Abstract
We previously observed that treatment of mice with a dominant negative form of cJun (dn-cJun) increased the expression of genes involved in lipid metabolism and modulated the expression of nine microRNAs (miR). To investigate the potential effect of these miRs on the expression of the genes of lipid metabolism, we performed studies in cultured HepG2 cells. Transfection of HepG2 cells with sense or antisense miR-370 or miR-122 upregulated and downregulated, respectively, the transcription factor sterol-regulatory element binding protein 1c (SREBP-1c) and the enzymes diacylglycerol acyltransferase-2 (DGAT2), fatty acid synthase (FAS), and acyl-CoA carboxylase 1 (ACC1) that regulate fatty acid and triglyceride biosynthesis. The other seven miRs identified by the miR array screening did not affect the expression of lipogenic genes. miR-370 upregulated the expression of miR-122. Furthermore, the effect of miR-370 on the expression of the lipogenic genes was abolished by antisense miR-122. miR-370 targets the 3′ untranslated region (UTR) of Cpt1α, and it downregulated the expression of the carnitine palmitoyl transferase 1α (Cpt1α) gene as well as the rate of β oxidation. Our data suggest that miR-370 acting via miR-122 may have a causative role in the accumulation of hepatic triglycerides by modulating initially the expression of SREBP-1c, DGAT2, and Cpt1α and, subsequently, the expression of other genes that affect lipid metabolism.
Keywords: microRNA-122, microRNA-370, carnitine palmitoyl transferase-1a, triglyceride biosynthesis, fatty acid biosynthesis, β oxidation
MicroRNAs (miR) constitute a new class of small, noncoding RNAs that regulate gene expression at the posttranscriptional level by binding, in most cases, to the 3′ untranslated region (UTR) of target genes and inhibiting translation or causing mRNA cleavage (1–7).
In humans, the precursors of miRs (called primiR) are transcribed from a single gene or from polycistronic sequence by the RNA polymerase II (8–11). PrimiRs are then cleaved in the nucleus by the action of the RNaseIII enzyme drosha and the double stranded RNA binding protein pasha to form an imperfect double-stranded stem loop ∼70 nucleotides long called premiR. The premiR is exported into the cytoplasm by the RAN GTP–dependent exportin 5 and is processed by the dicer initially to a ∼22 nucleotide miR duplex and subsequently to mature single stranded miR. The mature miR is incorporated into the RNA induced silencing complex (RISC) that includes the argonaute proteins (5, 6, 12, 13). The miR associated with the RISC complex binds to the 3′ UTR of the target mRNA and either inhibits translation (14–17) or may promote RNA degradation (18, 19).
Bioinformatic analyses of known miRs suggest that the majority of mRNAs can be controlled by miRs and that a single miR could target several hundred mRNAs, including mRNAs encoding for transcription factors (20–22). miRs have been implicated in important cellular processes, such as apoptosis (23–25), cell proliferation and differentiation (22, 26–28), tumor suppression (6, 29), development (30), and metabolism. At present, the role of miRs in controlling the genes involved in metabolism is in early stages of investigation, and our knowledge of the underlying mechanism is limited (31, 32). Known examples include the regulation of insulin secretion by miR-375, which targets myotrophin mRNA (28); miR-9, which targets granulophilin and other pancreatic and brain mRNAs involved in exocytosis (20, 33); and miR-29b and miR-122, which control genes that affect amino acid (34) and lipid (35, 36) metabolism, respectively. miR expression signatures have been associated with well-defined clinico-pathological features and disease outcome (37–42).
The observation that the dominant negative cJun (dn-cJun) increased plasma cholesterol and triglyceride levels in mice (43) prompted us to investigate its effects on the regulation of genes implicated in hepatic fatty acid and triglyceride metabolism and the potential involvement of miRs in this process.
In the present study, we find that miR-370 targets the 3′ UTR of the carnitine palmitoyl transferase 1α (Cpt1α) and downregulates the expression of this enzyme that is important to fatty acid β oxidation. Furthermore, miR-370 and miR-122 upregulate the expression of the lipogenic genes, initially activating SREBP-1c and DGAT2 and, subsequently, FAS and ACC1. miR-122 promotes lipogenesis directly, whereas miR-370 promotes lipogenesis indirectly via upregulation of miR-122. Our findings indicate that extracellular stimuli that trigger upregulation of miR-370 and/or miR-122 may have a causative role in the accumulation of hepatic triglycerides by promoting lipogenesis and inhibiting β oxidation. Previous studies have shown that excess hepatic fat is associated with insulin resistance (44, 53) and metabolic syndrome (45) and that it promotes oxidative (47) and endoplasmic reticulum stress (48, 49), mitochondrial dysfunction (50, 53), inflammation (52), and fibrosis (54).
EXPERIMENTAL PROCEDURES
Animal studies
C57BL/6 and apoE−/− mice 8–12 weeks old were used in these studies. ApoE−/− and C57BL/6 mice were infected intravenously through the tail vein with adenoviruses at a dose of 2 × 109 pfu as described previously (43). Livers were excised 4 days postinfection for lipid and RNA determinations as described (43). Three-month-old C57BL/6J (Jackson Laboratory) were fed either chow diet or 60% high-fat diet (Research Diets, Diet #D12492). After 2, 5, and 8 weeks of diet, mice were sacrificed, and livers were excised for miR and lipid determination. Animal studies were performed according to the National Institutes of Health guidelines following protocols approved by the Institutional Animal Care and Use Committee of Boston University.
miR expression studies
Hepatic miR levels were evaluated with miR TLDA microarray assays as previously described (55) using RNU48 and RNU44 as an internal control. Validation of these results was performed using the mirVana qRT–PCR miRNA detection kit (Ambion Inc., TX). The U6 small nuclear RNA was used as an internal control.
miR target prediction methods
TargetScan version 4.0 (http://www.targetscan.org/index.html) and RNA22 (http://cbcsrv.watson.ibm.com/rna22_targets.html) databases were used to identify potential miR targets.
Oligonucleotide transfections
HepG2 cells seeded in 6-well plates were transfected with 50 nM of specific miRs, inhibitors of miRs expression, or small interference RNAs (siRNA) using siPORT NeoFX transfection agent (Ambion, Inc.). This reagent did not cause cell toxicity. RNA was extracted at different time points post-transfection, and real-time PCR analysis was performed as described below.
Real-time PCR analysis
Transcription of 0.1 μg RNA to cDNA was performed using the AMV Kit (Roche, Indianapolis, IN). LightCycler-FastStart DNA master SYBR Green, which contains Taq DNA polymerase, dNTP mix, SYBR Green 1 dye, and MgCl2 (Roche) was used as a reaction mix. PCR was performed as described (56). The oligonucleotide primers used for real-time PCR are shown in Table 1. All samples were analyzed in triplicate, and the average value of the triplicates was used for quantification. The data were expressed relative to GAPDH, which was used as an internal control.
TABLE 1.
Primers used for RT-PCR quantitation of mRNA levels
| Species | Gene | Sense Primer | Antisense Primer |
|---|---|---|---|
| Homo sapiens | Srebp-1c | 5′-GGAGGGGTAGGGCCAACGGCCT-3′ | 5′-CATGTCTTCGAAAGTGCAATCC-3′ |
| Fas | 5′-ACAGGGACAACCTGGAGTTCT-3′ | 5′-CTGT GGTCCCACTTGATGAGT-3′ | |
| Acc1 | 5′-GTTGCACAAAAGGATTTCAG-3′ | 5′-CGCATTACCATGCTCCGCAC-3′ | |
| Dgat2 | 5′-GAATCGTGGATCCCAAA-3′ | 5′-GTCTTCATATAACCAAAGCGGG-3′ | |
| Srebp-2 | 5′-CAAGATGCACAAGTCTGGCG -3′ | 5′-GCTTCAGCACCATGTTCTCCTG-3′ | |
| Apoe | 5′-CCCAGGTCACCCAGGAACT-3′ | 5′-TTCCGATTTGTAGGCCTTCAA-3′ | |
| ApoA-I | 5′-ATCGAGTGAAGGACCTGGC-3′ | 5′-AGCTTGCTGAAGGTGGAGGT-3′ | |
| Cpt1a | 5′-TGCTTTACAGGCGCAAACTG-3′ | 5′-TGGAATCGTGGATCCCAAA-3′ | |
| Gapdh | 5′-CCCATCACCATCTTCCAGGAG-3′ | 5′-CTTCTCCATGGTGGTGAAGACG-3′ | |
| Mus musculus | Srebp-1c | 5′-ATCGGCGCGGAAGCTGTCGGGGTAGC GTC-3′ | 5′-ACTGTCTTGGTTGTTGATG AGCTGGAGCAT-3′ |
| Cpt1a | 5′-CGCACGGAAGGAAAATGG-3′ | 5′-TGTGCCCAATATTCCTGG-3′ | |
| Acc1 | 5′-ATTGGGCACCCCAGAGCTA-3′ | 5′-CCCGCTCCTTCAACTTGCT-3′ | |
| Dgat2 | 5′- ACTCTGGAGGTTGGCACCAT-3′ | 5′-GGGTGTGGCTCAGGAGGAT-3′ | |
| Fas | 5′-CATGACCTCGTGATGAACGTGT-3′ | 5′-CGGGTGAGGACGTTTACAAAG-3′ | |
| Srebp-2 | 5′-CCGAGATGCAGGGCAAAG-3′ | 5′-GATGAAAGAACAATGAACAA GGCTTA-3′ | |
| Gapdh | 5′-CCTGCACCACCAACTGCTTA-3′ | 5′-TCATGAGCCCTTCCACAATG-3′ |
Abbreviations: ACC1, acyl-CoA carboxylase 1; dn-cJun, dominant negative cJun; Cpt1α, carnitine palmitoyl transferase 1α; DGAT2, diacylglycerol acyltransferase-2; miR, microRNA; SREBP-1c, sterol-regulatory element binding protein 1c; UTR, untranslated region.
Protein extraction from HepG2 cells
HepG2 cell pellets were lysed using Nonidet P-40 lysis buffer containing 30 mM Tris (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Nonidet P-40, and a cocktail of protease inhibitors. Cell pellets were obtained by centrifugation. Supernatant was collected and stored at −20°C. Nuclear protein extracts were prepared from HepG2 cells using NE-PER nuclear and cytoplasmic extraction reagents (Pierce) according to the manufacturer's protocol.
Western blot analysis and quantification
The cell lysates or nuclear extracts of HepG2 cells were electrophoresed and separated on a 4–20% Tris-HCl gel (Bio-Rad, Hercules, CA) and transferred to a Hybond-ECL nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ) as described (56). The nitrocellulose membrane was probed with anti-SREBP-1c (1:500 dilution) (K10); anti-FAS (1:500 dilution) (H-300); anti-ACC1 (1:1,000 dilution) (FL-114); and anti-GAPDH (1:5,000 dilution) (Santa Cruz Biotechechnology, Inc., Santa Cruz, CA). Signals were detected using anti-rabbit immunoglobulin IgG conjugated with horseradish peroxidase (1:5,000 dilution). The nitrocellulose membranes were then exposed to photographic film, which was scanned, and the intensities of the protein bands, which were expressed as arbitrary units, were determined by computerized densitometry. Protein levels were normalized to GAPDH protein levels. Protein expression levels of Western blots were quantified using the Image Quant 6 program analysis.
Luciferase assays
HepG2 cells were cotransfected in 12-well plates with 0.5 μg of firefly luciferase reporter vector including the 3′ UTR of Cpt1α (pEZX, GeneCopoeia, Inc.) together with 0.05 μg of the phRL-SV40 control vector (Promega) and 100 nM miR-370 (Ambion) using Lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activities were measured consecutively by using the Dual Luciferase Assay (Promega) 24 h after transfection. Each reporter plasmid was transfected at least twice (on different days), and each sample was assayed in triplicate.
Palmitic acid (PA) oxidation
Analysis was performed as described (57). HepG2 cells were transfected with 50 nM miR-122 or miR-370, and 24 h later, were washed with PBS prior to labeling in DMEM (1 ml/well) containing 0.4 mM PA/1.5% BSA and [14C]PA (1μCi/ml) (Perkin Elmer) for 16 h. The medium was collected and centrifuged, and the supernatant was transferred to a 25 ml flask with a center well (Fisher Scientific) containing filter paper saturated with 100 μl of 1M KOH. The flask was sealed with a stopper (Fisher Scientific), and 200 μl of 69–72% perchloric acid (Fisher Scientific) was added to the medium through the top of the stopper to release the [14C]CO2. The flask was then incubated in a shaking incubator at 37°C for 1 h. The radioactivity of [14C]CO2 trapped on the filter paper was determined by liquid scintillation counting. The results were normalized with the protein content of the cell culture plates determined by the Bradford method.
Statistical analysis
Data are presented as the mean ± S.E. Comparison of data from different groups of mice or different groups of HepG2 cells was performed using the paired two-sample-for-means and two-tailed t-test. Statistically significant differences between two groups were defined as those giving a value of P < 0.05.
RESULTS
Effect of Ad-dn-cJun on expression of miRs and lipogenic genes in the liver
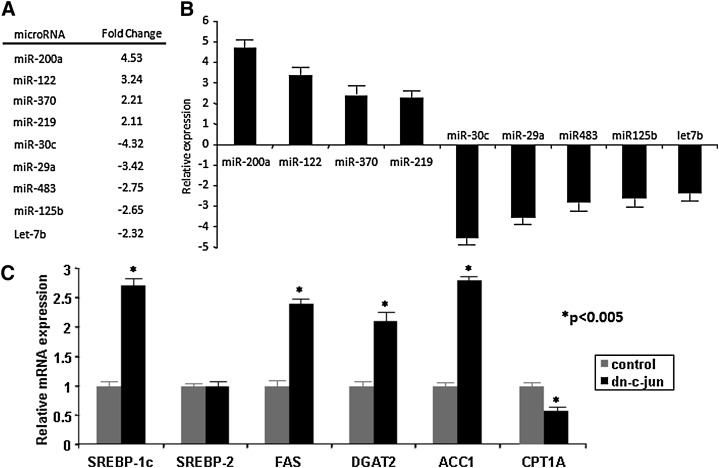
We assessed the expression levels of 365 miRs by miR arrays and identified nine miRs differentially expressed in the livers of Ad-dn-cJun-infected apoE−/− mice as compared with Ad-GFP–infected mice (Fig. 1A and supplementary Fig. I). Four miRs were upregulated (miR-200a, miR-122, miR-370, miR-219) while five were downregulated (miR-30c, miR-29a, miR-483, miR-125b, let-7b). Validation of these results using a real-time SYBR Green miR assay (Fig. 1A, B) confirmed that dn-cJun had altered the expression levels of these nine miRs. Real-time PCR analysis of the hepatic mRNA of genes related to lipid metabolism showed that the SREBP-1c gene expression was upregulated by 2.7-fold, DGAT2 by 2.1-fold, FAS by 2.4-fold, and ACC1 by 2.75-fold. The Cpt1α gene was downregulated by 40%, whereas the expression of the SREBP-2 gene did not change (Fig. 1C).
Fig. 1.
Effect of dn-cJun on the hepatic lipid metabolism and miR gene expression in apoE−/− mice. A: miR changes detected by analysis of a microRNA TaqMan array using hepatic RNA obtained from five dn-cJun-treated mice vs. five control mice. B: Validation of the miR data by real-time SYBR Green PCR assay. C: Real-time PCR analysis of genes involved in fatty acid and triglyceride biosynthesis and catabolism in the liver of dn-cJun-treated vs. control mice. Statistically significant changes in gene expression levels between control and dn-cJun treated mice are indicated by asterisks, P < 0.005. ACC1, acyl-CoA carboxylase 1; dn-cJun, dominant negative cJun; Cpt1α, carnitine palmitoyl transferase 1α; DGAT2, diacylglycerol acyltransferase-2; miR, microRNA; SREBP-1c, sterol-regulatory element binding protein 1c.
Increased expression of miR-370 is associated with increased expression of genes that are involved in hepatic lipid metabolism
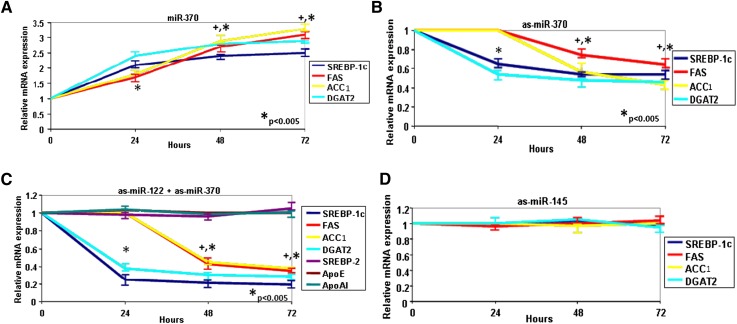
To assign a causative regulatory role to the nine miRs that were up- or downregulated by Ad-dn-c-Jun in vivo for the expression of genes associated with the fatty acid and triglyceride metabolism, HepG2 cells were transfected with the nine miRs. Real-time PCR analysis showed that miR-370 upregulated the SREBP-1c and DGAT2 gene expression 2.1- and 2.4-fold, respectively, 24 h post-transfection, and the FAS and ACC1 gene expression 3.1- and 3.3-fold, respectively, 72 h post-transfection (Fig. 2A). Similar upregulation of SREBP-1c, DGAT2, FAS, and ACC1 genes was achieved by miR-122, whereas miR-29a, miR-30C, miR-125b, miR-219, miR-200a, miR-483, and let7a did not affect the expression of the lipogenic genes (supplementary Fig. IIA–D,F). Transfection experiments in HepG2 cells also showed that miR-370 and miR-122 upregulated 3.2- and 3-fold, respectively, the expression of LXRα (supplementary Fig. IIIA).
Fig. 2.
Modulation of expression of SREBP-1c and genes involved in fatty acid and triglyceride biosynthesis by miR-370 and miR-122 in HepG2 cells. Real-time PCR analysis of SREBP-1c, FAS, ACC1, and DGAT2 after treatment of HepG2 cells with miR-370 at 0, 12, 24, 48, and 72 h post-transfection. Increases in gene expression at all time points were statistically significant (P < 0.005) A: Real-time PCR analysis of SREBP-1c, FAS, ACC1, and DGAT2 after liposomal transfection (50 nM) in HepG2 cells with anti-sense oligonucleotides for inhibition of (B) miR-370; (C) miR-122 and miR-370; and (D) miR-145 (as a control). Statistically significant changes in gene expression at different time points in panels B and C are indicated by * P < 0.005. The symbol + in panels A–C indicates that the difference between 48 h and 72 h post-transfection in FAS and ACC1 mRNA levels were statistically significant (P < 0.05). The difference observed in the expression of FAS and ACC1 between single antisense microRNA (B) or combination of both antisense microRNAs (C) is statistically significant (P < 0.05). There is no statistically significant difference (P > 0.1) in FAS and ACC1 expression levels in as-miR-145 treated HepG2 cells. ACC1, acyl-CoA carboxylase 1; dn-cJun, dominant negative cJun; Cpt1α, carnitine palmitoyl transferase 1α; DGAT2, diacylglycerol acyltransferase-2; miR, microRNA; SREBP-1c, sterol-regulatory element binding protein 1c.
Silencing miR-370 with antisense miR-370 in HepG2 cells reduced SREBP-1c and DGAT2 mRNA levels 24 h post-transfection (35% and 46% reduction, respectively) (Fig. 2B). Antisense miR-370 did not affect FAS or ACC1 mRNA levels 24 h post-transfection; however, it caused a time-dependent decrease of these mRNA levels over 24–48 h and a further slow decrease over 48–72 h post-transfection. Antisense miR-370 reduced the mRNA levels of FAS by 26% and 36% and those of ACC1 43% and 52%, respectively, 48 h and 72 h post-transfection, respectively. The kinetics of induction or inhibition of the lipogenic genes by sense or antisense miR-370 indicate that SREBP-1c and DGAT2 are early responders to miR-370 treatment (regulated within 24 h post-transfection), while FAS and ACC1 are late responders (regulated 24–72 h post-transfection). Similar time-dependent reduction of SREBP-1c, DGAT2, ACC1, and FAS expression were obtained by antisense miR-122 treatment of HepG2 cells (supplementary Fig. IIE). Combined inhibition by both antisense miR-370 and antisense miR-122 had a greater effect in the reduction of SREBP-1c (84%), DGAT2 (71%), FAS (65%), and ACC1 (62%) mRNA levels 72 h post-transfection (Fig. 2C). This treatment did not affect the expression apoE and apoA-I genes (Fig. 2C and supplementary Fig. IIIB). To validate the specificity of miR-370, we treated HepG2 cells with antisense miR-145. This treatment did not affect the expression of SREBP-1 or the genes associated with fatty acid and triglyceride biosynthesis (Fig. 2D).
miR-370 and miR-122 affect FAS and ACC-1 expression via modulation of SREBP-1c and DGAT2 expression
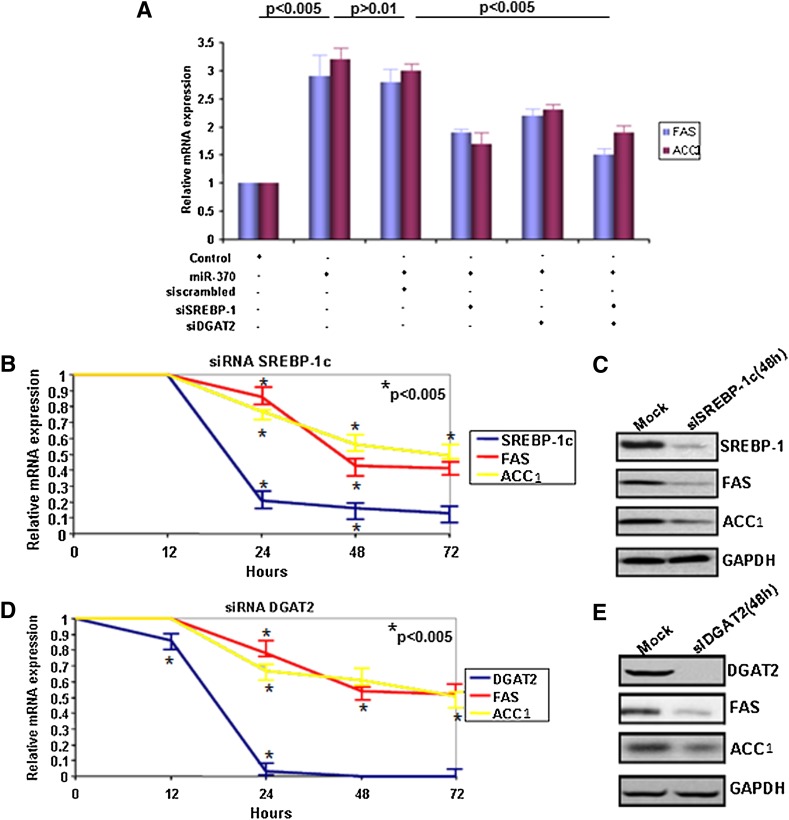
Our results strongly suggest that miR-370 regulates SREBP-1c, DGAT2, FAS, and ACC1 mRNA levels. However, the exact molecular pathway was not delineated. To address this question, we performed real-time PCR analysis following treatment of HepG2 cells with miR-370 in the presence of siRNA for SREBP-1c, DGAT2, or both. The miR-370 treatment alone or in the presence of scrambled siRNA increased FAS and ACC1 mRNA levels approximately 3-fold (Fig. 3A, columns 2, 3). siRNA-mediated inhibition of SREBP-1c (Fig. 3A, column 4) or DGAT2 (Fig. 3A, column 5) reduced the miR-370–mediated increase in expression of FAS by 34% and 23%, respectively, and of ACC1 by 45% and 28%, respectively. Combined inhibition of SREBP-1c and DGAT2 by the corresponding siRNAs, reduced the miR-370–mediated increase of FAS by 47% and ACC1 by 44%, respectively (Fig. 3A, compare columns 2, 6). Similar results were obtained by treatment of HepG2 cells with miR-122 in the presence of siRNA for SREBP-1c, DGAT2, or both (supplementary Fig. IIG).
Fig. 3.
Regulation of SREBP-1c and DGAT2 by miR-370 precedes the regulation of FAS and ACC1 genes. A: Real-time PCR analysis of FAS and ACC1 genes after treatment with miR-370 and downregulation of SREBP-1c or DGAT2 or both by treatment with the corresponding siRNA (50 nM) for 48 h. Asterisks indicate statistical significance for both FAS and ACC1 expression levels between untreated and miR-370–treated cells (P < 0.005) as well as miR-370/siscrambled–treated and siSREBP-1c/siDGAT2–treated cells. There is no statistical significance between miR-370 and miR-370/siscrambled–treated cells. B: Real-time PCR analysis of SREBP-1c, FAS, and ACC1 genes after treatment of HepG2 cells with siRNA against SREBP-1c (50 nM) for 48 h. C: Western blot analysis of FAS and ACC1 proteins after treatment of HepG2 cells with siRNA against SREBP-1c (50 nM) for 48 h. D: Real-time PCR analysis of FAS and ACC1 genes after treatment of HepG2 cells with siRNA against DGAT2 (50 nM) for 48 h. E: Western blot analysis of FAS and ACC1 proteins after treatment of HepG2 cells with siRNA against DGAT2 (50 nM) for 48 h. GAPDH protein levels were used as control in both panels C and E. ACC1, acyl-CoA carboxylase 1; dn-cJun, dominant negative cJun; Cpt1α, carnitine palmitoyl transferase 1α; DGAT2, diacylglycerol acyltransferase-2; miR, microRNA; SREBP-1c, sterol-regulatory element binding protein 1c.
Control experiments showed that treatment of HepG2 cells with SREBP-1c siRNA downregulated SREBP-1c mRNA levels by 81% and 91% at 24 h and 72 h, respectively, and resulted in 55% and 45% decrease in the mRNA levels of FAS and ACC1 72 h post-transfection (Fig. 3B). Western blot analysis revealed a strong reduction of SREBP-1c, FAS, and ACC1 protein levels 72 h post-transfection (Fig. 3C). Similar experiments showed that treatment of HepG2 cells with DGAT2 siRNA downregulated DGAT2 mRNA levels by 95% and 100% at 24 h and 72 h, respectively, and resulted in 55% and 50% decrease in the mRNA levels of FAS and ACC1 72 h post-transfection (Fig. 3D). Western blot analysis revealed absence of DGAT2 and strong reduction of FAS and ACC1 protein levels 72 h post-transfection (Fig. 3E) (58). The combined data (Figs. 2A–D; 3A–E) suggest that the effect of miR-370 on the lipogenic genes are manifested by upregulation of SREBP-1c and DGAT-2. The increase in SREBP-1c directly upregulates the expression of FAS and ACC1 by binding to their SREs (59, 60), whereas the increase in DGAT-2 appears to upregulate these genes indirectly (61).
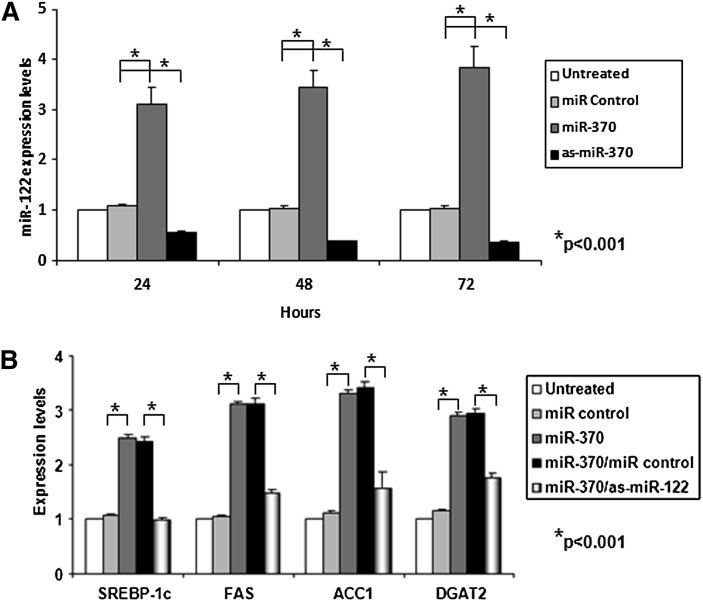
miR-370 induces the lipogenic genes indirectly by upregulating miR-122
The relationship between miR-370 and miR-122 was investigated in two sets of experiments. Transfection of HepG2 cells with miR-370 upregulated the expression of miR-122 3.1-, 3.5-, and 3.9-fold at 24, 48, and 72 h post-transfection, respectively, compared with untransfected or control transfected cells (Fig. 4A). The complementary experiment showed that transfection of HepG2 cells with antisense miR-370 decreased miR-122 levels by 45%, 60%, and 65% at 24, 48 h, and 72 post-transfection, respectively, relative to the controls (Fig. 4A). This finding prompted us to investigate whether the effect of miR-370 on the activation of the lipogenic genes was direct or indirect. Cotransfection of HepG2 cells with miR-370 and antisense miR-122 abolished the miR-370–mediated increase of SREBP-1c and reduced by 70–80% the miR-122–mediated activation of FAS, ACC1, and DGAT-2 (Fig. 4B). This finding indicates that miR-370 activates lipogenic genes indirectly via miR-122.
Fig. 4.
Effect of miR-370 on the expression of miR-122 in HepG2 cells. A: Real-time PCR analysis of miR-122 following treatment of HepG2 cells with sense and antisense miR-370 or with miR-negative control. B: Real-time PCR analysis of SREBP-1c FAS, ACC1, and DGAT2 gene expression following treatment of HepG2 cells with miR-370 alone or combination of miR-370 and miR-122. Asterisks indicate statistically significant changes between miR control and miR-370–treated HepG2 cells (P < 0.001) and between miR-370/miR control and miR-370/as-miR-122–treated HepG2 cells (P < 0.001). ACC1, acyl-CoA carboxylase 1; dn-cJun, dominant negative cJun; Cpt1α, carnitine palmitoyl transferase 1α; DGAT2, diacylglycerol acyltransferase-2; miR, microRNA; SREBP-1c, sterol-regulatory element binding protein 1c.
Bioinformatic and molecular analysis revealed that miR-370 regulates directly Cpt1α expression levels
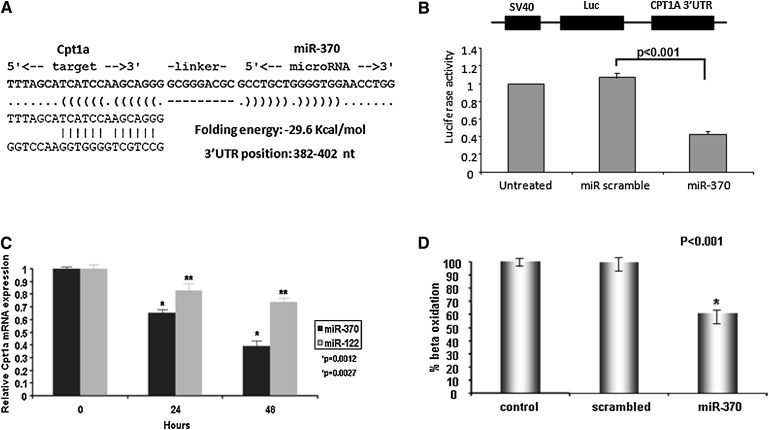
Using the ENSEMBL database, we identified complementary binding sites for miR-122a and miR-370 on chromosomes 18 and 14, respectively (supplementary Fig. IV). Furthermore, to identify whether miR-122 and miR-370 directly regulate expression of genes for lipid metabolism, we searched for their direct targets using TargetScan and RNA22 miR gene target prediction programs. This analysis identified five genes that may be direct targets of miR-122, but none of them were related to lipid metabolism (35, 62). On the other hand, we found that miR-370 has complementarity with the 3′ UTR of Cpt1α (Fig. 5A). To confirm the direct interaction between miR-370 and the predicted binding site on the 3′ UTR of the Cpt1α gene, we cotransfected HepG2 cells with a luciferase-containing construct driven by the SV40 promoter and linked to the 3′ UTR of Cpt1α and miR-370. This treatment decreased the activity of the luciferase promoter construct by 60% compared with a scrambled miR control (Fig. 5B). These data strongly suggest that Cpt1α is a direct target of miR-370.
Fig. 5.
Effect of miR-370 on Cpt1α expression in HepG2 cells. A: Target sequence in the 3′ UTR of Cpt1α for miR-370. B: Effect of miR-370 on the activity of the SV40 luciferase construct linked to the 3′ UTR of Cpt1α. C: Cpt1α mRNA levels assessed by real-time PCR analysis 24 h and 48 h post-transfection of HepG2 cells with 50 nM miR-370 or miR-122. D: Fatty acid β oxidation levels following transfection of HepG2 cells with 50 μM of miR-370. Statistically significant differences relative to the controls are indicated by asterisks in panels B, C, and D. ACC1, acyl-CoA carboxylase 1; dn-cJun, dominant negative cJun; Cpt1α, carnitine palmitoyl transferase 1α; DGAT2, diacylglycerol acyltransferase-2; miR, microRNA; SREBP-1c, sterol-regulatory element binding protein 1c; UTR, untranslated region.
We further tested the effect of miR-370 on the expression of Cpt1α in HepG2 cells. Transfection of HepG2 cells with miR-370 reduced by 62% Cpt1α mRNA levels 48 h post-transfection (Fig. 5C). A similar treatment of HepG2 cells with miR-122 showed a 26% reduction in Cpt1α mRNA 48 h post-transfection (Fig. 5C). These findings are consistent with the real-time PCR analysis of the hepatic RNA of apoE−/− mice treated with Ad-dn-cJun, which showed an upregulation of both miR-370 and miR-122 (Fig. 1B). These analyses also indicated a significant decrease (40%) of Cpt1α mRNA levels compared with control mice treated with Ad-GFP (Fig. 1C). To assess functional consequences of the miR-370–driven downregulation of Cpt1α mRNA levels, we determined the rate of β oxidation of 14C-palmitate following transfection of HepG2 cells with miR-370. This analysis showed that miR-370 reduced the β oxidation capacity of the cells by 40% (Fig. 5D). Overall, the data of Fig. 5A–D indicate that miR-370 directly downregulates Cpt1α, which controls the rate-limiting step in fatty acid β oxidation.
Correlation between miR-370 and miR-122 levels and hepatic triglyceride accumulation in C57BL/6 mice
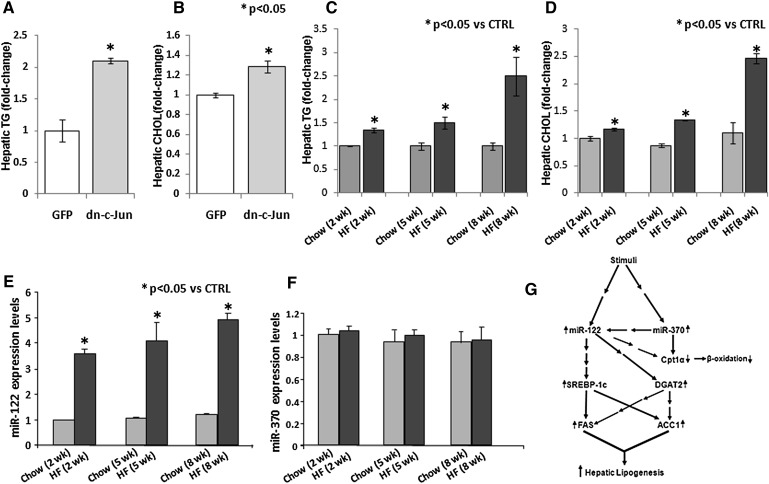
Infection of ApoE−/− mice with 2 × 109 pfu of adenovirus expressing the dn-cJun resulted in a 3.34- and 2.21-fold increase in miR-122 and miR-370 levels, respectively (Fig. 1B). Hepatic lipid analysis showed that this treatment of C57BL/6 mice with Ad-dn-cJun also led to a 2.1- and 1.25-fold increase in hepatic triglyceride and cholesterol levels, respectively (Fig. 6A, B). This treatment also led to a 3.34- and 2.21-fold increase in miR-122 and miR-370 levels, respectively (Fig. 1B). We explored whether other stress conditions that increase hepatic lipid accumulation are associated with alterations in hepatic miR-122 and miR-370 levels. Feeding C57BL/6 mice with 60% fat diet for 2, 5, or 8 weeks caused approximately 2.5- and 3-fold increase in hepatic triglyceride and cholesterol levels, respectively, in the 8th week of dietary treatment and smaller but statistically significant increases in the 2nd and 5th week of treatment (Fig. 6C, D). The increased hepatic lipid levels were associated with a progressive 3.6- to 4.9-fold increase in miR-122 expression on the 2nd, 5th and 8th week of the diet, but it had no effect regarding miR-370 expression (Fig. 6E, F).
Fig. 6.
Correlation of hepatic lipid levels with the levels of miR-122 and miR-370 in C57BL/6 mice. A, B: Effect of dn-cJun on the hepatic triglyceride (A) and cholesterol (B) levels, 4 days postinfection with adenoviruses expressing dn-cJun or GFP. C, D: Effect of 2-, 5-, and 8-week high-fat diet on the hepatic triglyceride (C) and cholesterol (D) levels of C57BL/6 mice. (E and F) Effect of 2-, 5-, and 8-week high-fat diets on the hepatic miR-122 (E) and miR-370 (F) levels of C57BL/6 mice. Statistically significant differences relative to the control are indicated by asterisks in panels A–E. G: Strength of miR-gene and gene-gene interactions related with the pathways of fatty acid triglyceride biosynthesis and catabolism. The figure depicts the interaction between miR-122 and miR-370 with their downstream targets. Both miRs indirectly affect initially SREBP-1c and DGAT2. In addition, miR-370 directly and miR-122 indirectly target Cpt1α gene expression. Downstream of the network, SREBP-1c directly and DGAT2 indirectly modulate the expression of FAS and ACC1 mRNA and protein levels. These changes result in increased fatty acid and triglyceride biosynthesis and decreased fatty acid catabolism and are expected to cause hepatic triglyceride accumulation. The line thickness indicates the strength of the correlation. A single arrow indicates direct interaction, while multiple arrows indicate indirect interaction. ACC1, acyl-CoA carboxylase 1; dn-cJun, dominant negative cJun; Cpt1α, carnitine palmitoyl transferase 1α; DGAT2, diacylglycerol acyltransferase-2; miR, microRNA; SREBP-1c, sterol-regulatory element binding protein 1c; UTR, untranslated region.
A putative mechanism regarding the effect of miR-370 and miR-122 on the regulation of the lipogenic genes and Cpt1α that is consistent with hepatic triglyceride accumulation is presented in Fig. 6G and discussed further below.
DISCUSSION
The initial experiments, which employed an unbiased screening method, showed that miR-370 and miR-122 were upregulated in mice treated with dn-cJun and that these changes were associated with upregulation of SREBP-1c and other hepatic genes of fatty acid and triglyceride biosynthesis. Previous studies showed that the transcriptional regulator SREBP-1c increases the transcription of genes of fatty acid biosynthesis, whereas SREBP-2 regulates the genes of cholesterol biosynthesis (59).
The question whether there was a causative relationship between the expression of miR-370 and miR-122 and the expression of the genes involved in fatty acid and triglyceride biosynthesis was addressed by a series of experiments in HepG2 cells. These analyses established that miR-370 and miR-122 upregulated SREBP-1c and the genes involved in fatty acid and triglyceride biosynthesis. Silencing with anti-miR-370 or anti-miR-122 (or both) led to proportionate downregulation of SREBP-1c, as well as the genes involved in fatty acid and triglyceride biosynthesis.
The kinetics of induction or inhibition of expression of the lipogenic genes by sense or antisense miR-370 and miR-122 showed that the induction or inhibition of SREBP-1c and DGAT2 expression precedes that of FAS and ACC1. Furthermore, the expression levels of ACC1 and FAS appear to depend on the levels of SREBP-1c and DGAT2. Thus, silencing of SREBP-1c or DGAT2 by siRNA inhibited the miR-370– and miR-122–mediated upregulation of ACC1 and FAS.
A significant, novel finding uncovered by our studies is that miR-370 upregulates the expression of miR-122 and that the effect of miR-370 on the upregulation of the lipogenic genes in HepG2 cells is greatly reduced by treatment with antisense miR-122.
Knowing that miRs are negative regulators of gene expression, the effects of miR-370 and miR-122 on their downstream targets is expected to involve an initial downregulation of one or more direct target genes. In most cases, this will be followed by a cascade of up- or downregulation of downstream target genes encoding for transcription factors and other proteins (20).
On the basis of these considerations, we propose the putative action mechanism of miR-370 and miR-122 on the expression of lipid metabolism genes shown in Fig. 6G. Intra- or extracellular stimuli upregulate the expression of miR-122 or miR-370 (or both) through intermediate steps depicted by arrows. Activation of miR-370 alone activates miR-122. Early indirect targets of miR-122 are SREBP-1c and DGAT2. At present, the intermediate steps in the signaling cascade that lead to the upregulation of SREBP-1c and DGAT2 by miR-370 and/or miR-122 are not known. LXRα, which is significantly upregulated by miR-122 and regulates SREBP-1c expression, may be involved in the process (59, 63, 64). This study also establishes a direct involvement of miR-370 in the regulation of the gene encoding Cpt1α, which controls β oxidation (65).
Computational analysis revealed complementarity between miR-370 sequence and the 3′ UTR region of Cpt1α. The functional interaction between miR-370 and the 3′ UTR region of Cpt1α was validated experimentally by downregulation of the activity of promoter luciferase construct linked to the 3′ UTR of Cpt1α. The inhibitory effect of miR-370 was confirmed by transfection of HepG2 cells with miR-370, which resulted in decreased Cpt1α expression and rate of β oxidation. Transfection experiments in HepG2 cells showed that miR-122 also downregulated by 26% the gene expression of Cpt1α 48 h post-transfection. As Cpt1α is not a direct target of miR-122, the observed effect must be indirect. The decrease in β oxidation, combined with the observed activation of SREBP-1c and DGAT2, is expected to lead to increased cellular storage of lipids. A previous study also showed that inhibition of Cpt1α by tetradecylglycidic acid (TGDA) caused hepatic triglyceride accumulation and led to steatosis (66).
The association between upregulation of miR-370 and miR-122 and/or hepatic lipid accumulation in vivo was demonstrated in two different ways. The first was adenovirus-mediated gene transfer of dn-Jun into mice that induced both miR-370 and miR-122 and caused hepatic lipid accumulation. The second was feeding experiments of mice with a high-fat diet for 2, 5, or 8 weeks. This treatment increased hepatic lipid accumulation and was associated with increased levels of miR-122 but not of miR-370. Others have also reported that fat feeding in rats increased miR-122 expression by 2.1- and 2.9-fold over a 4- and 12-week period, respectively (67). Thus, although the two treatments altered miR levels, their effects were different. It is possible that in mice treated with the dn-cJun, signaling pathways upregulate miR-370, which then upregulates miR-122. Alternatively both miRs may be activated independently. With fat feeding, upregulation of miR-122 is independent of miR-370 and may originate from a different signaling pathway. In this case, miR-122 induction by the high-fat diet may be required to promote lipogenesis via upregulation of SREBP-1c, DGAT-2, FAS, and ACC1. On the other hand, the lack of induction of miR-370 will not impede β oxidation and will facilitate catabolism of the fatty acids that are abundant in the high-fat diet. Future studies involving liposome-mediated or adenovirus-mediated transfer of miR-122 and miR-370 to the liver, in combination with microarray analysis, may reveal the signaling mechanisms linked to these miRs that lead to hepatic lipid accumulation.
Our findings, reported in the supplementary material, on the effects of sense or antisense miR-122 on hepatic lipogenesis are complementary to those reported previously using antisense miR-122 in HepG2 cells (35). This treatment resulted in downregulation of the genes involved in fatty acid synthesis, increased β oxidation, and phosphorylation of AMP-activated protein kinase α1 (AMPKα1), which increases β oxidation (35, 68, 69). Antisense miR-122 treatment also moderately decreased plasma lipid levels in normal and cholesterol-fed mice and reversed hepatic steatosis (35).
At variance with the findings of Esau et al. (35) and our data, a recently published study suggested that nonalcoholic steatosis in humans was associated with decreased levels of miR-122 (36). The miR-122 was underexpressed by 20% based on microarray data and by 63% based on qRT-PCR data (36). The control group, however, consisted of individuals who were diagnosed with clinical features of metabolic syndrome. The same study also indicated that treatment of HepG2 with either sense or antisense miR-122 transiently increased and then decreased the SREBP-1c mRNA levels and gradually decreased FAS expression (36). Several previous miR studies showed that overexpression of sense or antisense miR has opposite effects on the expression of the target genes.
CONCLUSION
Our study provides novel data on the role of miR-370 in the upregulation of miR-122, which ultimately controls the expression of lipogenic genes, and the direct role of miR-370 on the inhibition of Cpt1α, which controls β oxidation. The data suggest a putative molecular mechanism linking miR-370 and miR-122 with hepatic triglyceride accumulation that can be explored in future studies.
Supplementary Material
Acknowledgments
The authors thank Anne Plunkett for preparing the manuscript.
Footnotes
Abbreviations:
- ACC1
- acyl-CoA carboxylase 1
- dn-cJun
- dominant negative cJun
- Cpt1α
- carnitine palmitoyl transferase 1α
- DGAT2
- diacylglycerol acyltransferase-2
- miR
- microRNA
- SREBP-1c
- sterol-regulatory element binding protein 1c
- UTR
- untranslated region
This work was supported by National Institutes of Health Grants HL-48739, HL-68216, and HL-45095. This work was also supported by the Sixth Framework Programme of the European Union (LSHM-CT-2006-037631). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures.
REFERENCES
- 1.He L., Hannon G. J. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5: 522–531. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. 2001. microRNAs: tiny regulators with great potential. Cell. 107: 823–826. [DOI] [PubMed] [Google Scholar]
- 3.Lai E. C. 2002. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30: 363–364. [DOI] [PubMed] [Google Scholar]
- 4.Hurst L. D. 2006. Preliminary assessment of the impact of microRNA-mediated regulation on coding sequence evolution in mammals. J. Mol. Evol. 63: 174–182. [DOI] [PubMed] [Google Scholar]
- 5.Wilfred B. R., Wang W. X., Nelson P. T. 2007. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 91: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A., Slack F. J. 2006. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 6: 259–269. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 8.Chung K. H., Hart C. C., Al Bassam S., Avery A., Taylor J., Patel P. D., Vojtek A. B., Turner D. L. 2006. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 34: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber M. J. 2005. New human and mouse microRNA genes found by homology search. FEBS J. 272: 59–73. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez A., Griffiths-Jones S., Ashurst J. L., Bradley A. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res. 14: 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying S. Y., Lin S. L. 2006. Current perspectives in intronic micro RNAs (miRNAs). J. Biomed. Sci. 13: 5–15. [DOI] [PubMed] [Google Scholar]
- 12.Hammond S. M. 2005. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 579: 5822–5829. [DOI] [PubMed] [Google Scholar]
- 13.Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C. 1998. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17: 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jabri E. 2005. P-bodies take a RISC. Nat. Struct. Mol. Biol. 12: 564. [DOI] [PubMed] [Google Scholar]
- 15.Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7: 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 309: 1573–1576. [DOI] [PubMed] [Google Scholar]
- 17.Liu J., Carmell M. A., Rivas F. V., Marsden C. G., Thomson J. M., Song J. J., Hammond S. M., Joshua-Tor L., Hannon G. J. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- 18.Chan S. P., Slack F. J. 2006. microRNA-mediated silencing inside P-bodies. RNA Biol. 3: 97–100. [DOI] [PubMed] [Google Scholar]
- 19.Jackson R. J., Standart N. 2007. How do microRNAs regulate gene expression? Sci. STKE. 2007: re1. [DOI] [PubMed] [Google Scholar]
- 20.Lewis B. P., Burge C. B., Bartel D. P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 21.Miranda K. C., Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., Lim B., Rigoutsos I. 2006. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 126: 1203–1217. [DOI] [PubMed] [Google Scholar]
- 22.Hackl H., Burkard T. R., Sturn A., Rubio R., Schleiffer A., Tian S., Quackenbush J., Eisenhaber F., Trajanoski Z. 2005. Molecular processes during fat cell development revealed by gene expression profiling and functional annotation. Genome Biol. 6: R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim L. P., Lau N. C., Weinstein E. G., Abdelhakim A., Yekta S., Rhoades M. W., Burge C. B., Bartel D. P. 2003. The microRNAs of Caenorhabditis elegans. Genes Dev. 17: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. 2003. Prediction of mammalian microRNA targets. Cell. 115: 787–798. [DOI] [PubMed] [Google Scholar]
- 25.Stark A., Brennecke J., Russell R. B., Cohen S. M. 2003. Identification of Drosophila MicroRNA targets. PLoS Biol. 1: E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C. Z., Li L., Lodish H. F., Bartel D. P. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science. 303: 83–86. [DOI] [PubMed] [Google Scholar]
- 27.Kajimoto K., Naraba H., Iwai N. 2006. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA. 12: 1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P. E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., et al. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 432: 226–230. [DOI] [PubMed] [Google Scholar]
- 29.He L., He X., Lowe S. W., Hannon G. J. 2007. MicroRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat. Rev. Cancer. 7: 819–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cobb B. S., Nesterova T. B., Thompson E., Hertweck A., O'Connor E., Godwin J., Wilson C. B., Brockdorff N., Fisher A. G., Smale S. T., et al. 2005. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 201: 1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson P., Kiriakidou M., Sharma A., Maniataki E., Mourelatos Z. 2003. The microRNA world: small is mighty. Trends Biochem. Sci. 28: 534–540. [DOI] [PubMed] [Google Scholar]
- 32.Ambros V. 2004. The functions of animal microRNAs. Nature. 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 33.Plaisance V., Abderrahmani A., Perret-Menoud V., Jacquemin P., Lemaigre F., Regazzi R. 2006. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 281: 26932–26942. [DOI] [PubMed] [Google Scholar]
- 34.Mersey B. D., Jin P., Danner D. J. 2005. Human microRNA (miR29b) expression controls the amount of branched chain alpha-ketoacid dehydrogenase complex in a cell. Hum. Mol. Genet. 14: 3371–3377. [DOI] [PubMed] [Google Scholar]
- 35.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3: 87–98. [DOI] [PubMed] [Google Scholar]
- 36.Cheung O., Puri P., Eicken C., Contos M. J., Mirshahi F., Maher J. W., Kellum J. M., Min H., Luketic V. A., Sanyal A. J. 2008. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology. 48: 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calin G. A., Croce C. M. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 6: 857–866. [DOI] [PubMed] [Google Scholar]
- 38.Jin P., Zarnescu D. C., Ceman S., Nakamoto M., Mowrey J., Jongens T. A., Nelson D. L., Moses K., Warren S. T. 2004. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7: 113–117. [DOI] [PubMed] [Google Scholar]
- 39.Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., et al. 2005. MicroRNA expression profiles classify human cancers. Nature. 435: 834–838. [DOI] [PubMed] [Google Scholar]
- 40.Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA. 103: 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R. M., Okamoto A., Yokota J., Tanaka T., et al. 2006. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 9: 189–198. [DOI] [PubMed] [Google Scholar]
- 42.Murakami Y., Yasuda T., Saigo K., Urashima T., Toyoda H., Okanoue T., Shimotohno K. 2006. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 25: 2537–2545. [DOI] [PubMed] [Google Scholar]
- 43.Drosatos K., Sanoudou D., Kypreos K. E., Kardassis D., Zannis V. I. 2007. A dominant negative form of the transcription factor c-Jun affects genes that have opposing effects on lipid homeostasis in mice. J. Biol. Chem. 282: 19556–19564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chitturi S., Abeygunasekera S., Farrell G. C., Holmes-Walker J., Hui J. M., Fung C., Karim R., Lin R., Samarasinghe D., Liddle C., et al. 2002. NASH and insulin resistance: insulin hypersecretion and specific association with the insulin resistance syndrome. Hepatology. 35: 373–379. [DOI] [PubMed] [Google Scholar]
- 45.Grundy S. M., Cleeman J. I., Daniels S. R., Donato K. A., Eckel R. H., Franklin B. A., Gordon D. J., Krauss R. M., Savage P. J., Smith S. C., Jr, et al. 2005. Diagnosis and management of the metabolic syndrome. An American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Executive summary. Cardiol. Rev. 13: 322–327. [PubMed] [Google Scholar]
- 46.Unger R. H. 2002. Lipotoxic diseases. Annu. Rev. Med. 53: 319–336. [DOI] [PubMed] [Google Scholar]
- 47.Marchesini G., Marzocchi R., Agostini F., Bugianesi E. 2005. Nonalcoholic fatty liver disease and the metabolic syndrome. Curr. Opin. Lipidol. 16: 421–427. [DOI] [PubMed] [Google Scholar]
- 48.Ota T., Gayet C., Ginsberg H. N. 2008. Inhibition of apolipoprotein B100 secretion by lipid-induced hepatic endoplasmic reticulum stress in rodents. J. Clin. Invest. 118: 316–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puri P., Mirshahi F., Cheung O., Natarajan R., Maher J. W., Kellum J. M., Sanyal A. J. 2008. Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology. 134: 568–576. [DOI] [PubMed] [Google Scholar]
- 50.Sreekumar R., Rosado B., Rasmussen D., Charlton M. 2003. Hepatic gene expression in histologically progressive nonalcoholic steatohepatitis. Hepatology. 38: 244–251. [DOI] [PubMed] [Google Scholar]
- 51.Letteron P., Fromenty B., Terris B., Degott C., Pessayre D. 1996. Acute and chronic hepatic steatosis lead to in vivo lipid peroxidation in mice. J. Hepatol. 24: 200–208. [DOI] [PubMed] [Google Scholar]
- 52.Crespo J., Cayon A., Fernandez-Gil P., Hernandez-Guerra M., Mayorga M., Dominguez-Diez A., Fernandez-Escalante J. C., Pons-Romero F. 2001. Gene expression of tumor necrosis factor alpha and TNF-receptors, p55 and p75, in nonalcoholic steatohepatitis patients. Hepatology. 34: 1158–1163. [DOI] [PubMed] [Google Scholar]
- 53.Sanyal A. J., Campbell-Sargent C., Mirshahi F., Rizzo W. B., Contos M. J., Sterling R. K., Luketic V. A., Shiffman M. L., Clore J. N. 2001. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 120: 1183–1192. [DOI] [PubMed] [Google Scholar]
- 54.Gramlich T., Kleiner D. E., McCullough A. J., Matteoni C. A., Boparai N., Younossi Z. M. 2004. Pathologic features associated with fibrosis in nonalcoholic fatty liver disease. Hum. Pathol. 35: 196–199. [DOI] [PubMed] [Google Scholar]
- 55.Thum T., Galuppo P., Wolf C., Fiedler J., Kneitz S., van Laake L. W., Doevendans P. A., Mummery C. L., Borlak J., Haverich A., et al. 2007. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 116: 258–267. [DOI] [PubMed] [Google Scholar]
- 56.Iliopoulos D., Fabbri M., Druck T., Qin H. R., Han S. Y., Huebner K. 2007. Inhibition of breast cancer cell growth in vitro and in vivo: effect of restoration of Wwox expression. Clin. Cancer Res. 13: 268–274. [DOI] [PubMed] [Google Scholar]
- 57.You M., Matsumoto M., Pacold C. M., Cho W. K., Crabb D. W. 2004. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology. 127: 1798–1808. [DOI] [PubMed] [Google Scholar]
- 58.Millar J. S., Stone S. J., Tietge U. J., Tow B., Billheimer J. T., Wong J. S., Hamilton R. L., Farese R. V., Jr., Rader D. J. 2006. Short-term overexpression of DGAT1 or DGAT2 increases hepatic triglyceride but not VLDL triglyceride or apoB production. J. Lipid Res. 47: 2297–2305. [DOI] [PubMed] [Google Scholar]
- 59.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14: 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griffin M. J., Wong R. H., Pandya N., Sul H. S. 2007. Direct interaction between USF and SREBP-1c mediates synergistic activation of the fatty-acid synthase promoter. J. Biol. Chem. 282: 5453–5467. [DOI] [PubMed] [Google Scholar]
- 61.Yu X. X., Murray S. F., Pandey S. K., Booten S. L., Bao D., Song X. Z., Kelly S., Chen S., McKay R., Monia B. P., et al. 2005. Antisense oligonucleotide reduction of DGAT2 expression improves hepatic steatosis and hyperlipidemia in obese mice. Hepatology. 42: 362–371. [DOI] [PubMed] [Google Scholar]
- 62.John M., Constien R., Akinc A., Goldberg M., Moon Y. A., Spranger M., Hadwiger P., Soutschek J., Vornlocher H. P., Manoharan M., et al. 2007. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature. 449: 745–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 14: 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshikawa T., Shimano H., Amemiya-Kudo M., Yahagi N., Hasty A. H., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., et al. 2001. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell. Biol. 21: 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McGarry J. D., Brown N. F. 1997. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur. J. Biochem. 244: 1–14. [DOI] [PubMed] [Google Scholar]
- 66.Grefhorst A., Hoekstra J., Derks T. G., Ouwens D. M., Baller J. F., Havinga R., Havekes L. M., Romijn J. A., Kuipers F. 2005. Acute hepatic steatosis in mice by blocking beta-oxidation does not reduce insulin sensitivity of very-low-density lipoprotein production. Am. J. Physiol. Gastrointest. Liver Physiol. 289: G592–G598. [DOI] [PubMed] [Google Scholar]
- 67.Jin X., Ye Y. F., Chen S. H., Yu C. H., Liu J., Li Y. M. 2009. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig. Liver Dis. 41: 289–297. [DOI] [PubMed] [Google Scholar]
- 68.Velasco G., Gomez del Pulgar T., Carling D., Guzman M. 1998. Evidence that the AMP-activated protein kinase stimulates rat liver carnitine palmitoyltransferase I by phosphorylating cytoskeletal components. FEBS Lett. 439: 317–320. [DOI] [PubMed] [Google Scholar]
- 69.Minokoshi Y., Kim Y. B., Peroni O. D., Fryer L. G., Muller C., Carling D., Kahn B. B. 2002. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 415: 339–343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.