these species for primer design. Using primers identified by such approach, a DNA fragment (213 bp) was amplified from hamster genomic DNA purified from normal hamster livers using a DNeasy blood and tissue kit (Qiagen, CA) and sequenced. Primers used were: TGGGGAGGGCGAGGCCGAAA (forward); GGCTCAGTCCTCTAGCCTCA (reverse).
Statistical analysis
Significant differences between control and treatment groups were assessed by one-way ANOVA and two-way ANOVA with a Bonferroni post test and Student's t-test. A P-value of <0.05 was considered statistically significant.
RESULTS
Resistance of dyslipidemic hamsters to rosuvastatin-induced LDL-cholesterol reduction
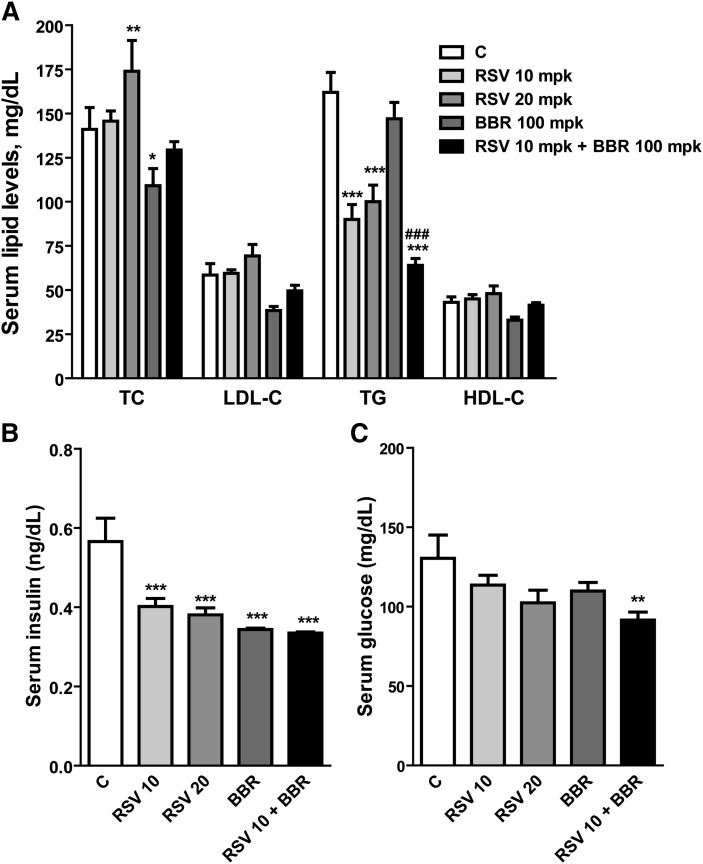
Forty-five hamsters were fed a fructose-enriched diet for 3 weeks, which increased plasma TC by 40% from 102 to 140 mg/dl (P < 0.05) and plasma TG by 60% from 103 to 162 mg/dl (P < 0.01) compared with hamsters fed a normal diet. After this initial feeding period, while continuously on the fructose diet, hamsters were randomly divided into control and four treatment groups of nine animals per group. One group was treated with rosuvastatin at a daily dose of 10 mg/kg; the second group was treated with 20 mg/kg of rosuvastatin, the third group was treated with BBR at a daily dose of 100 mg/kg, and the last group was treated with the combination of 10 mg/kg of rosuvastatin and 100 mg/kg of BBR. The control group received an equal amount of vehicle. Figure 1A shows the plasma lipid levels of different groups after 7 days of drug treatment. Plasma TC was unchanged by rosuvastatin at 10 mg/kg and was increased from 141 mg/dl to 174 mg/dl by rosuvastatin at the dose of 20 mg/kg (23% increase, P < 0.01) compared with vehicle control. BBR alone reduced TC by 23% (P < 0.05). In the presence of rosuvastatin, the TC lowering effect of BBR was not significant.
Fig. 1.
Differential modulation of lipid parameters by rosuvastatin and BBR. Hamsters on the fructose diet were administered vehicle (C), 10 mg/kg of rosuvastatin, 20 mg/kg of rosuvastatin, 100 mg/kg of BBR, and the combined treatment of rosuvastatin 10 mg/kg plus BBR at 100 mg/kg for 7 days. The serum samples were collected 24 h after the last treatment. TC, TG, LDL-C, HDL-C, and insulin were measured using commercial kits. Serum glucose was measured by a clinical chemistry analyzer (Olympus AU5431). Values are mean ± SEM of nine animals. *P < 0.05, ** P < 0.01, and *** P < 0.001 compared with the vehicle control group; ### P < 0.001 compared with rosuvastatin 10 mg/kg group (two-way ANOVA). mpk, mg/kg.
The change in plasma LDL-C levels mirrored the TC change. Rosuvastatin at 10 mg/kg did not lower LDL-C and at 20 mg/kg rosuvastatin caused an 18% increase in LDL-C despite not reaching a statistical significance. BBR treatment reduced LDL-C by 34.5%. The combination of BBR and rosuvastatin resulted in a modest reduction of LDL-C compared with each drug alone.
In contrast to TC and LDL-C, plasma TG was strongly reduced by rosuvastatin at both doses by 45% and 38%, respectively. Interestingly, BBR alone did not lower the TG, but it enhanced the TG lowering effect of rosuvastatin, reaching a 60.5% reduction (P < 0.01 compared with statin alone). The plasma level of HDL-C was not changed by rosuvastatin but was reduced by BBR to 25% of vehicle control.
In addition to the TG lowering effect, rosuvastatin treatment at both doses lowered the plasma level of insulin by 29% and 32%, respectively (Fig. 1B). BBR alone lowered insulin by 40%. While the combination of BBR with statin did not further reduce insulin level, it produced a statistically significant effect in decreasing fasting glucose (Fig. 1C).
We further performed HPLC analysis of lipoprotein-cholesterol and TG profiles in pooled serum of untreated hamsters and hamsters treated with different regimens. The results showed a prominent increase in LDL-associated cholesterol by rosuvastatin treatment in a dose-dependent manner (Fig. 2, upper panel). The HPLC analysis also demonstrated that the TG lowering effect of rosuvastatin was confined to VLDL and chylomicron fractions of the lipoproteins (Fig. 2, lower panel), which was consistent with previous studies (7). All together, these results demonstrated that the efficacy of rosuvastatin in hamsters fed the high-fructose diet is limited to TG and insulin reduction but not TC and LDL-C reductions, which were in line with previous reports (7, 8).
Fig. 2.
Demonstration of a dose-dependent increase in LDL-C by rosuvastatin treatment by HPLC analysis of plasma lipoprotein cholesterol profiles. Serum from the vehicle and drug-treated groups were pooled, and the pooled sera were subjected to HPLC analysis of lipoprotein profiles associated with TC (upper panel) and TG (lower panel).
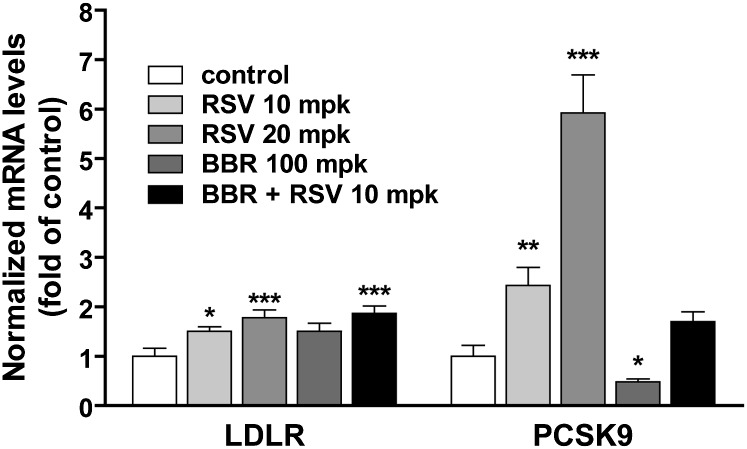
Correlation of highly elevated PCSK9 mRNA levels with reduced LDLR protein abundance in livers of rosuvastatin-treated hamsters
To seek a better understanding of the confounding LDL-C modulating activity of rosuvastatin in these animals at the molecular level, hamsters from control and treated groups were euthanized at the end of treatment and levels of liver LDLR and PCSK9 mRNA were individually assessed by quantitative real-time RT-PCR using hamster-specific primers. As shown in Fig. 3, the expression of hepatic LDLR mRNA was increased 1.5-fold (P < 0.05) by rosuvastatin at the dose of 10 mg/kg and 3.7-fold (P < 0.001) at the dose of 20 mg/kg. However, the increase in PCSK9 mRNA levels far exceeded those of LDLR with 2.4-fold (P < 0.01) and 5.9-fold (P < 0.001) by the rosuvastatin treatment at these doses, respectively. Consistent with the in vitro effect, BBR treatment led to a 1.5-fold increase in the amount of LDLR mRNA and a 50% decrease in PCSK9 mRNA. The inducing effect of rosuvastatin on PCSK9 mRNA expression was mitigated by BBR concomitant treatment to a level not significantly above the control. In contrast to PCSK9 mRNA, the increase in LDLR mRNA by rosuvastatin and BBR was additive.
Fig. 3.
Quantitative real-time PCR analysis of the liver mRNA expression of PCSK9 and LDLR. Twenty-four hours after the last treatment, all animals were euthanized and liver total RNA was isolated. Individual levels of LDLR mRNA or PCSK9 mRNA were assessed by quantitative real-time PCR using hamster-specific PCR primers as described in the Methods section. After normalization with GAPDH mRNA levels, the relative PCSK9 or LDLR mRNA levels are presented, and the results are means ± SEM of eight to nine animals per group. *P < 0.05, ** P < 0.01, and *** P < 0.001 compared with the vehicle control group.
The stronger induction of PCSK9 mRNA expression by rosuvastatin versus that of LDLR could lead to a reduced LDLR protein level due to the protein degradation mediated by PCSK9. Therefore, we examined LDLR protein abundance in individual liver samples as well as in pooled liver protein extracts of different treatment groups by Western blot analysis using an antibody recognizing the mature form of hamster LDLR. The results shown in Fig. 4A are a representative Western blot in which each sample consisted of equal amounts of pooled protein extracts from three livers of the same treatment group. Quantitative analyses of the results of Western blot of LDLR protein using individual liver samples are presented in Fig. 4B. Both analyses show that the amount of LDLR protein in the liver was reduced by rosuvastatin dose dependently by 21% and 42% (P < 0.05) compared with the vehicle control. In contrast to rosuvastatin, LDLR protein level was increased 59% by BBR treatment (P < 0.001). The tendency to reduce LDLR protein by rosuvastatin treatment was not apparent when BBR was coadministered and the LDLR protein level remained at the level of untreated vehicle control, thus indicating a weak counteraction of BBR on the statin effect in vivo.
Fig. 4.
Western blot analysis of liver LDLR protein abundance. Individual liver protein extracts were prepared and protein concentrations were determined. A: Equal amount of homogenate proteins from three liver samples of the same treatment group were pooled together and a total of 15 pooled samples from all five treatment groups were resolved by SDS-PAGE and LDLR protein was detected by immunoblotting using a rabbit anti-LDLR antibody. The membrane was reprobed with an anti-γ-tubulin antibody. B: Individual liver samples were analyzed by Western blotting and the expression levels of LDLR were quantified with the KODAK Molecular Imaging Software with normalization by γ-tubulin. Values are mean ± SEM of nine samples per group. P < 0.05 and *** P < 0.001 compared with the vehicle control group.
Induction of HNF1α expression by rosuvastatin in vivo
Our recent studies in liver-derived HepG2 cells have demonstrated that HNF1α has an important trans-activating role in PCSK9 gene transcription and works cooperatively with SREBP2 to stimulate PCSK9 promoter activity (26). To determine whether rosuvastatin treatment could alter HNF1α gene expression, we performed real-time PCR to determine the mRNA levels of HNF1α along with SREBP2 and SREBP1 in liver samples of different treatment groups. The results showed that the amount of HNF1α mRNA was increased 1.4-fold and 1.9-fold (P < 0.01) by rosuvastatin at 10 mg/kg and 20 mg/kg doses, respectively (Fig. 5A). As expected, rosuvastatin also increased the expression of SREBP2 mRNA, whereas the level of SREBP1 mRNA was not affected by the statin treatment.
Fig. 5.
Induction of HNF1α mRNA and protein expression in hamster livers by rosuvastatin. A: Individual levels of HNF1α, SREBP2, or SREBP1 mRNA were assessed by quantitative real-time PCR using hamster-specific PCR primers as described in the Methods section. Results are means ± SEM of nine animals per group. *P < 0.05, ** P < 0.01, and *** P < 0.001 compared with the vehicle control group. B: Six individual liver protein extracts from the control group and from the rosuvastatin 20 mg/kg-treated group were analyzed for HNF1α and SREBP2 protein expression by Western blotting. The target protein signals were normalized to the signal intensities of γ-tubulin individually. Values are means ± SEM. *P < 0.05 and *** P < 0.001 compared with the vehicle group (C). C: Sequence comparison of HNF1 and SRE-1 sites in proximal regions of PCSK9 promoter of human, mouse, rat, and hamster. Yellow-highlighted letters indicate HNF1 site, green-highlighted letters indicate SRE-1 site, and gray-highlighted letters indicate divergent nucleotides among the four promoter sequences. D: A double-stranded oligonucleotide (HNF1-wt) corresponding to human PCSK9 promoter region −400 to −362 was radiolabeled and incubated with 10 μg of hamster liver tissue nuclear extracts of untreated (lanes 1–6) and the rosuvastatin 20 mpk group (7–12) for 10 min at 22°C. In lane 13, the nuclear extract of lane 12 was incubated with 32P-labeled mutated probe (designated as HNF1-mu). In lane 14, the binding reaction mixture contained nuclear extracts of lane 12 with the labeled wt probe and 100-fold molar amounts of unlabeled wt probe as competitor. The reactions in lane 13 and lane 14 were designed to demonstrate the binding specificity.
These real-time PCR results are the first to show the inducing effect of statins on HNF1α expression in vivo. To confirm this important new finding, we performed Western blot analysis to detect HNF1α protein expression along with SREBP2 in liver samples of untreated and rosuvastatin 20 mg/kg-treated hamsters. Figure 5B shows that the amount of hepatic HNF1α protein was increased 1.6-fold by rosuvastatin treatment (P < 0.05). The protein abundance of SREBP2 was increased 2.3-fold by rosuvastatin treatment (P < 0.001).
To further demonstrate a functional role of HNF1α in rosuvastatin-induced upregulation of PCSK9 gene transcription, we isolated nuclear extracts from individual liver samples of untreated and rosuvastatin 20 mg/kg-treated hamsters and performed gel mobility shift assays using a 32P-labeled oligonucleotide probe (HNF1-wt) containing HNF1 motif of human PCSK9 promoter that is highly homologous to the HNF1 site of hamster PCSK9 promoter with only one nucleotide difference in the extreme 5′ end of the motif (Fig. 5C). Results in Fig. 5D showed that the signal intensity of the complex formed with 32P-labeled PCSK9-HNF1 wild-type probe was clearly higher in rosuvastatin-treated livers (lanes 7–12) as compared with untreated control samples (lanes 1–6). This DNA/protein complex was not detected in a binding reaction that contained 32P-labeled HNF1 mutated probe (lane 13) or in the reaction that contained a 100× excess amount of the wild-type unlabeled probe (lane 14). The quantification of the signal intensity of the complex indicated a 2-fold increase (P < 0.05) in the binding of HNF1α to the HNF1-PCSK9 sequence (Fig. 5D, lower panel). Altogether, these results suggest that the mechanism underlying statin-induced PCSK9 transcription in hamsters involves two trans-activators, HNF1α and SREBP2, which is different from the mechanism of statin-induced LDLR transcription involving only the SREBP pathway.
Rosuvastatin-mediated stimulation of PCSK9 expression in HepG2 cells occurring in the absence of HNF1α induction
To discern whether HNF1α induction by rosuvastatin also occurs in a human cellular model, we treated HepG2 cells for 24 h with a broad concentration range of rosuvastatin and isolated both RNA and total cell protein to examine the mRNA and protein expression of LDLR, PCSK9, SREBP2, and HNF1α. Figure 6A shows that rosuvastatin produced modest effects in upregulating LDLR and SREBP2 mRNA expression in HepG2 cells. Rosuvastatin increased PCSK9 mRNA up to 3-fold at the maximal dose (10 µM), which was slightly higher than that of LDLR. In contrast to the effect observed in hamsters, rosuvastatin did not induce HNF1α mRNA expression over this dose range. This finding was further corroborated by Western blot analysis that detected elevated LDLR, PCSK9, and SREBP2 protein expression without an appreciable increase in the cellular abundance of HNF1α. We obtained similar results in Huh7 cells, another human hepatoma-derived cell line (data not shown). The lack of inducing effect of rosuvastatin on HNF1α expression likely accounts for the modest induction of PCSK9 in HepG2 and Huh7cells. Our data thus provide one possible mechanistic explanation for the opposite effects of rosuvastatin on LDLR protein levels between human and hamster systems through the differential regulation of HNF1α.
Fig. 6.
Rosuvastatin induces PCSK9 expression without induction of HNF1α in HepG2 cells. HepG2 cells were seeded in 6-well culture plate for 24 h. Cells were cultured in medium containing 10% lipoprotein deficient serum overnight prior to the addition of rosuvastatin at the indicated doses for 24 h. The expression levels of mRNA of interest were assessed by real-time quantitative PCR after normalization with GAPDH. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with untreated control cells. The abundances of the gene products were analyzed by Western blot and quantified after normalization with β-actin as in Fig. 5. PCSK9-P and PCSK9-M represent the proprotein and cleaved mature form of PCSK9, respectively. SREBP2-P represents the precursor form (∼120 kDa) of SREBP2.
DISCUSSION
It has been well documented by several studies (5–7) that the efficacy of statins in rodent models such as rats and hamsters is measured by its TG reduction instead of its primary drug action of LDL-C reduction due to the observed resistance of these animals to statins. To gain a better insight into this complex cholesterol homeostatic regulation in these species, in this current study, we explored a possible link between this resistance to LDL reduction and elevated PCSK9 expression in rosuvastatin-treated hamsters fed a fructose-enriched diet. This effort led to the discovery of HNF1α being a new molecular mediator for the drug action of statins in modulating PCSK9 gene transcription in the dyslipidemic hamster model.
While plasma TG was efficaciously reduced in rosuvastatin-treated hamsters, we observed a modest and dose-dependent increase in LDL-C by rosuvastatin treatment in these animals. Reciprocally, the abundance of LDLR protein in the liver of rosuvastatin-treated hamsters was reduced in a similar dose-dependent fashion as demonstrated by Western blot analysis. Our examination of hepatic mRNA expression of LDLR and PCSK9 by real-time PCR analysis revealed that while rosuvastatin treatment increased LDLR gene expression 2–3-fold, its effect on PCSK9 transcription was stronger, leading to a 6-fold increase in PCSK9 mRNA level at a dose of 20 mg/kg. Although we were unable to confirm this induction at the protein level due to the unavailability of a specific antibody recognizing hamster PCSK9, it has been demonstrated that PCSK9 protein expression correlates with its mRNA abundance and is primarily regulated at the gene transcription level (20, 29). Thus, our data strongly suggest that the higher induction of PCSK9, which functionally mediates LDLR protein degradation, is responsible for the reduction of liver LDLR and the increased circulating LDL-C in this animal model.
The above-described results were quite different from human studies in which statins robustly lowered plasma LDL-C and raised an important question as to whether different mechanisms were involved in the regulation of PCSK9 transcription by statins in hamsters versus humans.
Previous studies have demonstrated that SREBP2 is one common mediator for LDLR and PCSK9 transcription through its binding sites embedded in the gene promoters (19, 20). Statin treatment increases SREBP2 expression and translocation to the nucleus where it activates LDLR and PCSK9 through direct interaction with the SRE-1 element. As expected, levels of mRNA and protein of SREBP2 in the hamster liver were increased by rosuvstatin treatment dose dependently. We did not detect a change in SREBP1 mRNA expression by statin treatment, which is consistent with the notion that in vivo SREBP2 is the primary activator for PCSK9 despite the observation that SREBP1c also activates PCSK9 gene transcription in cell cultures (29).
More importantly, our recent work has identified HNF1α as a pivotal transcription activator for the PCSK9 gene. HNF1α binds to a highly conserved HNF1 motif located 28 bp upstream of the SRE-1 site of the PCSK9 promoter. This binding is critical to the basal transcriptional activity as well as to the SREBP2-mediated transactivation of the PCSK9 promoter. The fact that rosuvastatin induced a higher level of PCSK9 mRNA than LDLR led us to examine the possible involvement of HNF1α in the statin drug action in hamsters. Indeed, HNF1α mRNA levels were increased 1.5- and 1.9-fold by rosuvastatin and this induction was corroborated at the protein level by Western blot. By conducting electrophoretic mobility shift assay with nuclear extracts prepared from livers of untreated and rosuvastatin 20 mg/kg-treated hamsters, we further demonstrated that the binding of HNF1α to the highly conserved HNF1 binding site of PCSK9 promoter was significantly increased by rosuvastatin treatment. To the best of our knowledge, this is the first report of modulation of HNF1α expression by statins.
The next important question is how this finding in hamsters is related to the action of statins in modulating PCSK9 transcription in humans. In an attempt to address this question, we examined the effects of rosuvastatin over a wide concentration range in human liver-derived HepG2 cells. Rosuvastatin induced a modest increase in LDLR up to ∼2-fold and PCSK9 up to ∼3-fold in mRNA expression and comparable increases in the corresponding protein expressions. However, we did not detect appreciable increases in HNF1α expression at both mRNA and protein levels. We repeated this study in another human liver cell line, Huh7, and identical results were obtained. These results suggest a species-specific regulation of HNF1α expression by statins.
Our previous studies and others have demonstrated that BBR, a plant-derived hypolipidemic compound, increases LDLR expression through dual mechanisms of enhancing LDLR mRNA stability (30–33) and blocking PCSK9 transcription (26, 34). HNF1α protein levels were reduced in BBR-treated HepG2 cells, which accounted for, in part, the inhibitory action of BBR on PCSK9 expression. When combined with statins, BBR counteracted the inducing effects of statins on PCSK9 expression to near baseline levels in HepG2 cells. Thus, we incorporated this antagonism into this in vivo study to examine the combined effects of rosuvastatin and BBR in regulating plasma lipid levels through their differential regulation on PCSK9. Our results confirmed the LDL-C lowering and LDLR upregulating effects of BBR in the absence of rosuvastatin. In the combined treatment, the undesirable effects of rosuvastatin on plasma LDL-C and LDLR expression were partially mitigated by the concomitant treatment with BBR. One possible mechanism accounting for the relative weak counteraction of BBR in hamsters as compared with its effects in HepG2 cells could be the lack of effect on HNF1α expression in vivo. We did not detect a significant decrease in HNF1α protein abundance in BBR-treated liver samples (data not shown). Combined with the difference observed in statin-mediated regulation of HNF1α between hamsters and HepG2 cells, the ineffectiveness of BBR in modulating HNF1α expression in hamster livers further implies that the regulatory mechanism governing HNF1α expression between humans and rodents could be different, which could be a contributing factor for different efficacies of statins and other lipid modulating drugs observed in various animal studies.
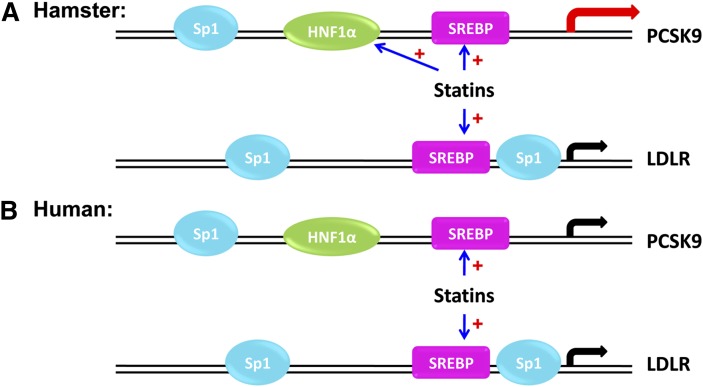
While the exact mechanisms underlying the differential effects of statins in regulating HNF1α expression in humans and hamsters await further investigations, results from the current study allow us to propose a model (Fig. 7) in which PCSK9 is more significantly upregulated by statins in hamsters where both SREBP2 and HNF1α are the driving force for gene expression in contrast to the use of only SREBP2 in LDLR induction. Thus, the net balance is in favor of PCSK9-induced degradation of LDLR over SREBP2 induction. The resulting net loss of LDLR leads to LDL-C accumulation in the plasma of hamsters, conferring the drug resistance phenomena. Interestingly, in human HepG2 or Huh7 cells, HNF1α is not induced by rosuvastatin and thus we see no net degradation of LDLR protein. This observation in HepG2 cells could have implications to statin efficacy in treating hypercholesterolemic patients. The question of whether HNF1α induction is involved in the elevation of PCSK9 plasma levels in patients treated with higher doses of statins needs to be thoroughly addressed with further investigations.
Fig. 7.
A model for statin-mediated differential regulations of PCSK9 and LDLR gene transcription in hamsters and humans. In hamsters, statin treatment increases the binding of HNF1α and SREBP2 to PCSK9 promoter to activate gene transcription, whereas only SREBP2 is induced by statin to activate LDLR gene transcription. In humans, statin treatment leads to the binding of SREBP2 to both PCSK9 and LDLR promoters and activate their gene transcription without an inducing effect on HNF1α.
Acknowledgments
We thank Dr. Michael R. Briggs for his critical review of the manuscript and other members of Liulab for their general support.
Footnotes
Abbreviations:
- BBR
- berberine
- HNF1
- hepatocyte nuclear factor 1
- LDL-C
- LDL-cholesterol
- LDLR
- LDL receptor
- PCSK9
- proprotein convertase subtilisin/kexin type 9
- SREBP
- sterol response element binding protein
- TC
- total cholesterol
- TG
- triglyceride
This study was supported by the Department of Veterans Affairs (Office of Research and Development, Medical Research Service, to J. Liu) and by grants (1RO1 AT002543-01A1 and 1R21AT003195-01A2 to J. Liu) from the National Center for Complementary and Alternative Medicine. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Steinberg D., Witztum J. L. 2009. Inhibition of PCSK9: a powerful weapon for achieving ideal LDL cholesterol levels. Proc. Natl. Acad. Sci. USA. 106: 9546–9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh K. M., Albassam M. A., Clarke D. E. 1996. Subchronic toxicity of atorvastatin, a hydroxymethylglutaryl-Coenzyme A reductase inhibitor, in beagle dogs. Toxicol. Pathol. 24: 468–476. [DOI] [PubMed] [Google Scholar]
- 3.Madsen C. S., Janovitz E., Zhang R., Nguyen-Tran V., Ryan C. S., Yin X., Monshizadegan H., Chang M., D'Arienzo C., Scheer S., et al. 2008. The guinea pig as a preclinical model for demonstrating the efficacy and safety of statins. J. Pharmacol. Exp. Ther. 324: 576–586. [DOI] [PubMed] [Google Scholar]
- 4.Ito B. R., Zhang B-H., Cable E. E., Song X., Fujitaki J. M., MacKenna D. A., Wilker C. E., Chi B., Poelje P. D., Linemeter D. L., et al. 2009. Thyroid hormone b receptor activation has additive cholesterol lowering activity in combination with atorvastatin in rabbits, dogs, and monkeys. B J Pharmacol. 156: 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krause B. R., Newton R. S. 1995. Lipid-lowering activity of atorvastatin and lovastatin in rodent species: triglyceride-lowering in rats correlates with efficacy in LDL animal models. Atherosclerosis. 117: 237–244. [DOI] [PubMed] [Google Scholar]
- 6.Roglans N., Verd J. C., Peris C., Alegret M., Vazquez M., Adzet T., Diaz C., Hernandez G., Laguna J. C., Sanchez R. M. 2002. High doses of atorvastatin and simuvastatin induce key enzymes involved in VLDL production. Lipids. 37: 445–454. [DOI] [PubMed] [Google Scholar]
- 7.Chong T., Naples M., Federico L., Taylor D., Smith G. J., Cheung R. C., Adeli K. 2006. Effect of rosuvastatin on hepatic production of apolipoprotein B-containing lipoproteins in an animal model of insulin resistance and metabolic dyslipidemia. Atherosclerosis. 185: 21–31. [DOI] [PubMed] [Google Scholar]
- 8.Naples M., Federico L., Xu E., Nelken J., Adeli K. 2008. Effects of rosuvastatin on insulin sensitivity in an animal model of insulin resistance: Evidence for statin-induced hepatic insulin sensitization. Atherosclerosis. 198: 94–103. [DOI] [PubMed] [Google Scholar]
- 9.Sanguino E., Roglans N., Alegret M., Sanchez R. M., Vazquez-Carrera M., Laguna J. C. 2005. Atorvastatin reverse age-related reduction in rat hepatic PPARa and HNF-4. B J Pharmacol. 145: 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meshkani R., Adeli K. 2009. Hepatic insulin resistance, metabolic syndrome and cardiovascular disease. Clin. Biochem. 42: 1331–1346. [DOI] [PubMed] [Google Scholar]
- 11.Horton J. D., Cohen J. C., Hobbs H. H. 2007. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem. Sci. 32: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson A. S., Fong L. G., Young S. G. 2008. PCSK9 function and physiology. J. Lipid Res. 49: 1595–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seidah N. G. 2009. PCSK9 as a therapeutic target of dyslipidemia. Expert Opin. Ther. Targets. 13: 19–28. [DOI] [PubMed] [Google Scholar]
- 14.Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Bélanger J., Stifani S., Basak A., Prat A., Chrétien M. 2003. The secretory proprotein convertase neutal apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA. 100: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. H. 2007. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282: 18602–18612. [DOI] [PubMed] [Google Scholar]
- 16.McNutt M. C., Kwon H. J., Chen C., Chen J. R., Horton J. D., Lagace T. A. 2009. Antagonism of secreted PCSK9 increases low density lipoprotein receptor expression in HepG2 cells. J. Biol. Chem. 284: 10561–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert G., Ancellin N., Charlton F., Comas D., Pilot J., Keech A., Patel S., Sullivan D. R., Cohn J. S., Rye K-A., et al. 2008. Plasma PCSK9 concentration correlates with LDL and total cholesterol in diabetic patients and are decreased by fenofibrate treatment. Clin. Chem. 54: 1038–1045. [DOI] [PubMed] [Google Scholar]
- 18.Grefhorst A., McNutt M. C., Lagace T. A., Horton J. D. 2008. Plasma PCSK9 preferentially reduces liver LDL receptors in mice. J. Lipid Res. 49: 1303–13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubuc G., Chamberland A., Wassef H., Davignon J., Seidah N. G., Bernier L., Prat A. 2004. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 24: 1454–1459. [DOI] [PubMed] [Google Scholar]
- 20.Jeong H. J., Lee H-S., Kim K-S., Kim Y-K., Yoon D., Park S. W. 2008. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatroy element binding protein-2. J. Lipid Res. 49: 399–409. [DOI] [PubMed] [Google Scholar]
- 21.Mayne J., Dewpura T., Raymond A., Cousines M., Chaplin A., Lahey K., Lahaye S. A., Mbikay M., Ooi T. C., Chrétien M. 2008. Plasma PCSK9 levels are significantly modified by statins and fibrates in humans. Lipids Health Dis. 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Careskey H. E., Davis R. A., Alborn W. E., Troutt J. S., Cao G., Konrad R. J. 2008. Atovastatin increases human serum levels of proprotein convertase subtilisin/kexin type 9. J. Lipid Res. 49: 394–398. [DOI] [PubMed] [Google Scholar]
- 23.Qian Y-W., Schmidt R. J., Zhang Y., Chu S., Lin A., Wang H., Wang X., Beyer T. P., Bensch W. R., Li W., et al. 2007. Secreted PCSK9 downregulates low density lipoprotein receptor through receptor-mediated endocytosis. J. Lipid Res. 48: 1488–1498. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt R. J., Beyer T. P., Bensch W. R., Qian Y-W., Lin A., Kowala M. 2008. Secreted proportein convertase subtilisin/kexin type 9 reduces both hepatic and extrahepatic low-density lipoprotein receptors in vivo. Biochem. Biophys. Res. Commun. 370: 634–640. [DOI] [PubMed] [Google Scholar]
- 25.Cao G., Qian Y-W., Kowala M. C., Konrad R. J. 2008. Further LDL cholesterol lowering through targeting PCSK9 for coronary artery disease. Endocr. Meatb. Immune Disord. Drug Targets. 8: 238–243. [DOI] [PubMed] [Google Scholar]
- 26.Li H., Dong B., Park S. W., Lee H-S., Chen W., Liu J. 2009. Hepatocyte nuclear factor 1α plays a critical role in PCSK9 gene transcription and regulation by the natural hypocholesterolemic compound berberine. J. Biol. Chem. 284: 28885–28895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Usui S., Hara Y., Hosaki S., Okazaki M. 2002. A new on-line dual enzymatic method for simutaneous quantification of cholesterol and triglycerides in lipiproteins by HPLC. J. Lipid Res. 43: 805–814. [PubMed] [Google Scholar]
- 28.Shih D. Q., Bussen M., Sehayek E., Ananthanaraynan M., Shneider B. L., Suchy F. J., Shefer S., Bollileni J. S., Gonalez F. J., Breslow J. L., et al. 2001. Hepatocyte nuclear factor-1α is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 27: 375–382. [DOI] [PubMed] [Google Scholar]
- 29.Costet P., Cariou B., Lambert G., Lalanne F., Lardeux B., Jarnoux A-L., Grefhorst A., Staels B., Krempf M. 2006. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatroy element-binding protein 1c. J. Biol. Chem. 281: 6211–6218. [DOI] [PubMed] [Google Scholar]
- 30.Kong W., Wei J., Abidi P., Lin M., Inaba S., Li C., Wang Y., Wang Z., Si S., Pan H., et al. 2004. Berberine is a promising novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat. Med. 10: 1344–1352. [DOI] [PubMed] [Google Scholar]
- 31.Abidi P., Jiang J., Liu J. 2005. ERK-dependent regulation of hepatic LDL receptor expression by herbal medicine berberine. Arterioscler. Thromb. Vasc. Biol. 25: 2170–2176. [DOI] [PubMed] [Google Scholar]
- 32.Abidi P., Chen W., Kraemer F. B., Li H., Liu J. 2006. Medicinal plant goldenseal is a natural LDL-lowering agent with multiple active components and new action mechanisms. J. Lipid Res. 47: 2134–2147. [DOI] [PubMed] [Google Scholar]
- 33.Li H., Chen W., Zhou Y., Abidi P., Sharpe O., Robinson W. H., Liu J. 2009. Identification of mRNA-binding proteins that regulate the stability of LDL receptor mRNA through AU-rich elements. J. Lipid Res. 50: 820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron J., Ranheim T., Kulseth M. A., Leren T. P., Berge K. E. 2008. Berberine decreases PCSK9 expression in HepG2 cells. Atherosclerosis. 201: 266–273. [DOI] [PubMed] [Google Scholar]