Abstract
Several potentially atherogenic LDL subfractions present low affinity for the LDL receptor, which result in impaired plasma clearance. Electronegative LDL [LDL(−)] is one of these minor subfractions and the molecular basis for its reduced receptor affinity is not well understood. In the present study, high-resolution 2D-NMR spectroscopy has been employed to characterize the surface-exposed lysine residues of the apolipoprotein (apo)B-100 protein in both LDL(−) and LDL(+) subfractions. LDL(+) showed two populations of lysine residues, similar to those previously described in total LDL. “Normal” Lys have a pka of 10.4 whereas “active” Lys have a pka of 8.8 and have been suggested to be involved in receptor binding. In contrast to LDL(+), the LDL(−) subfraction presented a third type of Lys, named as “intermediate” Lys, with a different microenvironment and higher basicity (pka 10.7). These intermediate Lys cannot be reliably identified by 1D-NMR. Because the abundance of normal Lys is similar in LDL(+) and LDL(−), the intermediate Lys in the apoB-100 molcule of LDL(−) should come from a group of active Lys in LDL(+) particles that have a less basic microenvironment in the LDL(−) particle. These differences between LDL(+) and LDL(−) are indicative of a distinct conformation of apoB-100 that could be related to loss of affinity of LDL(−) for the LDL receptor.
Keywords: electronegative low density lipoprotein, apolipoprotein B-100, two-dimensional-nuclear magnetic resonance
LDL is the main transporter of the cholesterol in blood and its endothelial transmigration is the main factor responsible for the accumulation of plasma cholesterol in the sub-endothelial space (1). LDL consists of one molecule of apolipoprotein B-100 (apoB-100) (550 KDa) and a variable number of lipids (2500–3500 molecules/particle). As a consequence, LDL is a heterogeneous group of particles that vary in density, size, electric charge, and composition, depending on the relative lipid content (2). Several minor modified forms of LDL with increased atherogenicity have been described in blood and include small, dense LDL (sdlDL) (3), oxidized LDL (oxLDL) (4), and electronegative LDL [LDL(–)] (5, 6).
LDL(–) is a minor subfraction of LDL with atherogenic, apoptotic, and inflammatory properties whose proportion in plasma is increased in patients with high cardiovascular risk, such as those with hyperlipemia and diabetes (5). LDL(–) differs from LDL(+) in its higher susceptibility to aggregation (7), its capability to form lipid clusters (8), its secondary structure content of apoB-100 (9), and in the propensity of apoB-100 to form amyloid-like structures (10). Another atherogenic property of LDL(–) is its low affinity for the LDL receptor (LDLr) (6). This results in increased time of residence in plasma that could favor further atherogenic modifications. However, the mechanism underlying such low affinity is not well understood. It has been suggested that lysines in apoB-100 of LDL(–) involved in receptor binding could be either chemically modified in LDL(–) or not available for recognition by the LDL receptor due to conformation differences (6).
NMR has proved to be a powerful tool for studying the physicochemical characteristics of modified LDL. Particle size, particle aggregation and fusion, phospholipid degradation, and other structural features of apoB-100 have been studied by different NMR approaches. Reductive methylation of the amino groups of lysine residues with [13C]formaldehyde allows selective observation of the exposed lysine residues in LDL by means of 13C-NMR spectroscopy (11). Using one-dimensional (1D) NMR methods, two classes of exposed lysines have been detected in total LDL and in LDL fractions of different densities (12, 13). We report here two-dimensional (2D) NMR measurements on isolated LDL(+) and LDL(–) particles that show a third group of lysines in LDL(–) with a different pka that could be involved in their different atherogenic properties.
MATERIALS AND METHODS
Materials
All reagents were purchased from Sigma (Madrid, Spain) unless otherwise stated.
Isolation of LDL subfractions
Total LDL was isolated by sequential ultracentrifugation from pooled plasma from healthy volunteers, as described (14). The study was approved by the institutional Ethics Committee and subjects gave their written informed consent. Three independent pools were used to obtain the amount of LDL necessary for NMR analysis. To avoid the possibility of contamination with lipoprotein(a) the density range was 1.019–1.050 g/ml. All steps were performed at 4°C in the presence of 1 mM EDTA and 2 μM BHT. The concentration of LDL was expressed as g/L of apoB-100 measured by immunoturbidimetry (Roche Diagnostics). Total LDL was fractionated in LDL(+) and (LDL(–) fractions by preparative anion exchange chromatography in an AKTA-FPLC system (GE Healthcare) using a High Load 26/10 Q-Sepharose HP column, as described (15). LDL fractions were collected and concentrated using Amicon Ultra-4 (10 kDa) concentrators (Millipore). LDL subfractions of each pool were characterized by measuring composition including apoB-100, total cholesterol, triglycerides (Roche Diagnostics), phospholipids, and nonesterified fatty acids (NEFAs) (Wako Chemicals) and by agarose gel electrophoresis (Midigel, Biomidi), as described (15). Malondialdehyde (MDA)-Lys reactivity (LDLs at 0.5 g apoB-100/L) was measured by a commercial ELISA (Mercodia) that uses the monoclonal antibody 4E6 directed against this conformational epitope in oxidized apoB-100. Trinitrobenzene sulphonic (TNBS) reactivity, which measures the number of free Lys, was determined as described (7), using LDL at 0.5 g apoB-100/L previously dialyzed against 4% NaHCO3 pH 8.4. NEFA-loaded LDL(+) (hereafter named NEFA-LDL) was prepared by incubation of LDL(+) with a mixture of NEFA at 0.5 mmol/L for 4 h at 37°C, as described (6).
Reductive methylation of Lys and isotopic labeling
The reductive methylation procedure described by Lund-Katz et al. (11) was used to introduce 13C-methyl groups in the amino groups of the exposed Lys in apoB-100. Reductive methylation with [13C]formaldehyde converted up to 225 Lys to the dimethylamino derivative, indicating that about 2/3 of the 357 total Lys in apoB-100 are accessible (11). These authors demonstrated that methylation of 11–13% of Lys residues is suitable for the characterization of apoB-100 without altering its conformation. To keep the total level of methylated lysines below 12%, the procedure was strictly standardized, as described below. LDL (final concentration 1 g apoB-100/L) dialyzed in 0.15 M NaCl, 1 mM EDTA, 2 μM BHT, and 0.02% sodium azide pH 7.4, was incubated with NaCNBH3 (final concentration 10 mM) and with 13C-formaldehyde (final concentration 0.167 μM). Because 1 mg of apoB-100 contains 0.65 × 10−6 mol of Lys, the ratio formaldehyde/Lys was 0.25. Higher ratios modified the electrophoretic mobility of LDL in agarose gels. The modification of all Lys residues exposed (60% of all Lys) was obtained at a formaldehyde/Lys molar ratio of 10/1, as described by Lund-Katz et al. (11). 13C-formaldehyde was doped with a minor amount of 14C-formaldehyde (0.5%) and 14C counts were used to monitor formaldehyde incorporation into apoB-100. This mixture was incubated overnight in gentle agitation at 4°C. Afterwards, samples were extensively dialyzed in 0.15 M NaCl, 1 mM EDTA, 2 μM BHT, and 0.02% sodium azide, pH 7.4, and concentrated using Amicon Ultra-4 (10 kDa) concentrators. The final concentration of LDL samples was 11.5 g of apoB-100/L. Agarose gel electrophoresis and composition of labeled LDL subfractions were performed to rule out the possibility of extensive modification of lipoproteins.
NMR spectroscopy
Five hundred μl of LDL subfractions (each 10% reductive-methylated) at 11 mg/ml of apoB-100 in 0.15 M NaCl, 1 mM EDTA, 0.1% sodium azide, pH 7.4 were mixed with 2H2O (5% v/v) and placed in 5 mm NMR tubes. External DSS (2,2-Dimethyl-2-silapentane-5-sulfonic acid) was used for the direct calibration of 1H chemical shifts. 13C chemical shifts in the 2D heteronuclear single quantum correlation spectra (HSQC) were calibrated indirectly, as described (16). NMR spectra were acquired at 37°C on a Bruker AVANCE III 800 MHz spectrometer with a xyz-gradient QXI probe. pH titration was performed by adding 1–5 μl of 0.5 N NaOH directly into the NMR tube, and the pH was measured with a thin electrode. The intensity of the signals was evaluated by volume integration. 1H-13C HSQC spectra were acquired with 40–300 scans, a repetition time of 2 s, and 32–64 complex points in the 13C dimension (total experiment time was 1.5–6 h). Spectra for quantification and for the titrations were acquired with a 13C-spectral width of 12 ppm, centered at 41 ppm, to increase the resolution for the lysine aminomethyl signals. The pulse sequence used gradients for coherence selection (echo-antiecho) and adiabatic 13C inversion pulses. Signal intensity was measured by cross-peak volume integration using the procedure implemented in Topspin (Bruker). The methyl 1H chemical shifts also differed for the distinct types of Lys, allowing sufficient separation between the cross-peaks for the individual quantification of their intensities without the need for deconvolution methods, and with less possibility of bias by unknown overlapping signals. Of note, the 13C-HSQC experiment derives its intensity from 1H[13C], rather than 13C, polarization. This affords a factor γH/γC of approximately 4-fold intrinsic sensitivity, and moreover, allows for shorter repetition times because the proton longitudinal time is smaller than the carbon one for slowly tumbling molecules. Therefore, relative 13C-HSQC signal intensities should be much less biased from possible differential T1 relaxation than 13C- spectra. The uncertainty in the signal intensity was evaluated by integration of three regions containing only noise, and was smaller than 1% of the sum of the intensities of all the signals observed in the corresponding amino-methyl regions of the spectra shown in Fig. 1.
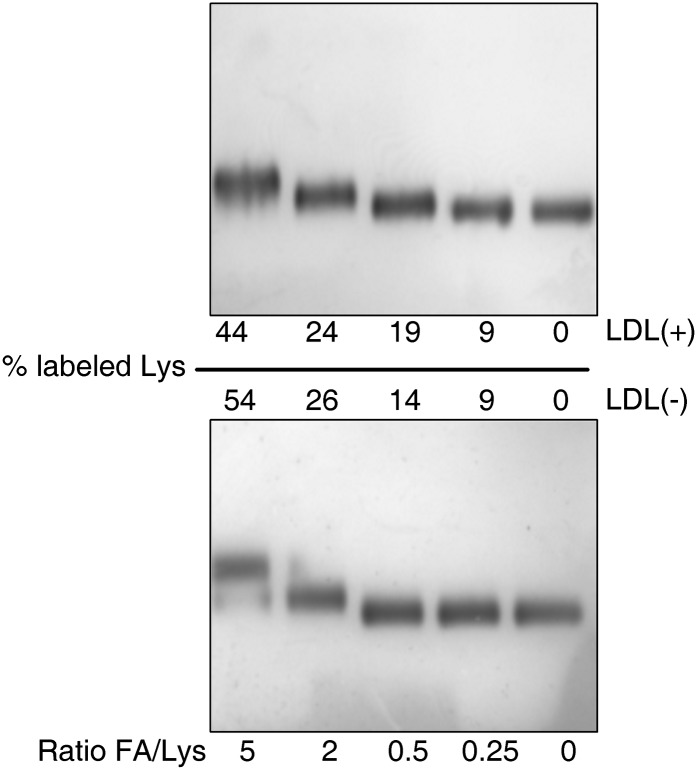
Fig. 1.
Representative electrophoresis in agarose gels of LDL subfractions [upper gel LDL(+); lower gel LDL(–)] labeled with increasing formaldehyde/lysine (FA/Lys) ratio. LDLs were labeled as indicated in Material and Methods. Electrophoresis was performed using commercial gels (Midigel, Biomidi) at 90 V for 90 min in a cold room with 5 μl of labeled LDLs (0.5 g apoB-100/L). Samples were stained with Sudan Black according to the manufacturer's instructions.
RESULTS
LDL composition
The composition of LDL subfractions isolated from each pool is shown in Table 1. Composition results and TNBS reactivity agreed with previously reported data (7, 14). LDL(–) contained more triglyceride, more NEFA, and less apoB-100 proportion. MDA-Lys and TNBS reactivity showed that Lys in both LDL subfractions were not differentially modified. NEFA-LDL had the same composition than LDL(+) (data not shown), except for the NEFA content, which was similar to that of LDL(–) (36.1 ± 6.5 mol NEFA/mol apoB, n = 3).
TABLE 1.
LDL subfraction composition
| LDL(+) | LDL(–) | |
|---|---|---|
| Cholesterol (%) | 41.3 ± 1.8 | 41.5 ± 2.3 |
| Triglyceride (%) | 6.9 ± 1.6 | 9.8 ± 1.3 |
| Phospholipid (%) | 27.8 ± 2.1 | 26.9 ± 2.1 |
| NEFA (mol/mol apoB) | 12.6 ± 3.0 | 34.1 ± 5.0 |
| ApoB-100 (%) | 24.9 ± 2.0 | 23.1 ± 2.6 |
| TNBS reactivity [% vs LDL(+)] | 100 | 98 ± 2 |
| MDA-Lys reactivity (mU/L) | 15.9 ± 2.3 | 17.0 ± 6.4 |
Data are the mean ± SD of three independent samples used for 2D-NMR analysis.
Isotopic labeling
Fig. 1 shows a representative agarose gel of LDL subfractions [upper gel, LDL(+); lower gel, LDL(–)] labeled with different formaldehyde/Lys ratio. The percentage of labeled Lys residues was quantified isotopically and is indicated in the figure. Increased formaldehyde/Lys ratio resulted in more extensive labeling and increased electrophoretic mobility, indicative of changes in the secondary structure of apoB (11). Samples used for 2D-NMR spectroscopy had less than 12% of labeled Lys, as recommended by Lund-Katz et al. (11), and both fractions were labeled to a similar extent [LDL(+) 8.9 ± 1.9%; LDL(–) 8.8 ± 2.2%, 14C counts measured in triplicate]. No increase of electrophoretic mobility (Fig. 1) and no change in LDL composition (data not shown) were observed after labeling with a formaldehyde/Lys ratio of 0.25.
2D-NMR spectroscopy
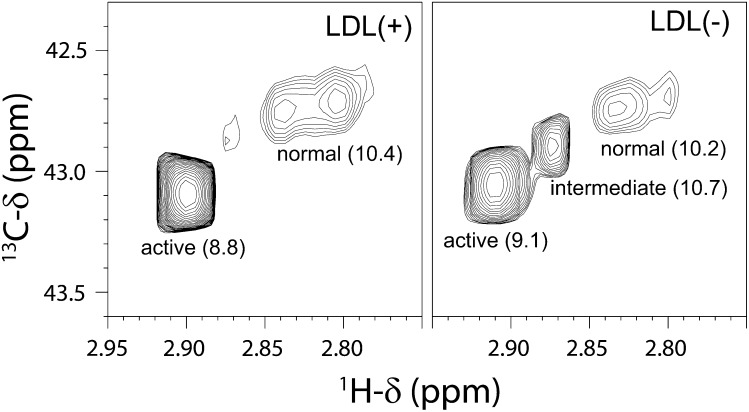
The 2D-1H-13C HSQC spectra of the LDL particles showed a number of cross-peaks with different intensities. Most of them can be attributed to protons bound to the 1% natural abundance 13C carbons of the most mobile parts of the abundant lipid chains. The cross-peaks belonging to the fraction of 13C-methylated exposed lysines of the apo-B100 protein were identified based on the expected 1H and 13C chemical shifts (around 2.9 and 43.0 ppm, respectively) considering its chemical structure and the previously reported carbon chemical shifts in 13C-methylated LDL particles (11). The spectrum of the LDL(+) subfraction (Fig. 2, left panel) shows a major peak in the amino-methyl region, with 13C chemical shift and pka value (Fig. 3, Table 2) almost identical to the signal previously assigned to “active” lysines in the 1D spectra of LDL particles (11). A pair of signals with the 13C chemical shift of the “normal” lysines is clearly resolved in the 1H dimension of the 2D spectrum. The pka of the signal with the largest 1H chemical shift is the same as reported for normal lysines, whereas the other signal changes frequencies with pH such that it overlaps and becomes indistinguishable from the former one. The relative proportion of each group of Lys differs from those measured by Lund-Katz et al. (11) on 1D spectra of total LDL. In the 2D spectra of LDL(+) subfraction we measured 11 and 89% of normal and active Lys, respectively. Assuming that only 9% of the exposed Lys were labeled (32 Lys, see Fig. 1), this corresponds to 4 normal Lys and 28 active Lys (Table 1). This is different from the published data where, at a similar labeling level, 14 normal and 10 active Lys were observed (11). This difference is not due to the LDL fractionation, as total LDL contains around 95% of LDL(+) component. Rather, these differences are probably due to the higher sensitivity, resolution, and reliability of the signal intensity measurement in the HSQC spectra as compared with the 1D-13C direct detection ones.
Fig. 2.
Contour plots of the amino-methyl regions of the 1H-13C HSQC NMR spectra of LDL(+) and LDL(–) subfractions. Left: LDL(+); right: LDL(–). The assignment to the Lys types and the pKa values (in parenthesis) are indicated for each signal. The two spectra are plotted just above the overall noise level.
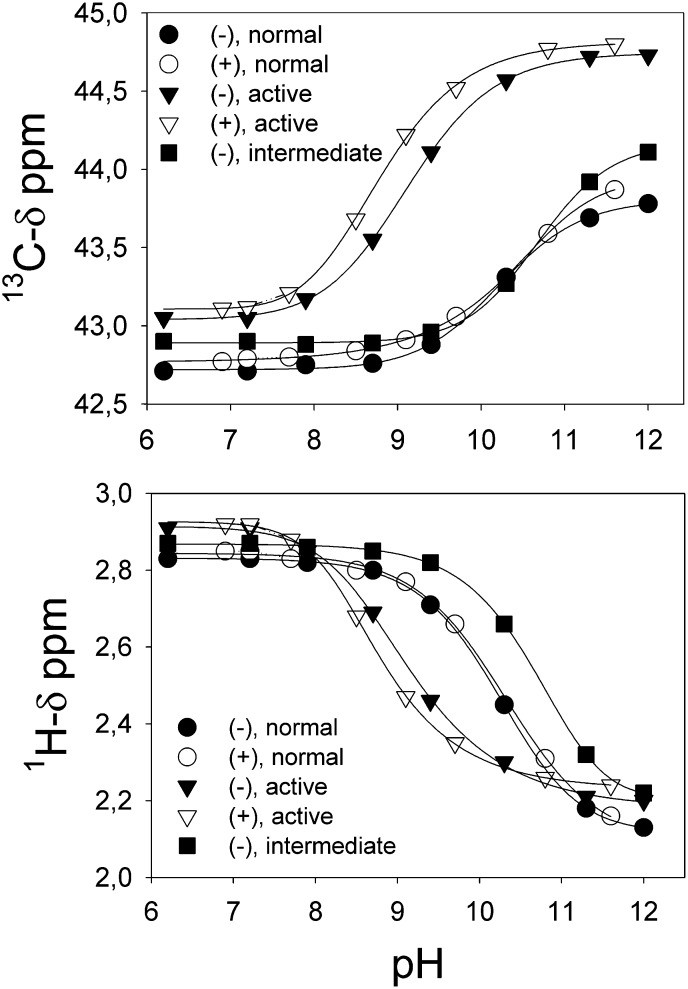
Fig. 3.
pH titrations of the different Lys types measured by the change in their corresponding 13C (left) and 1H (right) chemical shifts. pH titration was performed by adding 1–5 μl of 0.5 N NaOH directly into the NMR tube, and the pH was measured with a thin electrode. Data were fitted to a four-parameter sigmoid equation using SigmaPlot 8.0.
TABLE 2.
Summary of chemical shifts (ppm), pKa values and number of labeled Lys of the three types identified in the 1H-13C-HSQC spectra of LDL subfractions after reductive methylation
| “Active” Lys |
“Normal” Lys |
“Intermediate” Lys |
||||
|---|---|---|---|---|---|---|
| ppma | 13C | 1H | 13C | 1H | 13C | 1H |
| LDL(+) | 43.09 | 2.90 | 42.73 | 2.83, 2.80 | — | — |
| LDL(–) | 43.05 | 2.91 | 42.74 | 2.83 | 42.90 | 2.87 |
| pKab | 13C | 1H | 13C | 1H | 13C | 1H |
| LDL(+) | 8.8 | 8.8 | 10.4 | 10.3 | — | — |
| LDL(–) | 9.1 | 9.1 | 10.2 | 10.2 | 10.7 | 10.7 |
| # Lys (%)c | ||||||
| LDL(+) | 28 (89%) | 4 (11%) | 0 (0%) | |||
| LDL(–) | 23 (75%) | 4 (10%) | 5 (15%) | |||
The estimated error in the chemical shifts is smaller than 0.006 ppm for protons and 0.009 ppm for carbons.
The errors in the pH measurements, and in the fitting of the curves are both smaller than 0.1 unit.
The uncertainty in signal intensity is estimated to be smaller than 1%.
Interestingly, the spectrum of LDL(–) shows a third cross-peak corresponding to a different type of Lys (Fig. 2, right panel). This group accounts for 15% of the total amino-methyl signal intensity and has a pka 10.7 (Fig. 3, Table 1). Because its cross-peak is between those for previously described Lys groups (Fig. 1), we call this type “intermediate” Lys. The other signals behave like the corresponding ones of LDL(+) subfraction but have different relative proportions (11% and 74% for active and normal lysines, respectively). The measured populations in LDL(–) correspond to 4 normal Lys, 23 active Lys, and 5 intermediate Lys. A summary of chemical shifts, pka values, populations, and number of Lys is shown in Table 2. It is worth mentioning that the pka values obtained from both 1H and 13C chemical shifts upon pH titration are identical within the experimental error. A very weak signal with chemical shifts similar to the intermediate Lys is observed in the spectrum of LDL(+), which could be a small population of this kind of lysine (less than 1%).
Because LDL(–) has a higher content of NEFA than LDL(+), the appearance of intermediate Lys in LDL(–) could be due to the presence of the carboxyl groups of NEFA in the vicinity of some of the exposed amino groups, changing their chemical environment and basicity. This possibility was evaluated by NMR analysis of NEFA-loaded LDL(+) with a content of NEFA similar to that of LDL(–). The spectrum of NEFA-LDL is, within error, the same as the spectrum of LDL(+) (supplementary Fig. I), with 87% active Lys, 12% normal Lys, and 1% intermediate Lys.
DISCUSSION
Lys residues in apoB-100 are involved in LDL recognition by its receptor (17) and their chemical modification by different mechanisms abolishes their binding. Acetylation, oxidation, or nonenzymatic glycosylation promote derivatization of Lys by acetate, MDA, or glucose, inducing a progressive loss of affinity between LDL and its receptor (18–20). Based on 1D-13C-NMR measurements of methylated LDL particles, Lund-Katz et al. (11) described two types of exposed Lys in apoB-100. These two types differed in their 13C-methyl resonance frequencies (42.8 ppm and 43.2 ppm), relative populations (70 and 30%) and basicity (with pka values of 10.5 and 8.9, respectively). The lower pka indicated a more basic microenvironment within the apoB-100 molecule. This group was termed active Lys, the other being normal Lys, and was suggested to be involved in LDLr recognition (11, 12). These authors demonstrated that plasma LDL subspecies with low affinity toward the LDL receptor, such as small, dense, or large, buoyant LDL subfractions (21), have altered ionization of basic amino acids with a reduced proportion of active Lys (13). Hence, a decrease in the number of active Lys in LDL would imply a loss of binding affinity toward LDLr. LDL(–) is a heterogeneous population of LDL particles that distributes mainly in the most dense and most buoyant fractions of total LDL (22, 23); thus, current results suggest that a similar mechanism underlies the poor binding affinity of LDL(–) to the LDLr.
Because the abundance of normal Lys is similar in LDL(+) and LDL(–), we hypothesize that the intermediate Lys in the apoB-100 molecule of LDL(–) come from a group of active Lys in LDL(+) particles that has a different microenvironment in the LDL(–) particle as a result of conformational differences. The higher pka value of this population of Lys indicates a stabilization of the -N+H3 group, reflecting a less basic microenvironment (11). It has been reported that active Lys are clustered in basic microenvironments, some of them being involved in LDLr recognition (11–13). These conformational differences between LDL(+) and LDL(–) agree with the observation of misfolded apoB-100 in LDL(–) (9), a property that increases its overall amyloidogenic propensity (10) and could also be involved in its higher susceptibility to particle aggregation (7, 8).
The fact that the new intermediate Lys type observed in this work has not been previously reported in small, dense LDL or large, buoyant LDL (12, 13) is probably due to the lower sensitivity and resolution of the 1D-13C-NMR method compared with our 2D-1H-13C HSQC method.
Another possible mechanism responsible for the partial loss of LDLr affinity is the chemical modification of Lys in apoB-100 of LDL(–). There is some controversy regarding the presence of mild oxidative modification in LDL(–) (5); however, the degree of oxidation reported by most authors is lower (23–25) than that necessary for extensive modification of Lys by MDA (17, 19). In agreement with these observations, current data of MDA-Lys reactivity showed that Lys in LDL(–) were not differentially modified by oxidation-related mechanisms compared with LDL(+). Indeed, the lack of differences in TNBS reactivity in the current and previous studies indicates that the proportion of free Lys is similar in both LDL subfractions (6, 26).
Because the content of NEFA is almost three times increased in LDL(–) compared with LDL(+) and the NEFA carboxyl groups are likely to be on the surface of the particle, NEFA content could be a major contributor to the increased heterogeneity of amino groups in LDL(–). This possibility is ruled out by the measurement of indistinguishable HSQC spectra of LDL(+) and NEFA-loaded LDL(+).
In summary, our results indicate that there are conformational differences between the apoB-100 protein in LDL(+) and LDL(–) particles that affect the basicity of its solvent-exposed Lys. These differences could be responsible for the partial loss of affinity of LDL(–) to the LDLr. Our results also show that 2D 1H-13C-HSQC spectra of labeled apoB-100 reliably identify different Lys types with high sensitivity and resolution, and could be used to uncover other hidden molecular signatures of LDL particles that might be relevant for their atherogenic properties.
Supplementary Material
Footnotes
Abbreviations:
- apoB-100
- apolipoprotein B-100
- BHT
- butylated hydroxytoluene
- DSS
- 2,2-Dimethyl-2-silapentane-5-sulfonic acid
- HSQC
- heteronuclear single quantum correlation spectra
- LDL(−)
- electronegative LDL
- LDL(+)
- native LDL
- LDLr
- LDL receptor
- MDA
- malondialdehyde
- NEFA
- nonesterified fatty acid
- oxLDL
- oxidized LDL
- sdLDL
- small, dense LDL
- TNBS
- trinitrobenzene sulphonic acid
This work was supported by grants from the Ministerio de Sanidad/Instituto de Salud Carlos III/FIS PI060500 and PI070148. S.V. acknowledges a grant from Fundación Mutua Madrileña (FMM-08). S.B. and J.L.S-Q. are recipients of personal grants CP040110 and CP060220 from Ministerio de Sanidad. C.B. is recipient of a personal grant from the Ministerio de Educación y Ciencia AP2004-1468. C.B., S.B., J.O-L. and J.L.S-Q. are members of the 2009-SGR-1205 Research Group from the Generalitat de Catalunya. S.V. is member of the 2009-SGR-00761 Research Group from the Generalitat de Catalunya. The Structural Biology Unit of CIC bioGUNE was partly supported by an ETORTEK-2008 grant from SPRI.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Tabas I., Williams K. J., Borén J. 2007. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications. Circulation. 116: 1832–1844. [DOI] [PubMed] [Google Scholar]
- 2.Segrest J. P., Jones M. K., De Loof H., Dashti N. 2001. Structure of apolipoprotein B-100 in low density lipoproteins. J. Lipid Res. 42: 1346–1367. [PubMed] [Google Scholar]
- 3.Krauss R. M. 1995. Dense low density lipoproteins and coronary artery disease. Am. J. Cardiol. 75: 53B–57B. [DOI] [PubMed] [Google Scholar]
- 4.Holvoet P. 2004. Oxidized LDL and coronary heart disease. Acta Cardiol. 59: 479–484. [DOI] [PubMed] [Google Scholar]
- 5.Sánchez-Quesada J. L., Benítez S., Ordóñez-Llanos J. 2004. Electronegative low-density lipoprotein. Curr. Opin. Lipidol. 15: 329–335. [DOI] [PubMed] [Google Scholar]
- 6.Benítez S., Villegas V., Bancells C., Jorba O., González-Sastre F., Ordóñez-Llanos J., Sánchez-Quesada J. L. 2004. Impaired binding affinity of electronegative low-density lipoprotein (LDL) to the LDL receptor is related to nonesterified fatty acids and lysophosphatidylcholine content. Biochemistry. 43: 15863–15872. [DOI] [PubMed] [Google Scholar]
- 7.Bancells C., Benítez S., Villegas S., Jorba O., Ordóñez-Llanos J., Sánchez-Quesada J. L. 2008. Novel phospholipolytic activities associated with electronegative low-density lipoprotein are involved in increased self-aggregation. Biochemistry. 47: 8186–8194. [DOI] [PubMed] [Google Scholar]
- 8.De Spirito M., Brunelli R., Mei G., Bertani F. R., Ciasca G., Greco G., Papi M., Arcovito G., Ursini F., Parasassi T. 2006. Low density lipoprotein aged in plasma forms clusters resembling subendothelial droplets: aggregation via surface sites. Biophys. J. 90: 4239–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parasassi T., Bittolo-Bon G., Brunelli R., Cazzolato G., Krasnowska E. K., Mei G., Sevanian A., Ursini F. 2001. Loss of apoB-100 secondary structure and conformation in hydroperoxide rich, electronegative LDL(–). Free Radic. Biol. Med. 31: 82–89. [DOI] [PubMed] [Google Scholar]
- 10.Parasassi T., De Spirito M., Mei G., Brunelli R., Greco G., Lenzi L., Maulucci G., Nicolai E., Papi M., Arcovito G., et al. 2008. Low density lipoprotein misfolding and amyloidogenesis. FASEB J. 22: 2350–2356. [DOI] [PubMed] [Google Scholar]
- 11.Lund-Katz S., Ibdah J. A., Letizia J. Y., Thomas M. T., Phillips M. C. 1988. A 13C NMR characterization of lysine residues in apolipoprotein B and their role in binding to the low density lipoprotein receptor. J. Biol. Chem. 263: 13831–13838. [PubMed] [Google Scholar]
- 12.Aviram M., Lund-Katz S., Phillips M. C., Chait A. 1988. The influence of the triglyceride content of low density lipoprotein on the interaction of apolipoprotein B-100 with cells. J. Biol. Chem. 263: 16842–16848. [PubMed] [Google Scholar]
- 13.Lund-Katz S., Laplaud P. M., Phillips M. C., Chapman M. J. 1998. Apolipoprotein B-100 conformation and particle surface charge in human LDL subspecies: implication for LDL receptor interaction. Biochemistry. 37: 12867–12874. [DOI] [PubMed] [Google Scholar]
- 14.Havel R. J., Eder H. A., Bragdon J. H. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sánchez-Quesada J. L., Camacho M., Antón R., Benítez S., Vila L., Ordóñez-Llanos J. 2003. Electronegative LDL of FH subjects: chemical characterization and induction of chemokine release from human endothelial cells. Atherosclerosis. 166: 261–270. [DOI] [PubMed] [Google Scholar]
- 16.Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 6: 135–140. [DOI] [PubMed] [Google Scholar]
- 17.Weisgraber K. H., Innerarity T. L., Mahley R. W. 1978. Role of lysine residues of plasma lipoproteins in high affinity binding to cell surface receptors on human fibroblasts. J. Biol. Chem. 253: 9053–9062. [PubMed] [Google Scholar]
- 18.Mahley R. W., Innerarity T. L., Weisgraber K. H. 1980. Alterations in metabolic activity of plasma lipoproteins following selective chemical modification of the apoproteins. Ann. N. Y. Acad. Sci. 348: 265–280. [DOI] [PubMed] [Google Scholar]
- 19.Henriksen T., Mahoney E. M., Steinberg D. 1983. Enhanced macrophage degradation of biologically modified low density lipoprotein. Arteriosclerosis. 3: 149–159. [DOI] [PubMed] [Google Scholar]
- 20.Klein R. L., Laimins M., Lopes-Virella M. F. 1995. Isolation, characterization, and metabolism of the glycated and nonglycated subfractions of low-density lipoproteins isolated from type I diabetic patients and nondiabetic subjects. Diabetes. 44: 1093–1098. [DOI] [PubMed] [Google Scholar]
- 21.Nigon F., Lesnik P., Rouis M., Chapman M. J. 1991. Discrete subspecies of human low density lipoproteins are heterogeneous in their interaction with the cellular LDL receptor. J. Lipid Res. 32: 1741–1753. [PubMed] [Google Scholar]
- 22.Sánchez-Quesada J. L., Benítez S., Otal C., Franco M., Blanco-Vaca F., Ordóñez-Llanos J. 2002. Density distribution of electronegative LDL in normolipemic and hyperlipemic subjects. J. Lipid Res. 43: 699–705. [PubMed] [Google Scholar]
- 23.Chen H. H., Hosken B. D., Huang M., Gaubatz J. W., Myers C. L., Macfarlane R. D., Pownall H. J., Yang C. Y. 2007. Electronegative LDLs from familial hypercholesterolemic patients are physicochemically heterogeneous but uniformly proapoptotic. J. Lipid Res. 48: 177–184. [DOI] [PubMed] [Google Scholar]
- 24.Chen C. H., Jiang T., Yang J. H., Jiang W., Lu J., Marathe G. K., Pownall H. J., Ballantyne C. M., McIntyre T. M., Henry P. D., et al. 2003. Low-density lipoprotein in hypercholesterolemic human plasma induces vascular endothelial cell apoptosis by inhibiting fibroblast growth factor 2 transcription. Circulation. 107: 2102–2108. [DOI] [PubMed] [Google Scholar]
- 25.Damasceno N. R., Sevanian A., Apolinário E., Oliveira J. M., Fernandes I., Abdalla D. S. 2006. Detection of electronegative low density lipoprotein (LDL-) in plasma and atherosclerotic lesions by monoclonal antibody-based immunoassays. Clin. Biochem. 39: 28–38. [DOI] [PubMed] [Google Scholar]
- 26.Demuth K., Myara I., Chappey B., Vedie B., Pech-Amsellem M. A., Haberland M. E., Moatti N. 1996. A cytotoxic electronegative LDL subfraction is present in human plasma. Arterioscler. Thromb. Vasc. Biol. 16: 773–783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.