Abstract
A large scale profiling and analysis of glycerophospholipid species in macrophages has facilitated the identification of several rare and atypical glycerophospholipid species. By using liquid chromatography tandem mass spectrometry and comparison of the elution and fragmentation properties of the rare lipids to synthetic standards, we were able to identify an array of ether-linked phosphatidylinositols (PIs), phosphatidic acids, phosphatidylserines (PSs), very long chain phosphatidylethanolamines (PEs), and phosphatidylcholines (PCs) as well as phosphatidylthreonines (PTs) and a wide collection of odd carbon fatty acid-containing phospholipids in macrophages. A comprehensive qualitative analysis of glycerophospholipids from different macrophage cells was conducted. During the phospholipid profiling of the macrophage-like RAW 264.7 cells, we identified dozens of rare or previously uncharacterized phospholipids, including ether-linked PIs, PSs, and glycerophosphatidic acids, PTs, and PCs and PTs containing very long polyunsaturated fatty acids. Additionally, large numbers of phospholipids containing at least one odd carbon fatty acid were identified. Using the same methodology, we also identified many of the same species of glycerophospholipids in resident peritoneal macrophages, foam cells, and murine bone marrow derived macrophages.
Keywords: lipidomics, ether-linked phospholipids, electrospray ionization, atypical lipids
Ether-linked phospholipids may contain either an alkyl ether or vinyl ether bond at the sn-1 position of the glycerol backbone. These lipids are mostly found in both phosphatidylcholine (PC) and phosphatidylethanolamine (PE) in a variety of mammalian cell types, including macrophages (1–3). It is widely accepted that in most tissues, ether-linked PC exists mostly as plasmanylcholine (1-O-alkyl-2-acyl-sn-glycero-3-phosphocholine), with the exception of myocardia, whereas PE exists as plasmenylethanolamine. (1-O-alk-1’-enyl-2-acyl-sn-glycero-3-phosphoethanolamine). Vinyl-ether bearing phospholipids are also known as plasmalogens. In plasmalogens, the sn-2 position is usually occupied by PUFAs. Typically, the sn-1 positions in plasmanyl and plasmenyl lipids are occupied by either 16 or 18 carbon ether or vinyl ether moieties. Plasmalogen phospholipids affect membrane fluidity and fusion and the ether lipids alterations are associated with several cellular dysfunctions and diseases such as Alzheimer's, Down syndrome, and cerebro-hepato-renal (Zellweger) syndrome (4–6). Responsive cell types such as macrophages have relatively high content of plasmalogen lipids. Due to the high levels of PUFA in plasmalogens they are considered a storage for long-chain PUFAs and especially arachidonic acid, which can be released by plasmalogen-specific PLA2(iPLA2) into free arachidonic and docosahexaenoic acids which are further metabolized to second messenger molecules like eicosanoids and prostaglandins (7).
Mass spectrometry has been the analytical method of choice for the characterization of lipid molecules, especially after introduction of the “soft” ionization techniques of MALDI (8) and ESI (9). The great potential of ESI-MS for analysis and characterization of nonvolatile and labile lipid molecules from biological extracts has been utilized extensively (9–12), including the detection of some rare and unusual lipids (13). By employing phospholipid class separation and tandem mass spectrometry (LC/MS/MS), we were able to identify a number of previously not reported ether-linked phosphatidylserine (PS) and phosphatidic acid (PA) together with detection of ether phosphatidylinositol (PI) (14) and phosphatidylthreonine (PT) (15) in extracts from RAW264.7 cells, foam cells, murine bone marrow derived macrophages (BMDM) and murine resident peritoneal macrophages (RPM). In the course of the analysis, we also identified some plasmanyl and plasmenyl PC and PE lipids containing very long-chain fatty acids (VLCFA) (24 C-atom and more). The motivation of this study was to identify novel and atypical lipid species in a variety of commonly used macrophage preparations. This study is not intended to provide a comprehensive accounting of all of the glycerophospholipid species in these cells, which continues as an ongoing project.
MATERIALS AND METHODS
Materials
37:4 PI (1-heptadecanoyl-2-(5Z,8Z,11Z,14Z-eicosatetraenoyl)-sn-glycero-3-phospho-(1’-myo-inositol)), 38:4e PI (1-octadecyl-2-(5Z,8Z,11Z,14Z-eicosatetraenoyl)-sn-glycero-3-phospho- (1’-myo-inositol)), 34:1e PC (1-hexadecyl-2-(9Z-octadecenoyl)- sn-glycero-3-phosphocholine), 34:1p PC (1-(1Z-octadecenyl)- 2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine), 34:1e PE (1-hexadecyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoethanolamine), and 34:1p PE (1-(1Z-octadecenyl)-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoethanolamine) were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL). HPLC grade solvents were purchased from VWR (West Chester, PA) and used without further purification. BMDM, foam, and RAW 264.7 cells were acquired as cell pellets prepared by established protocols. Briefly, BMDMs were harvested from tibias and femurs of 2-month-old C57BL6 male mice. They were suspended in Bone Marrow-Derived Macrophage Growth Medium (BMDMGM), plated on 100 mm Petri dishes and maintained at 37°C in a humidified incubator for 4 days. On day 4, cells were washed with RPMI medium and maintained at 37°C in BMDMGM for 2 more days, after which the macrophages were plated on 100 mm tissue culture dishes at a density of 5 × 106 per plate. Foam cells were elicited from male mice (B6.129S7-LDLrtm1 Her/J) (3–5 weeks of age; Jackson Laboratory) after being placed on a high-cholesterol diet (# TD96121, Harlan Teklad) for 10 weeks. Mice were injected with 2.5 ml thioglycollate intraperitoneally and foam cells were harvested 4 days post injection. RAW 264.7 cells were maintained essentially as described elsewhere (12).
RPM culture
All studies involving animals were conducted with the approval of the Institutional Animal Care and Use Committee of Vanderbilt University. Female ICR (CD-1) mice (25–30g) were obtained form Harlan (Indianapolis, IN). Cells were obtained by peritoneal lavage as described previously (16) and suspended at a density of 2–3 × 106 cells/ml (cells from one mouse per 2 ml) in Minimal Essential Alpha Medium supplemented with GlutaMax (Gibco), 10% heat-inactivated fetal calf serum (Atlas Biologicals, Norcross, GA), and 100 units/ml penicillin and 0.10 mg/ml streptomycin (Sigma, St.Louis, MO) (α-MEM/FCS). The cell suspension was plated on 60 mm tissue culture plates at a density of 6ml/plate and incubated for 2 h at 37°C in a humidified 5% CO2 atmosphere. Nonadherent cells were removed by washing the plates four times with PBS, and the cultures were then incubated overnight in fresh α-MEM/FCS.
Extraction of macrophage lipids
Phospholipids were extracted using a modified Bligh and Dyer procedure (17). Typically, between 1 and 3 × 106 cells per sample were used in this protocol. The method is suitable for extraction from cell culture plates after aspirating the medium and washing the adhered cells twice with 5 ml ice cold 1× PBS. Cells are then scraped in 1 ml of 1× PBS, and centrifuged (600 g, 4°C, 5 min). PBS is aspirated and the cell pellet is extracted with 800 μl of cold 0.1 N HCl: MeOH (1:1) and 400 μl of cold CHCl3 with vortexing (1 min) followed by centrifugation (5 min, 4°C, 18,000 g). The lower organic phase is then isolated and solvent evaporated (Labconco Centrivap Concentrator, Kansas City, MO). The extraction procedure includes acidification (0.05N HCl) in the aqueous phase that aids in the lysolipids and acidic lipids recovery.
LC/MS
Class separation of glycerophospholipids was achieved by the use of a previously published LC/MS technique (18). After extraction and solvent evaporation (as described above) the resulting lipid film is dissolved in 100 μl of IPA:Hexane:100 mM NH4CO2H(aq) 58:40:2 (mobile phase A). For the lipid screens, we utilized an Applied Biosystems/ MDS SCIEX 4000 Q TRAP hybrid triple quadrupole/ linear ion trap mass spectrometer (Applied Biosystems, Foster City, CA). Coupled to this instrument were a Shimadzu (Shimadzu Scientific Instruments, Inc., Columbia, MD) HPLC system consisting of a SCL 10 AVP controller, two LC 10 ADVP pumps and a CTC HTC PAL autosampler (Leap Technologies, Carrboro, NC). All samples shown were separated on a Phenomenex (Phenomenex, Torrance, CA) Luna Silica column (2 × 250 mm, 5 micron particle size) using a 20 μl sample injection. Lipids were separated using a binary gradient program consisting of IPA:Hexane:100 mM NH4CO2H(aq) 58:40:2 (mobile phase A), and IPA:Hexane:100 mM NH4CO2H(aq) 50:40:10 (mobile phase B). The following LC gradient was used: 0–5min, B = 50%; 5–30min, B = 50%–100%; 30–40min, B = 100%; 40–41min, B = 100%–50%; 41–50min, B = 50%. The mobile phase was infused at a flow rate of 0.3 ml/min. The MS spectra were acquired in negative ionization mode using a turbo spray source operated at 450°C with an ion voltage of –3500V, and nitrogen as curtain and nebulizer gas. The curtain gas was 30 L/h, and ion source gas 1 and 2 were both 50 L/h. The declustering potential was −110 V and the collision energy was −5 V. Scan type: EMS, unit resolution for Q1; Scan rate: 1000 amu/s; Scan range from m/z 350–1200, with the ion trap set for dynamic fill time.
Lipid identification by LC/MS/MS
Large scale screening and identification of glycerophospholipids was accomplished by LC/MS/MS analysis using the information dependent analysis (IDA) software option within the Analyst software package (Applied Biosystems) and individual fragmentation. In conjunction with the above mentioned LC/MS phospholipid separation protocol, MS/MS experiments were run over the following m/z regions: 350-600 m/z, 600-800 m/z, 800-1000 m/z, and 1000-1200 m/z. Using this technique, upwards of 500 MS/MS data files can be collected per injection (or 2000 per sample if all four spectra regions are analyzed). Identification of lipid species is then made by analysis of retention time and comparison to previously published fragmentation patterns (19–22) and chemically defined standards obtained from Avanti Polar Lipids (Avanti Polar Lipids, Inc., Alabaster, AL).
Direct infusion electrospray mass spectrometry
Direct infusion mass spectral analysis was performed on an Applied Biosystems/ MDS SCIEX 4000 Q TRAP hybrid triple quadrupole linear ion trap mass spectrometer (Applied Biosystems, Foster City, CA). The instrument was equipped with a Harvard Apparatus syringe pump and an electrospray ion source. Samples were analyzed at an infusion rate of 10 μl/min in negative ionization mode over the range of m/z 350 to 1200. Data were collected with the Analyst software package (Applied Biosystems). Collision induced fragmentation of targeted lipid species was accomplished using a collision energy appropriate for the specific lipid class (−35 to −45 V).
RESULTS AND DISCUSSION
Ether-linked PI, PA, PS
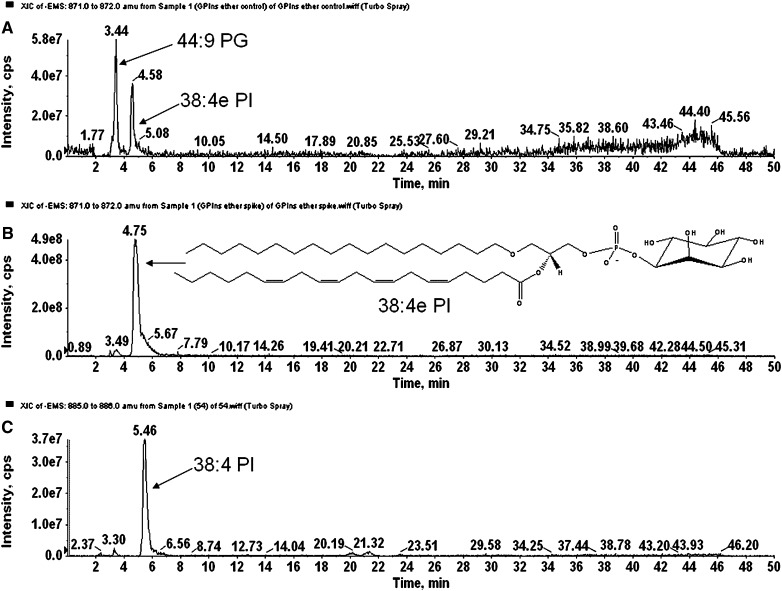
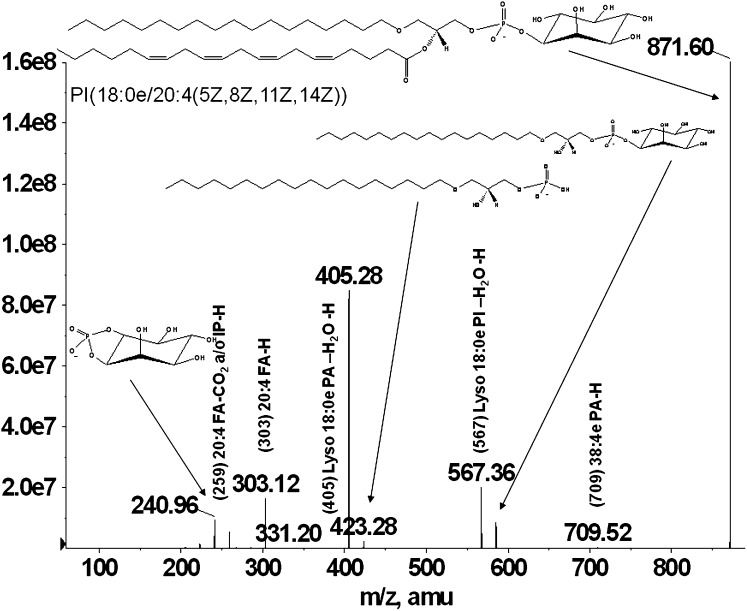
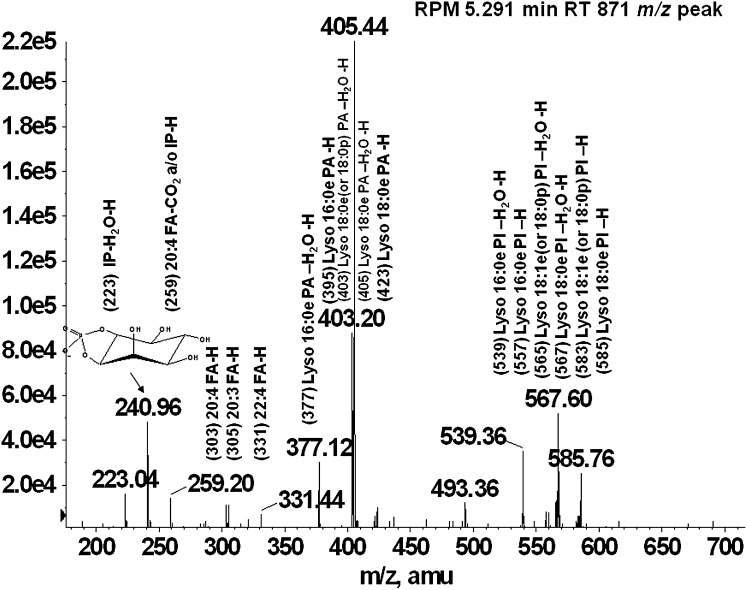
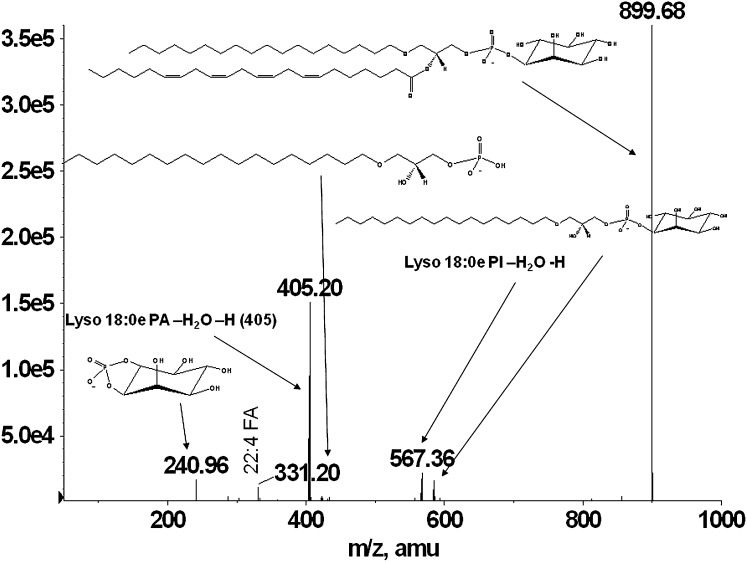
Glycerophospholipids are separated using normal-phase chromatography by class according to their polarity. Ether-linked phospholipids elute faster than their diacyl counterparts and thus are separated on the Luna silica column. Under the described chromatographic conditions, the phospholipids elute in the following order: PG< PI, PE< PA< PS/PT< PC. Figure 1A, B, and C shows the extracted ion chromatograms for PG and PI, showing the faster elution times of the ether-linked PI (Fig. 1A) compared with diacyl-PI (Fig. 1C). Odd carbon fatty acid containing 37:4 PI elutes after 38:4 PI, thus being distinguished from ether PI with the same mass (37:4 PI would have the same mass as 38:4e PI). Because the presence of ether-PI in macrophages has not been previously reported, the identification was confirmed by comparison of retention time and fragmentation spectra to a chemically defined synthetic standard (38:4e PI). Retention time confirmation was achieved by addition of synthetic 38:4e PI standard to an RPM cell extract and analyzed by LC/MS. Figure 1 shows the extracted ion chromatograms of the [M-H]− ion at m/z 871.6 characteristic for the 38:4ePI from the RPM extract (Fig. 1A) compared with the one from an RPM extract with the addition of the 38:4e PI standard (Fig. 1B). Structural identification was accomplished by LC/MS/MS. The synthetic standard with a formula of C47H85O12P has a molecular ion [M-H]− at m/z 871.57 and its fragmentation spectrum is shown on Fig. 2. The major identified peaks correspond to the 38:4e PA, 18:0e lysoPA and its dehydrated form at m/z 709.5, 423.3, and 405.3, respectively, due to loss of the headgroup. Another key fragment belongs to 18:0e lysoPI at m/z 585.7 and its dehydrated form at m/z 567.6 from losses of 20:4 fatty acid at sn-2 as a ketene and as an acid, respectively. These ions reflect the 20:4 substituent at sn-2, including the carboxylate ion detected at m/z 303.1. Fragments corresponding to the headgroup are also present at m/z 259.3 and 241. As shown in Figs. 3 and 4, fragmentation spectra from naturally occurring ether-PI are detected in RPM and foam cell extracts. The MS/MS spectrum of the molecular ion [M-H]− at m/z 871.6 from RPM cell extracts (Fig. 3) has a very similar pattern, but contains more fragment peaks, clearly revealing the presence of more than one pair of fatty acid combinations for the 38:4e PI. According to the fragmentation pattern, this peak (at m/z 871.6) is composed of 18:0e/20:4; 18:1e/20:3; and 16:0e/22:4 PI. Similarly, ether PI species of different chemical composition were detected in the other investigated cell types. An example is shown in Fig. 4, depicting the MS/MS data from a naturally occurring 40:4e PI in foam cells.
Fig. 1.
LC/MS separation of 38:4e PI. A: 871 m/z Extracted ion chromatogram (XIC) of an RPM cell extract. In this scan, the naturally occurring isobaric lipids 44:9 PG and 38:4e PI are shown. B: 871 m/z XIC of an RPM cell extract spiked with synthetic 38:4e PI. The retention times of the spiked and naturally occurring lipid were identical. C: 885 m/z XIC from the same RPM cell extract as in (A). The retention time of the diacyl 38:4 PI was longer than the 38:4e PI variant. This retention time pattern was observed across lipid classes analyzed. The odd-carbon diacyl 37:4 PI (mass 871) elutes after 38:4 PI (data not shown) making the identification of isobaric ether-linked and odd-carbon containing lipids trivial when using LC/MS/MS.
Fig. 2.
MS/MS spectra of a 38:4e PI (18:0e/20:4) synthetic standard. Fragmentation of the chemically defined PI standard yielded a plethora of lyso PA, lyso PI, and fatty acid fragments.
Fig. 3.
MS/MS spectra of naturally occurring 38:4e PI lipid from RPM cell extract. Fragmentation of the 871 m/z peak eluting prior to 38:4 PI yielded fragments consistent with the pattern observed in the synthetic standard. Additional fragments were also identified that were assigned to fatty acid combinations other than 18:0e/20:4.
Fig. 4.
MS/MS spectra of a naturally occurring foam cell 40:4e PI lipid. MS/MS analysis of the 899 m/z peak eluting prior to the 40:X series of diacyl PI lipids was shown to be primarily composed of a 18:0e and 22:4 ether/fatty acid combination.
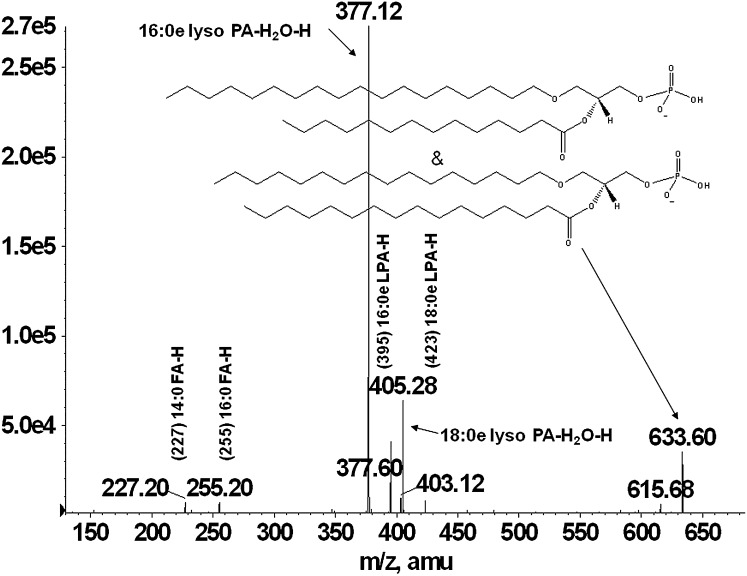
During the comprehensive lipid profiling of these cells, another two classes of ether-containing glycerophospholipids were identified, which have not been reported previously in macrophages, but have been identified in other biological tissues (23, 24). The presence of ether-PA was established in the chromatographic peak fraction from RAW 264.7 and foam cells. A representative MS/MS spectrum of a naturally occurring 32:0e PA is shown in Fig. 5. The spectrum contains a pair of prominent ions at m/z 395.1 and 377.1 arising from the loss of 16:0 fatty acid as a ketene and acid, respectively. Another pair of ions at m/z 423.2 and 405.3 corresponds to the loss of 14:0 fatty acid as a ketene and acid. These ions reflect the 14:0 and 16:0 fatty acid substituents at sn-2, and the ions reflecting the radyl group at sn-1 are not available. The presence of carboxylate ions at m/z 255.2 and 227.2 corresponding to 16:0 and 14:0 fatty acids, respectively, confirm the presence of 18:0e/14:0 and 16:0e/16:0 PA in the molecular ion at m/z 633.6. Proposed ether-linked PA species detected in RAW 264.7 and one in foam cell extracts are presented in Table 1.
Fig. 5.
MS/MS spectra from a RAW cell extract 32:0e PA lipid. Fragmentation of the 633 m/z peak revealed that this lipid species was primarily composed of 18:0e/14:0 and 16:0e/16:0 ether/fatty acid combinations.
TABLE 1.
Proposed ether- and vinyl ether-linked PA and PS molecular species detected in RAW, BMDM, and foam cells
| m/z | RAW | BMDM | Foam |
|---|---|---|---|
| PA | |||
| 633 | 16:0e/16:0 | ||
| 18:0e/14:0 | |||
| 659 | 16:0e/18:1 | ||
| 18:0e/16:1 | |||
| 661 | 16:0e/18:0 | ||
| 18:0e/16:0 | |||
| 681 | 18:0e/20:4 | ||
| 687 | 18:0e/18:1 | ||
| 707 | 16:0e/22:5 | 38:4p/38:5e | |
| 737 | 20:0e/20:4 | ||
| PS | |||
| 796 | 16:0e/22:4 | 18:0e/20:4 | |
| 18:0e/20:4 | |||
| 798 | 16:0e/20:3 | ||
| 18:0e/20:3 | |||
| 824 | 18:0e/22:4 | 18:0e/22:4 | 18:0e/22:4 |
| 20:0e/20:4 | |||
More species are identified in RAW 264.7 cell extracts due to the extensive work on these cells compared with others.
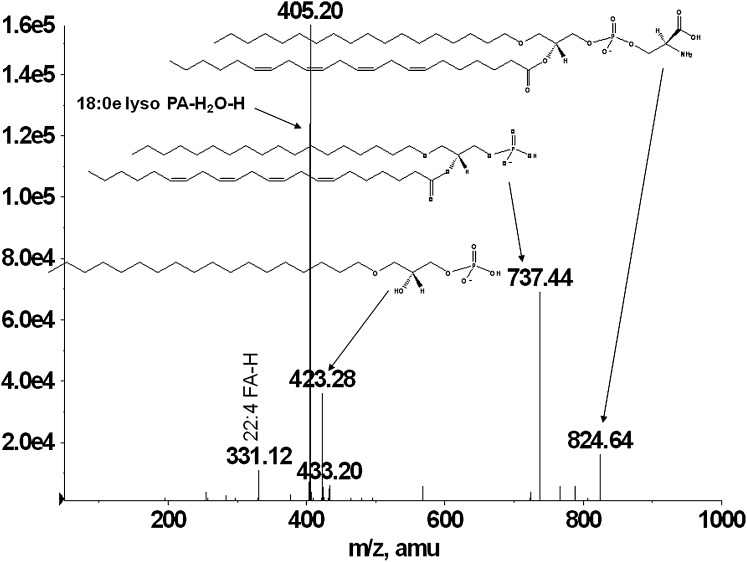
Similarly, a number of ether-PS species were identified in other macrophage extracts (Table 1). The representative MS/MS spectrum on Fig. 6 shows fragments consistent with 18:0e/22:4 PS. Phosphatidylserine shows a strong molecular ion [M-H]− at m/z 824.6, as well as a prominent [M-H-87]− at m/z 737.4, indicating the loss of the serine headgroup and identical to 40:4e PA. The ion at m/z 423.3 reflects the loss of 22:4 fatty acid at sn-2 position as a ketene [M-H-87-R2CH = C = O]−, whereas the ion at m/z 405.2 indicates the loss of 22:4 fatty acid as an acid [M-H-87-R2COOH]−. 22:4 Carboxylate ion at m/z 331.1 is also present. Here again, the ions corresponding to the radyl substituents at sn-1 are not detected in agreement with previously shown fragmentation of plasmanyl phospholipids (21).
Fig. 6.
MS/MS spectra of a BMDM 40:4e PS. Analysis of the lyso lipid and fatty acid fragments from the 824 m/z peak eluting prior to the 40:x series of PS lipids was found to be mostly attributed to 18:0e/22:4 PS.
We hypothesize that the ether and vinyl ether PA, PI, and PS glycerophospholipids reported above are direct or indirect products from the biosynthesis of plasmanyl and plasmenyl PC and PE glycerophospholipids. This reaction pathway is a combination of the traditional biosynthesis routes for plasmanyl and plasmenyl lipids (25) and the Kennedy pathway (26).
Glycerophosphothreonine detection
PT is a phospholipid that has been identified and reported as present in significant amounts in some cultured cells (15, 27, 28). Reports of these lipids have not been published for macrophage cell extracts. Structural analogy with PS makes its detection impossible without careful class separation via LC/MS/MS. The method we employ for phospholipid LC analysis allows its separation from PS and other glycerophospholipids. Using the gradient and the silica column described above, PT elutes after PS, although some ether-linked PS species coelute with PT species, but differs in its fragmentation spectrum used for the identification. As in the case of PS, PT ionizes well and is analyzed in negative ionization mode. The fragmentation spectrum shown in Fig. 7 reveals the major peaks identified in the molecular ion peak at m/z 850.5 (from BMDM). The most prominent ion present in the spectrum is the one from a neutral loss of the headgroup ([M-H-101]−) yielding the corresponding phosphatidic acid at m/z 749.4, which is consistent with [M-H]− ion of 18:0/22:5 PA. The ions at m/z 483.4 ([M-H-101-R1CH = C = O]−) and 437.3 ([M-H-101-R2CH = C = 0]−) are a result of losing fatty acid substituents at sn-1 and sn-2 as ketenes, whereas the ions at m/z 465.3 ([M-H-101-R1COOH]−) and 419.3 ([M-H-101-R2COOH]−) illustrate the loss of the corresponding substituents as acids. The carboxylic ions at m/z 283.1 and 329.4 are characteristic for the two fatty acid substituents at sn-1 and sn-2. As mentioned before, the molecular ion peak at m/z 850.5 also contains some PS (ether-linked) and the peak assignable to a PA fragment (m/z 763.52) coming from PS is noted by an asterisk in Fig. 7. All of the above peak assignments are in good correlation with previously reported data for PS and PT mass spectral analyses (27–29). Over 30 PT species were identified in different macrophages (proposed phospholipid species presented in Table 2). Differences in the number of identified species are mainly due to the large number of RAW 264.7 cell extracts analyzed during the course of the work compared with the other types of macrophages.
Fig. 7.
MS/MS spectra from a representative PT lipid. Fragmentation of the 850 m/z peak present in BMDM cell extracts yielded fatty acid and lyso PA ions consistent with 40:5 PT. This PT lipid coeluted with a PS under our LC/MS separation protocol. A PA fragment (m/z 763.52) assignable to PS is noted by an asterisk.
TABLE 2.
Proposed PT molecular species detected in RAW, BMDM, foam, and RPM cells
| m/z | RAW | BMDM | Foam | RPM |
|---|---|---|---|---|
| 760 | 16:1/17:0 | 16:1/17:0 | ||
| 16:0/17:1 | 16:0/17:1 | 16:0/17:1 | ||
| 788 | 17:0/18:1 | 17:0/18:1 | ||
| 17:1/18:0 | 17:1/18:0 | |||
| 800 | 18:0/18:2 | |||
| 18:1/18:1 | ||||
| 802 | 18:0/18:1 | 18:0/18:1 | 18:0/18:1 | |
| 17:0/19:1 | ||||
| 804 | 18:0/18:0 | 18:0/18:0 | ||
| 812 | 17:0/20:3 | 17:0/20:3 | ||
| 17:1/20:2 | 17:1/20:2 | |||
| 824 | 18:1/20:3 | |||
| 18:0/20:4 | 18:0/20:4 | |||
| 826 | 18:0/20:3 | |||
| 834 | 17:0/22:6 | 17:0/22:6 | 17:0/22:6 | |
| 836 | 17:0/22:5 | 17:0/22:5 | 17:0/22:5 | |
| 838 | 17:0/22:4 | 17:0/22:4 | ||
| 848 | 18:0/22:6 | |||
| 850 | 18:0/22:5 |
VLCFA plasmanyl and plasmenyl lipids
A significantly large group of PC and PE species containing VLCFA with different degrees of unsaturation were identified in all of the analyzed macrophages. The presence of these fatty acids is usually associated with peroxisomal β-oxidation defects and neurodegenerative disorders (30). These species have never been detected in macrophages before. Multiple species of PC from RAW 264.7 cell extracts contain 24:X and 26:X fatty acids in diacyl, plasmanyl, and plasmenyl phosphatidylcholines, whereas VLCFA PC from the other types of macrophages mostly consist of 24:X fatty acids. 24:X and 26:X fatty acids are present in diacyl, plasmanyl, and plasmenyl PE from all types macrophages (Tables 3 and 4). Here again, the majority of identified species are within RAW264.7 cell extracts as a result of the most number of samples analyzed. During the process of identification by tandem mass spectrometry, multiple ions corresponding to the fatty acid substituents and the lyso components of the molecules were observed all in agreement with previously published data (21, 31, 32). As with all plasmanyl PE and PC, there were no fragments corresponding to the alkyl moiety, but just fragments arising from the loss of the acyl moiety, thus creating ether-containing lyso lipid fragments and their dehydrated forms. Both PE and PC were analyzed in negative ionization mode. PCs were identified as formate adducts. Analysis in negative mode ionization affords information on the fatty acid composition as well as the headgroup. Not only were VLCFA-containing PC and PE identified, but also the presence of unusual ether moieties was detected. Usually, the sn-1 positions in plasmanyl and plasmenyl lipids are occupied by either 16 or 18 carbon ether or vinyl ether moieties, whereas in these cell extracts, we detected species containing 14, 16, 18, 20, 22, and 24 carbon ether or vinyl ether substituents. Mass spectral analysis confirmed their structure following analysis of fragmentation ions corresponding to the acyl substituent as a carboxylate ion and only the loss of the acyl group as acid or ketene (and therefore forming a ether-lysoPC and ether-lysoPE).
TABLE 3.
Proposed very long-chain diacyl, plasmanyl, and plasmenyl PC molecular speciesa
| m/z | RAW | BMDM | Foam | RPM |
|---|---|---|---|---|
| 864 | 16:0e/24:6 | 16:0e/24:6 | ||
| 866 | 16:0e/24:5 | 16:0e/24:5 | 16:0e/24:5 | |
| 20:0e/20:5 | 20:0e/20:5 | |||
| 868 | 16:0e/24:4 | 16:0e/24:4 | 16:0e/24:4 | 16:0e/24:4 |
| 20:0e/20:4 | 20:0e/20:4 | 20:0e/20:4 | 20:0e/20:4 | |
| 870 | 16:0e/24:3 | |||
| 20:0e/20:3 | ||||
| 872 | 16:0e/24:2 | 16:0e/24:2 | ||
| 20:0e/20:2 | ||||
| 874 | 16:0e/24:1 | 16:0e/24:1 | ||
| 20:0e/20:1 | 20:0e/20:1 | |||
| 22:0e/18:1 | ||||
| 876 | 16:1/24:6 | |||
| 878 | 16:0/24:6 | 16:0/24:6 | 16:0/24:6 | |
| 16:1/24:5 | ||||
| 880 | 16:0/24:5 | 16:0/24:5 | 16:0/24:5 | 16:0/24:5 |
| 16:1/24:4 | 16:1/24:4 | 16:1/24:4 | ||
| 882 | 16:0/24:4 | 16:0/24:4 | 16:0/24:4 | 16:0/24:4 |
| 16:1/24:3 | ||||
| 892 | 18:0e/24:6 | |||
| 20:0e/22:6 | ||||
| 894 | 18:0e/24:5 | 18:0e/24:5 | 18:0p/24:4 | |
| 20:0e/22:5 | 20:0e/22:5 | 20:0e/22:5 | 22:0p/20:4 | |
| 896 | 16:0e/26:4 | |||
| 18:0e/24:4 | 18:0e/24:4 | 18:0e/24:4 | ||
| 20:0e/22:4 | 20:0e/22:4 | 20:0e/22:4 | ||
| 22:0e/20:4 | 22:0e/20:4 | |||
| 898 | 18:0e/24:3 | |||
| 20:0e/22:3 | ||||
| 22:0e/20:3 | ||||
| 900 | 16:0e/26:2 | |||
| 18:0e/24:2 | ||||
| 902 | 18:0e/24:1 | |||
| 920 | 22:0e/22:6 | |||
| 922 | 18:0e/26:5 | |||
| 20:0e/24:5 | ||||
| 22:0e/22:5 | 22:0e/22:5 | |||
| 924 | 18:0e/26:4 | |||
| 20:0e/24:4 | 20:0e/24:4 | |||
| 22:0e/22:4 | ||||
| 24:0e/20:4 | ||||
| 926 | 20:0e/24:3 | |||
| 22:0e/22:3 | ||||
| 24:0e/20:3 | ||||
| 948 | 22:0e/24:6 | |||
| 24:0e/22:6 | ||||
| 950 | 22:0e/24:5 | |||
| 966 | 22:4/24:4 |
PC lipids were identified as formate adducts.
TABLE 4.
Proposed very long chain diacyl, plasmanyl, and plasmenyl PE molecular species
| m/h | RAW | BMDM | Foam | RPM | m/z | RAW | BMDM | Foam | RPM |
|---|---|---|---|---|---|---|---|---|---|
| 778 | 16:0e/24:5 | 804 | 20:0e/22:6 | 20:0e/22:6 | 20:0e/22:6 | ||||
| 18:0e/22:5 | 16:0p/24:4 | 16:0p/26:5 | 16:0p/26:5 | 16:0p/26:5 | |||||
| 16:0p/24:4 | 18:0p/22:4 | 16:0p/24:4 | 18:0p/24:5 | 18:0p/24:5 | 18:0p/24:5 | ||||
| 18:0p/22:4 | 20:0p/20:4 | 20:0p/22:5 | 20:0p/22:5 | 20:0p/22:5 | 20:0p/22:5 | ||||
| 20:0p/20:4 | 806 | 18:0e/24:5 | 18:0e/24:5 | 18:0e/24:5 | |||||
| 780 | 16:0e/24:4 | 20:0e/22:5 | 20:0e/22:5 | 20:0e/22:5 | 20:0e/22:5 | ||||
| 18:0e/22:4 | 16:0p/26:4 | ||||||||
| 20:0e/20:4 | 20:0e/20:4 | 18:0p/24:4 | 18:0p/24:4 | ||||||
| 16:0p/24:3 | 20:0p/22:4 | 20:0p/22:4 | 20:0p/22:4 | 20:0p/22:4 | |||||
| 18:0p/22:3 | 22:0p/20:4 | 22:0p/20:4 | 22:0p/20:4 | ||||||
| 782 | 16:0e/24:3 | 810 | 17:2/24:1 | ||||||
| 18:0e/22:3 | 816 | 16:1/26:6 | |||||||
| 20:0e/20:3 | 18:2/24:5 | ||||||||
| 16:0p/24:2 | 18:1/24:6 | ||||||||
| 18:0p/22:2 | 818 | 18:2/24:4 | 18:2/24:4 | 18:2/24:4 | |||||
| 784 | 16:0e/24:2 | 16:0e/24:2 | 18:1/24:5 | 18:1/24:5 | 18:1/24:5 | ||||
| 18:0e/22:2 | 18:0e/22:2 | 18:0/24:6 | 18:0/24:6 | 18:0/24:6 | |||||
| 16:0p/24:1 | 16:0p/24:1 | 16:1/26:5 | |||||||
| 18:0p/22:1 | 18:0p/22:1 | 820 | 16:1/26:4 | 18:1/24:4 | |||||
| 20:0p/20:1 | 20:0p/20:1 | 16:0/26:5 | 18:0/24:5 | ||||||
| 786 | 14:0e/26:1 | 18:2/24:3 | |||||||
| 16:0e/24:1 | 18:1/24:4 | 18:1/24:4 | |||||||
| 790 | 16:1/24:5 | 18:0/24:5 | 18:0/24:5 | ||||||
| 16:0/24:6 | 822 | 16:1/26:3 | |||||||
| 792 | 16:1/24:4 | 16:0/26:4 | |||||||
| 16:0/24:5 | 16:0/24:5 | 18:1/24:3 | 18:1/24:3 | ||||||
| 794 | 16:1/24:3 | 18:0/24:4 | 18:0/24:4 | 18:0/24:4 | |||||
| 16:0/24:4 | 16:0/24:4 | 16:0/24:4 | 824 | 16:1/26:2 | |||||
| 796 | 16:1/24:2 | 18:2/24:1 | |||||||
| 16:0/24:3 | 18:1/24:2 | ||||||||
| 800 | 16:1/24:1 | 18:0/24:3 | |||||||
| 16:0/24:0 | 828 | 16:1/26:0 | |||||||
| 802 | 16:0p/26:6 | 18:1/24:0 | |||||||
| 18:0p/24:6 | 18:0p/24:6 | 18:0/24:1 | |||||||
| 20:0p/22:6 | 850 | 18:1/26:4 | |||||||
| 17:1/24:6 | 18:1/26:3 | ||||||||
| 20:3/24:1 | |||||||||
| 20:4/24:0 |
Despite the widely held notion that odd carbon fatty acids only exist in plants and some lower organisms, we detected multiple species in almost all of the analyzed glycerophospholipid classes in all types of macrophages (also in multiple human tissue, astrocytoma, and other cancer cell extracts, data not shown). Their molecular weight is equal to the alkyl ether species of the same phospholipid class, but can be distinguished from them by their elution properties and the fragmentation spectra. Unlike ether glycerophospholipids that do not produce carboxylate anions for the sn-1 (ether-bound) moiety, odd-carbon containing phospholipids show prominent carboxylate anions for the corresponding fatty acids in addition to the odd carbon lyso glycerophospholipid fragments. In most cell types analyzed, lipids containing at least one odd carbon fatty acid account for approximately 15% (by number) of the total number of phospholipids identified by MS/MS fragmentation.
The evidence presented here for the existence of these unusual glycerophospholipid classes in macrophages will initiate new endeavors to determine the biological functions of these species. The presence of a relatively large number of ether-containing phospholipid species (from most classes) could contribute to the ability of these cells to produce potent biological mediators like platelet-activating factor, free PUFAs, or very long-chain fatty acids upon stimulation. In addition, the increased presence of plasmalogen phospholipids in tissues has been correlated with malignancy and metastatic properties of human cancers (33).
Acknowledgments
The authors thank Dr. Carol Rouzer for providing RPM cells and Andrew Goodman for excellent technical assistance. We thank members of the LIPID MAPS consortium for providing some of the foam and BMDM cells used for the characterization of lipid species. This work was supported in part by a large scale consortium grant from the National Institutes of Health U54 GM069338.
Footnotes
Abbreviations:
- BMDM
- murine bone marrow derived macrophages
- IPA
- isopropyl alcohol
- LMAPS
- Lipid Metabolites and Pathways Strategy initiative
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- PT
- phosphatidylthreonine
- RAW
- RAW 264.7 macrophage cell line
- RPM
- murine resident peritoneal macrophages
- VLCFA
- very long-chain fatty acid
This work was partially supported by National Institutes of Health Grant U54 GM69338 and support from the Vanderbilt Institute for Chemical Biology. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Sugiura T., Nakajima M., Sekiguchi N., Nakagawa Y., Waku K. 1983. Different fatty chain compositions of alkenylacyl, alkylacyl and diacyl phospholipids in rabbit alveolar macrophages: high amounts of arachidonic acid in ether phospholipids. Lipids. 18: 125–129. [Google Scholar]
- 2.Akoh C. C., Chapkin R. S. 1990. Composition of mouse peritoneal macrophage phospholipid molecular species. Lipids. 25: 613–617. [DOI] [PubMed] [Google Scholar]
- 3.Gaposchkin D. P., Zoeller R. A. 1999. Plasmalogen status influences docosahexaenoic acid levels in a macrophage cell line: insights using ether lipid-deficient variants. J. Lipid Res. 40: 495–503. [PubMed] [Google Scholar]
- 4.Brites P., Waterham H. R., Wanders R. J. A. 2004. Functions of plasmalogens in health and disease. Biochim. Biophys. Acta. 1636: 219–231. [DOI] [PubMed] [Google Scholar]
- 5.Gorgas K., Teigler A., Komljenovic D., Just W. W. 2006. The ether lipid-deficient mouse: tracking down plasmalogen functions. Biochim. Biophys. Acta. 1763: 1511–1526. [DOI] [PubMed] [Google Scholar]
- 6.Paltauf F. 1994. Ether lipids in biomembranes. Chem. Phys. Lipids. 74: 101–139. [DOI] [PubMed] [Google Scholar]
- 7.Farooqui A. A., Yang H. C., Horrocks L. A. 1995. Plasmalogens, phospholipase A2 and signal transduction. Brain Res. Brain Res. Rev. 21: 152–161. [DOI] [PubMed] [Google Scholar]
- 8.Karas M., Hillenkamp F. 1988. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60: 2299–2301. [DOI] [PubMed] [Google Scholar]
- 9.Fenn J. B., Mann M., Meng C. K., Wong S. F., Whitehouse C. M. 1989. Electrospray ionization for mass spectrometry of large biomolecules. Science. 246: 64–71. [DOI] [PubMed] [Google Scholar]
- 10.Pulfer M., Murphy R. C. 2003. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22: 332–364. [DOI] [PubMed] [Google Scholar]
- 11.Han X., Yang J., Cheng H., Ye H., Gross R. W. 2004. Toward fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal. Biochem. 330: 317–331. [DOI] [PubMed] [Google Scholar]
- 12.Rouzer C. A., Ivanova P. T., Byrne M. O., Milne S. B., Marnett L. J., Brown H. A. 2007. Lipid profiling reveals glycerophospholipid remodeling in zymosan-stimulated macrophages. Biochemistry. 46: 6026–6042. [DOI] [PubMed] [Google Scholar]
- 13.Guan Z., Shengrong L., Smith D. C., Shaw W. A., Raetz C. H. R. 2007. Identification of N-acylphosphatidylserine molecules in eukaryotic cells. Biochemistry. 46: 14500–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee T-C., Malone B., Buell A. B., Blank M. L. 1991. Occurrence of ether-containing inositol phospholipids in bovine erythrocytes. BBRC. 175: 673–678. [DOI] [PubMed] [Google Scholar]
- 15.Mark-Malchoff D., Marinetti G. V., Hare G. D., Meisler A. 1978. Characterization of phosphatidylthreonine in polyoma virus transformed fibroblasts. Biochemistry. 17: 2684–2688. [DOI] [PubMed] [Google Scholar]
- 16.Rouzer C. A., Marnett L. J. 2005. Glycerylprostaglandin synthesis by resident peritoneal macrophages in response to a zymosan stimulus. J. Biol. Chem. 280: 26690–26700. [DOI] [PubMed] [Google Scholar]
- 17.Milne S., Ivanova P., Forrester J., Brown H. A. 2006. Lipidomics: an analysis of cellular lipids by ESI-MS. Methods. 39: 92–103. [DOI] [PubMed] [Google Scholar]
- 18.Ivanova P. T., Milne S. B., Byrne M. O., Xiang Y., Brown H. A. 2007. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 432: 21–57. [DOI] [PubMed] [Google Scholar]
- 19.Hsu F-F., Turk J. 2000. Charge-driven fragmentation processes in diacyl glycerophosphatidic acids upon low-energy collisional activation. A mechanistic proposal. J. Am. Soc. Mass Spectrom. 11: 797–803. [DOI] [PubMed] [Google Scholar]
- 20.Hsu F-F., Turk J. 2000. Charge-remote and charge-driven fragmentation processes in diacyl glycerophosphoethanolamine upon low-energy collisional activation: a mechanistic proposal. J. Am. Soc. Mass Spectrom. 11: 892–899. [DOI] [PubMed] [Google Scholar]
- 21.Hsu F-F., Turk J. 2007. Differentiation of 1-O-alk-1’-enyl-2-acyl and 1-O-alkyl-2-acyl glycerophospholipids by multiple-stage linear ion-trap mass spectrometry with electrospray. J. Am. Soc. Mass Spectrom. 18: 2065–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu F-F., Turk J. 2000. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry: a mechanistic study. J. Am. Soc. Mass Spectrom. 11: 986–999. [DOI] [PubMed] [Google Scholar]
- 23.Deeley J. M., Thomas M. C., Truscott R. J. W., Mitchell T. M., Blanksby S. J. 2009. Identification of abundant alkyl ether glycerophospholipids in the human lens by tandem mass spectrometry techniques. Anal. Chem. 81: 1920–1930. [DOI] [PubMed] [Google Scholar]
- 24.Kaneshiro E. S., Guo Z., Sul D., Kallam K. A., Jayasimhulu K., Beach D. H. 1998. Characterization of Pneumocystis carinii and rat lung lipids: glyceryl ethers and fatty alcohols. J. Lipid Res. 39: 1907–1917. [PubMed] [Google Scholar]
- 25.Lee T. C. 1998. Biosynthesis and possible biological functions of plasmalogens. Biochim. Biophys. Acta. 1394: 129–145. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy E. P. 1992. Sailing to Byzantium. Annu. Rev. Biochem. 61: 1–28. [DOI] [PubMed] [Google Scholar]
- 27.Heikinheimo L., Somerharju P. 2002. Translocation of phosphatidylthreonine and–serine to mitochondria diminishes exponentially with increasing molecular hydrophobicity. Traffic. 3: 367–377. [DOI] [PubMed] [Google Scholar]
- 28.Mitoma J., Kasama T., Furuya S., Hirabayashi Y. 1998. Occurrence of an unusual phospholipid, phosphatidyl-L-threonine, in cultured hippocampal neurons. J. Biol. Chem. 273: 19363–19366. [DOI] [PubMed] [Google Scholar]
- 29.Hsu F-F., Turk J. 2005. Studies on phosphatidylserine by tandem quadrupole and multiple stage quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes. J. Am. Soc. Mass Spectrom. 16: 1510–1522. [DOI] [PubMed] [Google Scholar]
- 30.Kemp S., Valianpour F., Denis S., Ofman R., Sanders R-J., Mooyer P., Barth P. G., Wanders R. J. A. 2005. Elongation of very long-chain fatty acids is enhanced in X-linked adrenoleukodistrophy. Mol. Genet. Metab. 84: 144–151. [DOI] [PubMed] [Google Scholar]
- 31.Kayganich K. A., Murphy R. C. 1992. Fast atom bombardment tandem mass spectrometric identification of diacyl, alkylacyl, and alk-1-enylacyl molecular species of glycerophosphoethanolamine in human polymorphonuclear leukocytes. Anal. Chem. 64: 2965–2971. [DOI] [PubMed] [Google Scholar]
- 32.Yang K., Zhao Z., Gross R. W., Han X. 2007. Shotgun lipidomics identifies a paired rule for the presence of isomeric ether phospholipid molecular species. PLoS One. 2: e1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith R. E., Lespi P., Di Luca M., Bustos C., Marra F. A., de Alaniz M. J. T., Marra C. A. 2008. A reliable biomarker derived from plasmalogens to evaluate malignancy and metastatic capacity of human cancers. Lipids. 43: 79–89. [DOI] [PubMed] [Google Scholar]