Abstract
Recently, we used the favorable properties of 9-aminoacridine (9-AA) as matrix for the quantitative analysis of acidic metabolites and glycerophospholipids from extracts of biological materials [Sun, G., Yang, K., Zhao, Z., Guan, S., Han, X., and Gross, R.W. (2007) A shotgun metabolomics approach for rapid analysis of negatively-charged water-soluble cellular metabolites from mouse heart tissue. Anal. Chem. 79: 6629–6640; Sun, G., Yang, K., Zhao, Z., Guan, S., Han, X., and Gross, R.W. (2008) Matrix-assisted laser desorption/ionization-time of flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal. Chem. 80: 7576–7585.] by MALDI-MS. Herein, we extend this discovery and identified the selective desorption/ionization of sulfatides over other examined anionic lipids present in lipid extracts of biological samples by MALDI-MS using 9-AA as matrix. Through this approach, a high throughput method for the quantitative analysis of low to very low abundance sulfatide molecular species directly from crude lipid extracts has been developed. This method possessed a linear dynamic range of over 1,000-fold, a detection limit at the high attomole level, and a reproducibility of approximately 10% deviation. Many potential factors that might affect the quantitation of sulfatide species employing the method were examined and their effects were found to be negligible within experimental error. Collectively, these results demonstrate a powerful high throughput method for the measurement of sulfatides directly from extracts of biological samples, facilitating the study of sulfatide metabolism, trafficking, and homeostasis in health and disease.
Keywords: shotgun lipidomics, sphingolipidomics, matrix-assisted laser desorption/ionization
Sulfatides are a class of sulfated galactocerebrosides that are enriched in both the central and peripheral nervous systems as well as in kidney, while a small amount of sulfatides are also present in other organs as well as in biofluids. Sulfatides play important roles in mediating diverse biological processes and in regulating numerous cell functions (1, 2). For example, sulfatides are able to induce the generation of oxygen radicals in leukocytes (3, 4) and neutrophils (5), regulate the cytosolic free calcium level (6, 7), and modulate platelet function (8, 9).
More importantly, numerous studies have demonstrated that sulfatides are associated with the pathogenesis of various human diseases (2). Sulfatides were substantially depleted in postmortem brain samples from individuals with very mild Alzheimer's dementia (10) and significantly accumulated in those with Parkinson's disease (11). Sulfatide content is markedly enriched in human patients and mouse models of inherited glycosphingolipid disorders (e.g., metachromatic leukodystrophy) (12–14) and causes hyperexcitability and axonal degeneration in a mouse model of metachromatic leukodystrophy (15). Studies have also demonstrated that altered sulfatide levels may be potential biomarkers or predictors for the diagnoses of Alzheimer's disease (16), cardiovascular diseases (17), colorectal adenocarcinoma (18), and metachromatic leukodystrophy (19). However, a high throughput approach for analysis of sulfatides has not been established. Further increasing the penetrance into sulfatide research requires an accurate and efficient approach for the rapid identification and accurate quantitation of sulfatide molecular species present in tissues or biofluids.
Various technologies have previously been applied for the analysis of sulfatides, among which MS has been the most powerful tool (10, 20–23). In comparison to chromatographic and enzymatic methods, which are only capable of determining the total mass of sulfatides, MS can unambiguously identify and quantify individual species of sulfatides. Although ESI-MS is commonly employed for the analysis of sulfatides (10, 20, 21, 24–26), ESI-MS has limitations with either time-consuming HPLC separation (24, 26) in LC-MS method or overlap of ion peaks of sulfatides with glycerophospholipids or other sphingolipids and the potential occurrence of ion suppression using shotgun lipidomics approaches if insufficient dilution is present (25). Alkaline hydrolysis of glycerophospholipids in a lipid extract can largely eliminate ion peak overlap and ion suppression (27) but introduces an additional sample preparation step for sulfatide analysis. In addition, the sensitivity of these previously developed methods is limited and unable to readily access the sulfatide content in peripheral tissue samples.
MALDI-MS, a widely used MS technology for analyses of protein and nucleotide polymers, possesses the potential for rapid analysis of sulfatides and has showed success in directly imaging and detecting sulfatide species from biological tissue samples (28–32). Quantitation of sulfatide species has previously been demonstrated by MALDI-MS after converting sulfatides to their lyso forms by saponification and sample purification using a C18 column or C18 tips (22, 23). Those methods, however, require large amounts of material, are labor intensive (due to saponification and purification), and are incapable of quantitation of intact sulfatide species.
Recently, we used the special properties of 9-aminoacridine (9-AA) as MALDI matrix for quantitative analysis of acidic metabolites (33) as well as glycero(phospho)lipids (34). By use of this matrix, multiple concerns that are usually associated with MALDI-MS analysis of lipids (e.g., the presence of high background resulting from ionization of matrix or matrix clusters, severe postsource decay, multiple adducts, and lipid aggregation) (35, 36) have been resolved. Utilization of 9-AA leads to a high sensitivity, minimal background and postsource decay, and quantitative analysis of many lipid classes and individual molecular species with relatively homogeneous distribution of lipids (34).
Following this line of reasoning, we examined if this matrix could also be used for the quantitative analysis of sulfatides with high sensitivity. Herein, we report the utility of 9-AA for the measurement of attomolar amounts of sulfatide molecular species by MALDI-MS. Moreover, an over 35-fold selective desorption/ionization of sulfatide versus anionic phospholipids was demonstrated. Furthermore, quantitation of individual sulfatide species was determined by ratiometric comparisons of individual molecular species to the internal standards (37). The linearity of the signal response, dynamic range, sensitivity, and reproducibility of quantitation were extensively investigated. After optimization of experimental conditions, the developed method was applied to directly identify and quantify sulfatide species in a variety of biological samples, many of which were validated by comparisons to previously validated ESI-MS analyses. Finally, high throughput analysis of 80 samples (four to six spots per sample) using an Opti-TOF® 384 well plate from lipid extracts either without any prior treatment or with a simple one-step alkaline treatment if necessary was achieved using data processing through our recently developed program (38). Collectively, we report a method for the quantitative and high throughput analysis of cellular sulfatide species directly from lipid extracts of biological samples by MALDI-MS using 9-AA as matrix.
MATERIALS AND METHODS
Materials
9-AA hemihydrate was purchased from Acros Organics USA (Morris Plains, NJ). 2,5-Dihydroxybenzoic acid (DHB) was purchased from Sigma-Aldrich Co. (St. Louis, MO). N-Lauroyl sulfatide (N12:0 sulfatide), bovine brain sulfatides, and soy phosphatidylinositol (PI) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). N-Palmitoyl sulfatide (N16:0 sulfatide) was obtained from Matreya LLC (Pleasant Gap, PA). The levels of sulfatides that were used as internal standards were quantified by nuclear magnetic resonance spectroscopy using choline chloride as standard. Mixtures comprised of these sulfatides were prepared from their stock solution in 1:1 (v/v) chloroform-methanol. All solvents used in extraction and MS analysis were purchased from Burdick and Jackson (Muskegon, MI). It should be noted that the prefix “N” denotes the fatty acyl amide chain and “d” represents the sphingoid base.
Preparation of lipid extracts
C57BL/6 wild-type male mice (4–6 months of age) were purchased from the National Institute on Aging (Bethesda, MD). All animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the Animal Studies Committee at Washington University. Mice were euthanized by asphyxiation with carbon dioxide. Mouse organs were perfused with phosphate-buffered saline plus heparin to remove possible plasma contamination before dissection. Brain cortex, spinal cord, sciatic nerves, plasma, kidney cortex, kidney medulla, liver, and skeletal muscle of mice were harvested immediately after perfusion. Tissue samples were individually freeze-clamped at the temperature of liquid nitrogen and pulverized into a fine powder with a stainless steel mortar and pestle. Protein assays for each individual sample were performed using a bicinchoninic acid protein assay kit (ThermoFisher Scientific, Rockford, IL) with BSA as a standard. Approximately 10 mg of sample powder was weighed from individual tissue sample and lipids were extracted by the modified method of Bligh and Dyer (39). A total of 100 μl of plasma sample from each animal was used to similarly prepare the plasma lipid extracts in the presence of internal standards (50 pmol/ml plasma). Either N12:0 or N16:0 or both sulfatides were used as the internal standard(s) for sulfatide analysis and the amounts of internal standards were varied with the samples as follows: sciatic nerve, spinal cord, brain cortex, kidney medulla, kidney cortex, skeletal muscle, and liver, 20, 7.5, 4, 0.1, 0.5, 0.025, and 0.025 nmol/mg protein, respectively. Only N12:0 sulfatide was used for analysis of peripheral tissues (e.g., kidney) due to the presence of a significant amount of endogenous N16:0 sulfatide. The internal standard(s) were selected because they represent <0.2% of the most abundant species of endogenous sulfatides as demonstrated by MALDI-MS analysis without addition of the internal standard(s). The final concentration of each individual lipid extract was reconstituted by resuspending the lipid content extracted from the amount of sample of 1 mg of original protein in 1 ml of 1:1 (v/v) chloroform-methanol or as specified (the original protein content of the samples was determined from protein assays). The lipid extracts were finally flushed with nitrogen, capped, and stored at −20°C for MALDI-time-of-flight (TOF)/TOF-MS or ESI-MS analyses.
Alkaline methanolysis of lipid extracts
Alkaline hydrolysis of phospholipids was performed as previously described (27) with minor modification. Briefly, a small portion of each individual crude lipid extract equivalent to the lipid extract from a tissue sample containing 0.1 mg protein was dried under a stream of nitrogen. Ice-cooled lithium methoxide solution (1 M, 200 μl) in methanol was added to the dried sample at 0°C. The reaction mixture was vortexed for 15 s followed by sonication for 5 min at room temperature. After incubation for 1 h in an ice bath, the reaction was quenched with 2 ml of 1.2% acetic acid solution. The pH of the quenched reaction solution should be adjusted to 4 ∼5 by addition of acetic acid solution if necessary. The aqueous phase was washed with hexane (2 ml) once and the hexane solution was discarded. The lipids located in the interface of the hexane layer and water layer as well as those dissolved in the aqueous phase were extracted by the modified Bligh and Dyer method. The combined chloroform phase was dried under a nitrogen stream. The residue was reextracted against a 10 mM aqueous lithium chloride solution. Each individual extract was reconstituted in 100 μl of chloroform-methanol (1:1, v/v)/mg of original sample protein after filtering through a 0.2 μm polytetrafluoroethylene membrane filter, flushed with nitrogen, capped, and stored at −20°C for MS analysis as specified.
MALDI-MS analyses of sulfatides
Five microliters of lipid extract was diluted to have a concentration of total sulfatides < 500 fmol/μl and/or a concentration of total lipids < 15 pmol/μl with methanol-water-chloroform solution (86:9:5, v/v/v). After mixing 10 μl of the diluted lipid solution with 10 μl of 9-AA (10 mg/ml) dissolved in a methanol-water solution (90:10, v/v) or of DHB (5 mg/ml) dissolved in water, 0.7 μl of the mixture was spotted on an Opti-TOF® 384 well plate (Applied Biosystems Inc., Foster City, CA). MS analysis was performed using a 4800 MALDI-TOF/TOF Analyzer. A mass spectrum of sulfatides was acquired in the negative ion reflector mode by averaging 3,000 consecutive laser shots (50 shots per subspectra and 60 subspectra) with default calibration. Tandem MS analyses of sulfatides were accomplished using collision-induced dissociation with the metastable suppressor on and the timed ion selector enabled. Ion peaks with S/N > 3 were considered to be potential lipid signals.
ESI-MS analysis of sulfatides
A triple-quadrupole mass spectrometer (Thermo Electron TSQ Quantum Ultra Plus; San Jose, CA) equipped with a TriVersa Nanomate device (Advion BioSciences, Ithaca, NY) and an Xcalibur operation system was utilized in the study. Lipid extracts after appropriate dilution in 1:2:4 (v/v/v) chloroform-methanol-isopropanol were infused at a flow rate of ∼150 nl/min as previously described (40). The temperature of the capillary tube was set at 150°C. Typically, a 2 or 5 min period of signal averaging in the profile mode was employed for each MS or MS/MS spectrum, respectively. All MS spectra and tandem MS spectra were automatically acquired by a customized sequence subroutine operated under Xcalibur software. Data processing of two-dimensional MS analyses was conducted as outlined previously (38).
RESULTS AND DISCUSSION
Selectively enhanced desorption/ionization of sulfatides using 9-AA as matrix
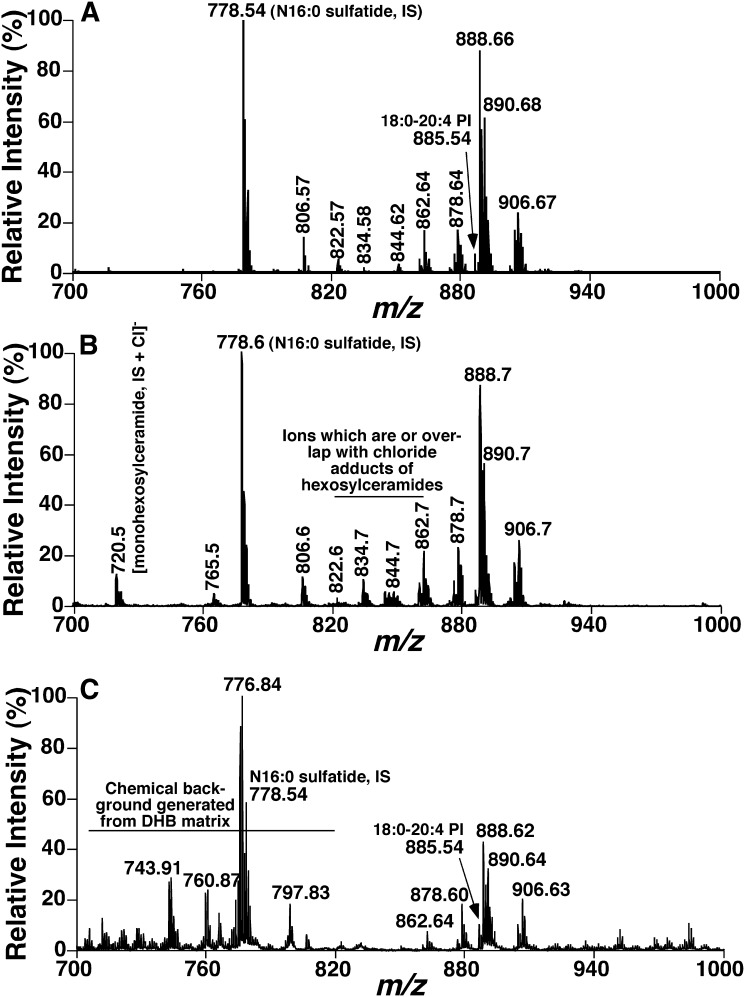
In an initial experiment, we tested the possibility of using 9-AA as matrix to directly analyze sulfatides from a lipid extract of mouse brain cortex. Commonly, MALDI-MS analysis using other matrices and ESI-MS analysis of mouse brain cortex demonstrate predominant ion peaks corresponding to deprotonated PI species in the spectra in the mass range of sulfatides (41, 42). To our surprise, and in contrast to the common observation using ESI-MS or MALDI-MS, negative-ion mass spectra acquired from lipid extracts of mouse brain cortices by MALDI-MS using 9-AA as matrix demonstrated abundant ions between m/z 750 and 950 (Fig. 1A), which all corresponded to deprotonated sulfatide species identified through accurate mass analysis and product-ion analyses (see next subsection). Moreover, this pattern of sulfatide species was well matched with that acquired from ESI-MS analysis of alkaline-treated lipid extracts of mouse cortices (Fig. 1B). It should be noted that the additional peaks or the ion peaks of which the relative intensities are different in the ESI-MS spectrum (Fig. 1B) from those in the MALDI-MS spectrum (Fig. 1A) represent the chloride adducts of galactosylceramide species alone or overlapping with sulfatide molecular species. The presence of galactosylceramide chloride adduct was confirmed by neutral loss of HCl as previously described (43).
Fig. 1.
Mass spectral comparison of sulfatide molecular species present in mouse brain cortical lipid extracts acquired by either MALDI-MS or ESI-MS. Lipid extracts of mouse brain cortices were prepared by a modified Bligh and Dyer procedure as described in “Materials and Methods.” A: A negative-ion mass spectrum of a crude lipid extract of mouse brain cortex acquired by MALDI-MS utilizing 9-AA as matrix dissolved in methanol-water solution (90:10, v/v). B: A negative-ion mass spectrum of an alkaline-treated lipid extract of mouse brain cortex acquired by ESI-MS as described in “Materials and Methods.” C: A negative-ion mass spectrum of a crude lipid extract of mouse brain cortex acquired by MALDI-MS utilizing DHB acid as matrix dissolved in water. IS, Internal standard.
To directly compare the matrix effects on lipid analysis, we also performed mass spectrometric analyses of the identical lipid extracts of mouse cortices used for Fig. 1A by using DHB. Although we also demonstrated a sulfatide-dominant profile using DHB as matrix (Fig. 1C), the ratio of the deprotonated PI signal to sulfatide signal (e.g., 18:0-20:4 PI at m/z 885.5 vs. N24:1 suflatide at m/z 888.7) is significantly higher than that in Fig. 1A using 9-AA as matrix, indicating a less selective ionization/desorption by using DHB in comparison to 9-AA. Moreover, mass spectra acquired using DHB clearly displayed a high chemical background resultant from matrix and low signal to noise ratios of sulfatides (Fig. 1C). In addition, the profiles of sulfatide species acquired by MALDI-MS using DHB were different from those acquired by using 9-AA (Fig. 1A) and by ESI/MS (Fig. 1B), likely resultant from the poor homogeneity of the sample spots using DHB. For example, the ratios of ion peaks at m/z 862.7, 878.7, and 906.7 to the major species at m/z 888.7 are much higher in Fig. 1C than those in Fig. 1A, B. Accordingly, these results from this initial study not only reveal the selective desorption/ionization of sulfatides but also indicate the potential for quantitative analysis of sulfatide molecular species (including both hydroxy and nonhydroxy species) by MALDI-MS using 9-AA as matrix (see below).
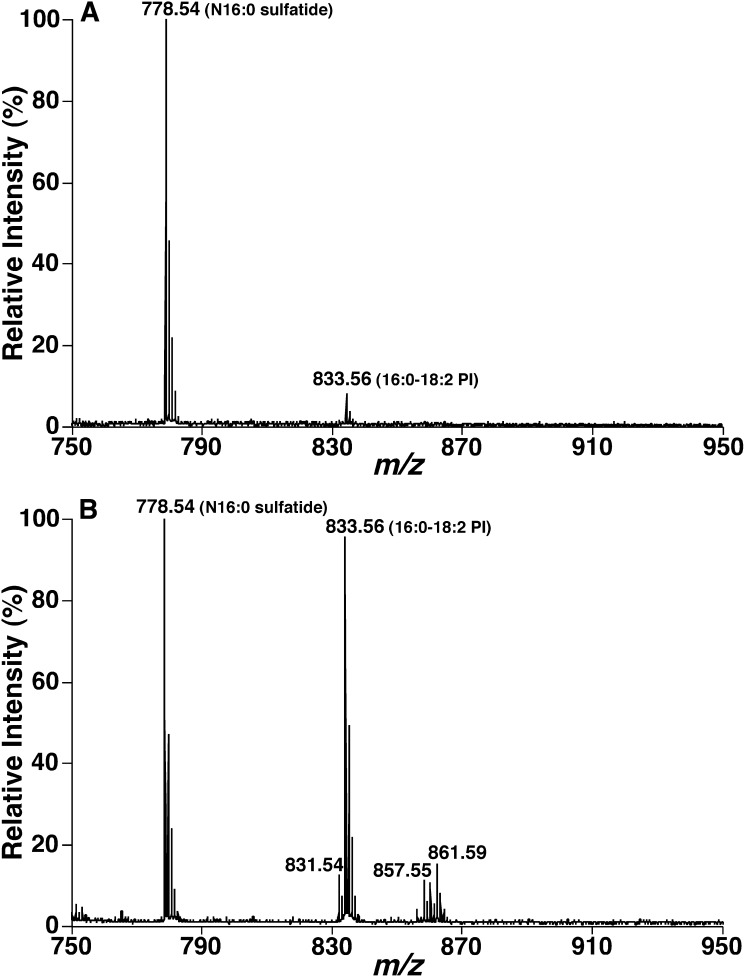
To further investigate the observed selective desorption/ionization of sulfatide species over other lipids by MALDI-MS using 9-AA, we analyzed the mixtures comprised of N16:0 sulfatide and soy PI at different molar ratios. The species of 16:0-18:2 PI accounts for 68 mol% of total mass of soy PI based on ESI-MS analysis. MALDI-MS analysis of mixtures consisting of N16:0 sulfatide and soy PI at ratios from 1:5 to 1:50 showed N16:0 sulfatide ion peak at m/z 778.54 as the base peak, 16:0-18:2 PI ion peaks at m/z 833.56 of a relative intensity of less than 10% for the mixture at the ratio of 1:5 (Fig. 2A) and of a relative intensity of 90% for the mixture at the ratio of 1:50 (Fig. 2B). These results indicate an at least 35-fold selectivity of desorption/ionization of sulfatides over 16:0-18:2 PI (50 × 0.68/0.9 = 38) (Fig. 2). Similar selectivity of desorption/ionization over other anionic phospholipids was also determined. It should be noted that the cerebroside species as chloride adducts that are present in the ESI-MS spectra (Fig. 1B) (43) was not detected in MALDI-MS analysis, which eliminates their potential overlapping with sulfatide species. It should also be noted that there exist large amounts of zwitterionic phospholipids in lipid extracts that can be analyzed by negative-ion MALDI-MS using 9-AA as matrix under certain conditions (44). However, these phospholipids did not affect the quantitative analysis of sulfatide species due to their lower ionization/desorption response in comparison to that of either PI or sulfatides.
Fig. 2.
Determination of enhanced desorption/ionization of sulfatides over PI species by MALDI-MS using 9-AA as matrix. Lipid mixtures comprised of N16:0 sulfatide (0.2 pmol/μl) and soy PIs [either 1 pmol/μl (A) or 10 pmol/μl (B)] were prepared and analyzed by MALDI-MS using 9-AA as matrix as described in “Materials and Methods.”
The selective detection of sulfatide species from other lipids present in brain lipid extracts using 9-AA in the negative-ion mode of MALDI-MS could possibly result from the minimized post source decay of sulfatide species compared with other anionic lipid species. Because we did not find apparent fragment ions of anionic phospholipids in the spectra, we do not believe that post source decay is the main reason for the selective detection of sulfatide species. Alternatively, this selectivity could be due to a specific interaction between 9-AA and sulfatide during laser irradiation that can profoundly facilitate the desorption/ionization efficiency of sulfatides. Investigation of the chemical mechanism underlying the enhanced desorption/ionization of sulfatides over other classes of lipids is intriguing, but because the precise factors resulting in ionization/desorption efficiency for most lipids is unknown, it is beyond the scope of the study.
Desorption/ionization of a specific lipid class with a high selectivity has many advantages. For example, it is not necessary to separate the lipid classes of interest by chromatography prior to analysis by MALDI-MS; ion suppression resulting from other lipid classes does not apparently exist and a high sensitivity of analysis can be easily achieved. However, when the content of PI species (and/or other anionic phospholipids or zwitterionic phospholipids) becomes much more abundant than that of sulfatides in a lipid extract of a biological sample, e.g., >10-fold, the inference of those lipid species to the quantitative analysis of sulfatides cannot be disregarded. In this case, alkaline treatment of lipid extracts of biological samples (e.g., mouse peripheral tissue samples; see below) should be performed as previously described (27) for sulfatide analysis by MALDI-MS using 9-AA.
Identification of sulfatide species by MALDI-TOF/TOF-MS
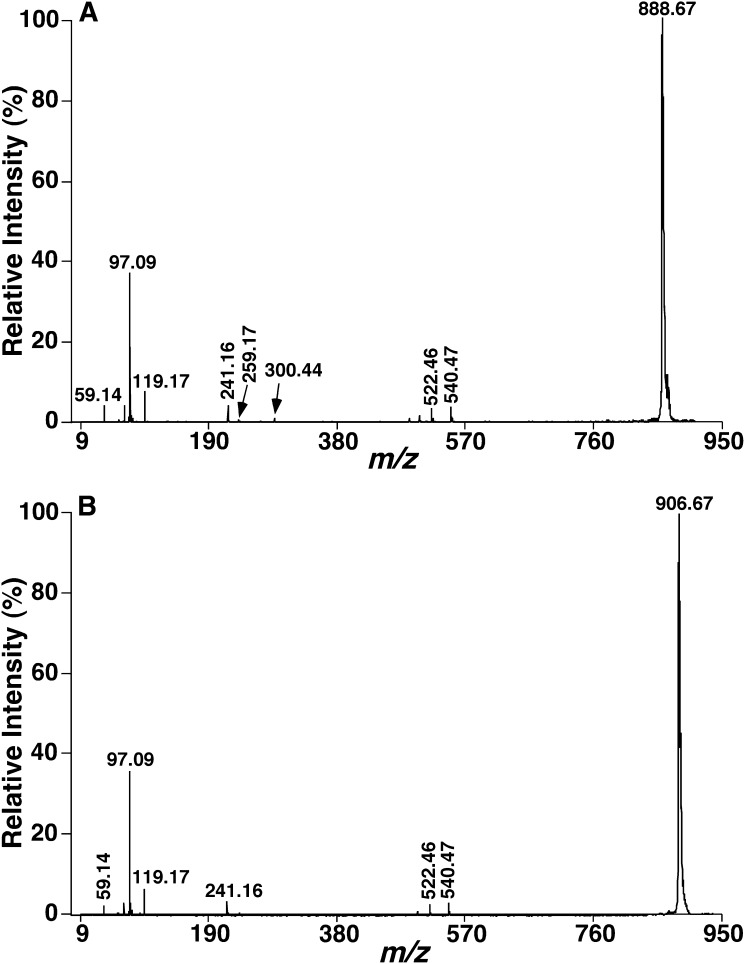
Product-ion analyses of the [M-H]− ions of sulfatide species by MALDI-TOF/TOF-MS demonstrated informative fragment ions for structural determination. For example, the product-ion spectrum of the [M-H]− ion at m/z 888 (Fig. 3A) shows a prominent ion at m/z 97, representing the HOSO3− ion and other fragment ions at m/z 259 and 241, reflecting the 3-sulfogalactosyl moiety as previously demonstrated (20). The m/z 259 ion may represent the galactose 3-sulfate anion (45). Further loss of water from the m/z 259 ion gives rise to the m/z 241 ion. These fragment ions indicate the presence of galactose sulfate. The direct loss of the fatty acyl chain as a ketene from [M-H]− via the NH-CO bond cleavage results in the m/z 540 ion, which undergoes a loss of water to yield the m/z 522 ion. The m/z 540 ion can also lead to the m/z 300 ion, corresponding to 1-O-2′-aminoethenyl galactosyl 3-sulfate ion, probably via the combined losses of the d18:1 sphingoid base as an aldehyde and H2. These fragment ions together with the m/z 97 ion permit confident identification of the sulfatide species as d18:1-N24:1 sulfatide. These common ions that identify the sphingoid base, galactose, and fatty acid moieties are present in product-ion mass spectra of sulfatides consisting of an α-hydroxy fatty acid substituent and a d18:1 sphingoid base (d18:1-OH N-FA sulfatide) (Fig. 3B) as previously described (20).
Fig. 3.
Product-ion analyses of sulfatide species in the negative ion refl ector mode using 9-AA as matrix. Tandem mass spectra of d18:1-N24:1 sulfatide at m/z 888.67 (A) and d18:1-[OH]N24:0 sulfatide at m/z 906.67 (B) present in mouse brain cortical lipid extracts acquired by MALDI-TOF/TOF-MS in the negative ion mode as described in “Materials and Methods.” Each tandem mass spectrum was recorded on a 4800 MALDI-TOF/TOF Analyzer in the negative ion refl ector mode using 9-AA as matrix using collision-induced dissociation with the metastable suppressor on and the timed ion selector enabled. The voltages of source 1, collision cell, and collision cell offset were 8.0, 7.0, and −0.035 kV, respectively. The tandem mass spectrum was obtained by averaging 3000 consecutive laser shots (50 shots/subspectra with 60 total subspectra).
Quantitation of sulfatide species by MALDI-MS with 9-AA
The identical patterns of sulfatide species of mouse cortical lipid extracts displayed in both MALDI-MS and ESI-MS spectra (Fig. 1) indicate that MALDI-MS using 9-AA as matrix can potentially be used for quantitative analysis of individual sulfatide species directly from lipid extracts of biological samples. This is due to the fact that quantification of sulfatide species by ESI-MS has been previously investigated (10, 27) and the mass levels of individual sulfatide species in mouse cortical lipid extracts cover a wide concentration range.
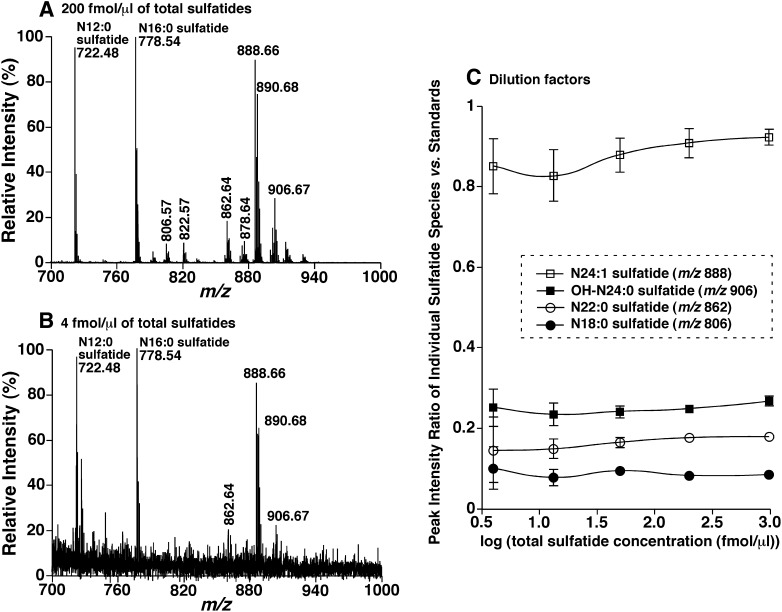
To further determine the possibility of negative-ion MALDI-MS analysis with 9-AA for quantitation of individual sulfatide species, three additional series of experiments were performed. First, MALDI-MS analyses of the mixtures comprised of N12:0, N16:0, and bovine brain sulfatides at a molar ratio of 1:1:2 at concentrations from 4 fmol/μl to 1 pmol/μl of total sulfatides when spotted onto the plate were performed (Fig. 4). The peak intensity ratio of each individual species of bovine brain sulfatides including hydroxy species versus both N12:0 and N16:0 sulfatide standards after 13C deisotoping was unchanged in the examined concentration range (Fig. 4C). This represents a linear dynamic range of over 200-fold. Moreover, the virtual linear dynamic range is much larger than the examined one, because the concentrations of individual bovine brain sulfatide species vary over 25-fold as demonstrated (Fig. 4A). Therefore, the limit of detection is in the high amol/μl range. In addition, we averaged the determined content of each species at all examined dilution levels using both N12:0 and N16:0 sulfatides as standards. The largest standard deviation for quantitative analysis of sulfatide species was unsurprisingly at the lowest concentration (i.e., 4 fmol/μl of total bovine brain sulfatides) (Fig. 4C) at which the effect of background noise was significant (Fig. 4B). The standard deviations for all sulfatide species at other concentrations were less than 10%. These results indicate a precision (i.e., reproducibility) of approximately 10% for quantitation of sulfatide species with the developed method.
Fig. 4.
Determination of the limit of detection or quantitation of MALDI-MS analysis of sulfatides using 9-AA as matrix. A mixture of N12:0, N16:0, and bovine brain sulfatides at a molar ratio of 1:1:2 was prepared and MALDI-MS analyses of this mixture after dilution were performed as described in “Materials and Methods.” A: A negative-ion mass spectrum of the mixture at a concentration of 200 fmol/μl. B: A negative-ion mass spectrum of the mixture at a concentration of 4 fmol/μl. C: The linear relationship of the ratios of ion peak intensities of representative bovine brain sulfatide species and each of the selected internal standards with different concentrations of the mixture. The data points represent the mean ± SD from four mixture preparations at each concentration. Note that some error bars are within the symbols. IS, Internal standard.
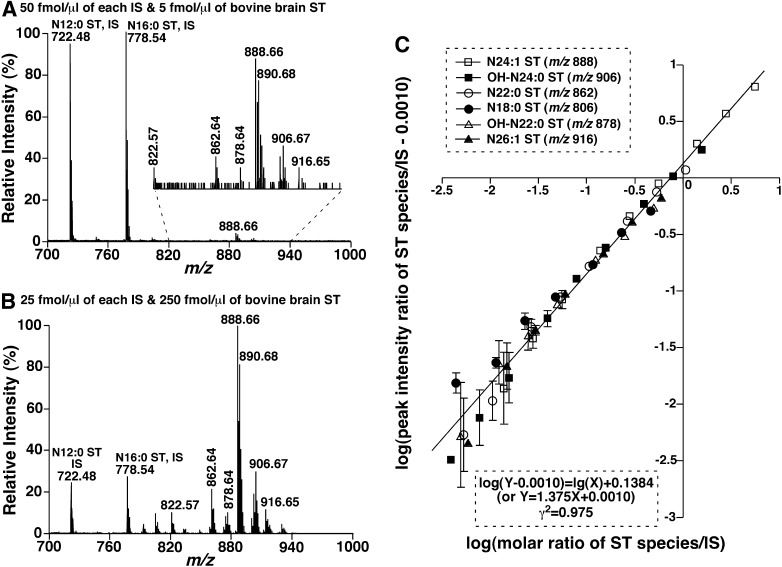
Next, we determined the linearity of quantitation using both N12:0 and N16:0 sulfatides as standards. We mixed both N12:0 and N16:0 sulfatides (2 pmol/μl of each) with different amounts of bovine brain sulfatides and diluted samples to a concentration of total sulfatides < 500 fmol/μl before mixing with the matrix solution. The mixtures were analyzed by negative-ion MALDI-MS in the reflector mode using 9-AA as matrix (Fig. 5A, B). The linearity of peak intensity ratios of individual bovine brain sulfatide species and standards after 13C deisotoping versus their corresponding molar ratios in the mixtures was analyzed by linear log plots [i.e., log(Y−b) = log(X) + c, which is derived from Y = aX + b] as discussed previously (46) (Fig. 5C). An essentially identical linear correlation was well obtained for all the species examined, including both nonhydroxy and hydroxy sulfatides (Fig. 5C), indicating that the desorption/ionization efficiency of this method is molecular species independent. It should be pointed out that the correlation coefficients of the linear regression for most of the individual sulfatide species are much higher than that of the assembled ones (Fig. 5C) in which the low abundance species generally possess a lower correlation.
Fig. 5.
Quantitative analyses of sulfatide species by MALDI-MS using 9-AA as matrix. Mixtures comprised of equimolar N12:0 and N16:0 sulfatides as internal standards and different amounts of bovine brain sulfatides were prepared and MALDI-MS analyses of the mixtures after proper dilution were performed as described in “Materials and Methods.” A, B: The negative-ion mass spectra of mixtures at the concentration of 5 and 250 fmol/μl, respectively. C: The linear log correlation between molar ratios and ion peak intensity ratios of representative bovine brain sulfatide species versus both of the selected internal standards. The data points represent the mean ± SD from four mixture preparations of each molar ratio of bovine brain sulfatides to the selected internal standards. Note that some error bars are within the symbols. IS, Internal standard; ST, sulfatide.
In the third set of experiments, we examined multiple factors that might influence accurate quantitation of individual sulfatide species (particularly those present in lipid extracts of biological samples) by negative-ion MALDI-MS using 9-AA as matrix. We first examined the effects of the proton concentration on the matrix and resultant alterations in sulfatide patterns. Through varying the pH values of the matrix solutions with the addition of formic acid (0.5%), ammonium acetate (0.1 mM), ammonium hydroxide (1%), or their mixtures before mixing with lipid samples, we did not find any apparent changes of the peak intensity patterns of the mixture comprised of N12:0, N16:0, and bovine brain sulfatides (spectra not shown). We also examined the effects of ion strength on sulfatide desorption/ionization and quantitation. In the study, we analyzed the profiles of sulfatide species of the N12:0, N16:0, and bovine brain sulfatide mixtures in the presence of different amounts of ammonium acetate or lithium chloride (varied from 0 to 1 mM of concentrations) and did not find significant changes of the molecular species patterns of sulfatides. Moreover, we examined the effects of other coexisting lipids on quantitative analysis of sulfatide molecular species. To this end, we mixed the sulfatide mixture comprised of N12:0, N16:0, and bovine brain sulfatides (total 100 fmol/μl) with either cholesterol (1.2 pmol/μl), galactosylceramide (300 fmol/μl), or mouse myocardial lipid extracts (3 pmol/μl). MALDI-MS analyses of these mixtures after dilution did not show any significant effects of the presence of other lipids or lipid extracts on quantitation of sulfatide species (data not shown). It should be pointed out that the amount of total lipids might affect the quantitative analysis of sulfatides, likely due to the effects of lipids on homogeneous distribution, because the reproducibility was affected if the ratios of total lipids to matrix were exceeded. Therefore, the total concentration of lipids should be kept as low as possible or at least < 15 pmol/μl for mass spectral determinations. The level of galactosylceramides, likely due to its structural similarity, gave the most significant effect on sulfatide analysis. Finally, we also examined the effects of solvents on the sulfatide analysis. In contrast to the positive-ion mode for the analysis of phospholipids where solvents are critical (34), we found that solvents used for crystallization of matrix/samples affect sulfatide analysis only moderately.
Quantitative analysis of sulfatide species in biological samples
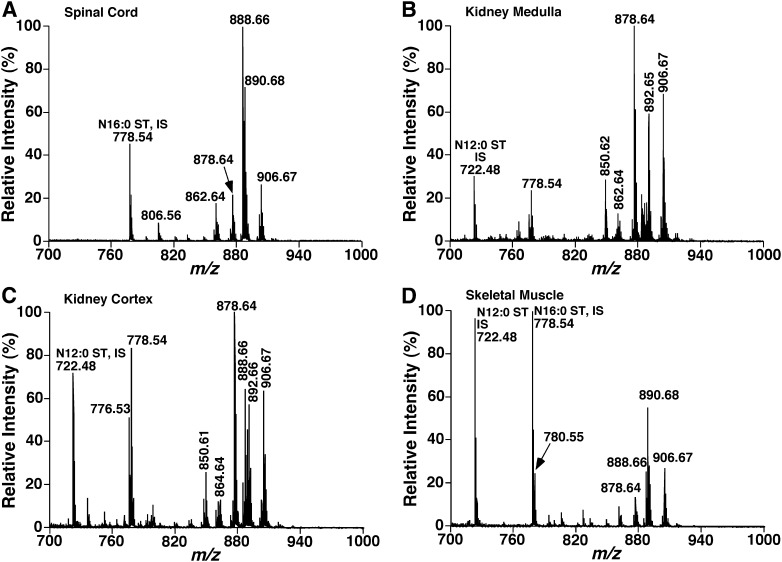
The developed methodology was employed to directly quantitate the contents of individual sulfatide species in lipid extracts of a variety of biological samples. These include spinal cord (Fig. 6A), kidney samples [e.g., medulla (Fig. 6B) and cortex (Fig. 6C)], skeletal muscle (Fig. 6D), brain tissues, sciatic nerve, and liver of mice as well as mouse plasma at 4–6 months of age. The quantitative results were tabulated (Table 1).
Fig. 6.
MALDI-MS mass spectra of sulfatides in lipid extracts of mouse spinal cord, kidney medulla, kidney cortex, and skeletal muscle. Total lipids of spinal cord, kidney medulla, kidney cortex, and skeletal muscle of mice at 4–6 months of age were extracted by using a modifi ed procedure of Bligh and Dyer against 50 mM LiCl as previously described (39). Part of the lipid extract of mouse kidney medulla, kidney cortex, or skeletal muscle was treated with lithium methoxide as previously described (27). MALDI mass spectra of lipid extracts of mouse spinal cord (A), kidney medulla (B), kidney cortex (C), and skeletal muscle (D) were acquired by averaging 3,000 consecutive laser shots (50 shots/subspectra with 60 total subspectra) using the 4800 MALDI TOF/TOF Analyzer with 9-AA as matrix. IS, Internal standard; ST, sulfatide.
TABLE 1.
Quantitative analysis of sulfatide species in the lipid extracts from a variety of mouse tissue samples and plasmaa
| Molecular species | m/z | Plasmab | Liverb | Muscleb | Kidney cortexb | Kidney medullab | Cortexc | Spinal cordc | Sciatic nervec |
|---|---|---|---|---|---|---|---|---|---|
| d18:0-N14:0 | 752.50 | 8.8 ± 0.7 | 22.4 ± 3.0 | ||||||
| d18:1-N16:1 | 776.50 | 63.3 ± 1.6 | 117.9 ± 14.5 | ||||||
| d18:1-N16:0 | 778.51 | 101.5 ± 1.2 | 282.5 ± 27.8 | ||||||
| d18:0-N16:0 | 780.53 | 7.5 ± 0.2 | 19.2 ± 2.1 | 0.5 ± 0.0 | 2.1 ± 0.1 | 2.8 ± 0.1 | |||
| d18:1-OHN16:1 | 792.49 | 0.5 ± 0.1 | 0.4 ± 0.1 | 3.6 ± 0.4 | 9.7 ± 1.9 | 0.1 ± 0.0 | 0.23 ± 0.02 | 0.3 ± 0.0 | |
| d18:1-OHN16:0 | 794.51 | 0.2 ± 0.0 | 0.9 ± 0.2 | 7.8 ± 1.1 | 50.7 ± 4.7 | 0.4 ± 0.0 | 1.4 ± 0.1 | 1.2 ± 0.1 | |
| d18:0-OHN16:0 | 796.52 | 0.1 ± 0.0 | 1.6 ± 0.1 | 2.7 ± 0.1 | 0.2 ± 0.0 | 0.3 ± 0.0 | |||
| d18:1-N18:1 | 804.53 | 2.4 ± 0.3 | 0.1 ± 0.1 | 0.1 ± 0.0 | 0.3 ± 0.0 | ||||
| d18:1-N18:0 | 806.55 | 1.0 ± 0.4 | 0.2 ± 0.1 | 1.2 ± 0.2 | 0.7 ± 0.1 | 3.5 ± 0.1 | 0.4 ± 0.0 | 1.7 ± 0.2 | 2.9 ± 0.1 |
| d18:0-N18:0 | 808.56 | 0.2 ± 0.0 | 0.5 ± 0.0 | 2.6 ± 0.1 | 0.1 ± 0.0 | 0.3 ± 0.0 | 0.9 ± 0.0 | ||
| d18:1-OHN18:1 | 820.52 | 0.2 ± 0.1 | 2.1 ± 0.2 | 5.1 ± 0.2 | 0.1 ± 0.0 | 0.1 ± 0.0 | |||
| d18:1-OHN18:0 | 822.54 | 0.2 ± 0.0 | 1.2 ± 0.1 | 11.7 ± 0.3 | 0.2 ± 0.0 | 0.4 ± 0.0 | 0.5 ± 0.0 | ||
| d18:1-N20:0 | 834.58 | 0.6 ± 0.1 | 3.6 ± 0.5 | 44.5 ± 2.0 | 0.1 ± 0.0 | 0.7 ± 0.1 | 2.9 ± 0.1 | ||
| d18:0-N20:0 | 836.59 | 0.2 ± 0.0 | 4.2 ± 0.1 | 19.5 ± 1.9 | 0.2 ± 0.1 | 1.1 ± 0.1 | |||
| d18:1-OHN20:1 | 848.56 | 1.6 ± 0.4 | 0.1 ± 0.0 | 16.3 ± 0.9 | 40.5 ± 2.3 | 0.3 ± 0.0 | |||
| d18:1-OHN20:0 | 850.57 | 0.3 ± 0.1 | 0.5 ± 0.1 | 35.9 ± 3.2 | 433.7 ± 39.4 | 0.1 ± 0.0 | 0.5 ± 0.1 | 1.6 ± 0.1 | |
| d18:0-OHN20:0 | 852.59 | 2.6 ± 0.2 | 29.7 ± 2.2 | 0.1 ± 0.0 | 0.7 ± 0.0 | ||||
| d18:1-N22:1 | 860.59 | 9.7 ± 0.2 | 24.2 ± 2.5 | 0.1 ± 0.0 | 0.9 ± 0.1 | 1.1 ± 0.1 | |||
| d18:1-N22:0 | 862.61 | 0.4 ± 0.1 | 0.4 ± 0.0 | 1.9 ± 0.3 | 15.8 ± 2.1 | 195.4 ± 4.3 | 0.7 ± 0.1 | 4.8 ± 0.4 | 9.0 ± 0.5 |
| d18:0-N22:0 | 864.62 | 0.7 ± 0.1 | 17.4 ± 1.0 | 142.3 ± 6.3 | 0.2 ± 0.0 | 1.8 ± 0.2 | 3.5 ± 0.2 | ||
| d18:1-OHN22:2 | 874.57 | 2.0 ± 0.5 | 6.3 ± 0.1 | 14.4 ± 3.2 | |||||
| d18:1-N23:0 /d18:1-OHN22:1 | 876.61 | 1.2 ± 0.3 | 0.1 ± 0.0 | 1.0 ± 0.2 | 16.6 ± 1.4 | 136.8 ± 3.5 | 0.4 ± 0.0 | 1.6 ± 0.1 | 2.2 ± 0.1 |
| d18:1-OHN22:0 | 878.60 | 1.6 ± 0.1 | 0.2 ± 0.0 | 2.8 ± 0.6 | 151.5 ± 16.1 | 2285.7 ± 224.6 | 1.8 ± 0.1 | 6.3 ± 0.4 | 8.5 ± 0.4 |
| d18:0-OHN22:0 | 880.62 | 0.8 ± 0.1 | 14.6 ± 1.3 | 215.5 ± 27.1 | 0.3 ± 0.0 | 2.1 ± 0.2 | 4.4 ± 0.3 | ||
| d18:1-N24:2 | 886.61 | 0.7 ± 0.1 | 0.8 ± 0.1 | 26.7 ± 1.2 | 61.0 ± 5.0 | 0.2 ± 0.0 | 1.0 ± 0.2 | 4.3 ± 0.4 | |
| d18:1-N24:1 | 888.62 | 2.2 ± 0.2 | 0.9 ± 0.1 | 6.2 ± 1.1 | 83.8 ± 2.4 | 202.4 ± 16.0 | 4.0 ± 0.3 | 24.6 ± 2.0 | 27.3 ± 2.3 |
| d18:1-N24:0 /d18:1-OHN23:1 | 890.64 | 0.4 ± 0.1 | 0.7 ± 0.1 | 11.8 ± 2.5 | 55.8 ± 4.7 | 339.3 ± 7.0 | 2.6 ± 0.3 | 17.5 ± 2.0 | 40.8 ± 2.9 |
| d18:0-N24:0 /d18:1-OHN23:0 | 892.65 | 0.3 ± 0.1 | 0.1 ± 0.0 | 3.0 ± 0.6 | 83.5 ± 13.1 | 1337.5 ± 86.4 | 1.0 ± 0.1 | 4.7 ± 0.5 | 9.3 ± 0.7 |
| d18:1-N25:1 /d18:1-OHN24:2 | 902.62 | 5.6 ± 0.5 | 21.3 ± 2.4 | 0.1 ± 0.0 | 0.4 ± 0.0 | 0.6 ± 0.0 | |||
| d18:1-N25:0 /d18:1-OHN24:1 | 904.62 | 1.1 ± 0.2 | 18.9 ± 2.8 | 144.6 ± 2.8 | 0.9 ± 0.1 | 3.1 ± 0.2 | 3.3 ± 0.3 | ||
| d18:1-OHN24:0 | 906.63 | 0.8 ± 0.2 | 0.2 ± 0.0 | 6.8 ± 1.6 | 99.6 ± 15.8 | 1751.3 ± 140.2 | 2.3 ± 0.2 | 8.6 ± 0.6 | 15.3 ± 1.2 |
| d18:0-OHN24:0 | 908.65 | 1.1 ± 0.3 | 12.8 ± 1.3 | 166.3 ± 13.5 | 0.2 ± 0.0 | 1.5 ± 0.2 | 3.9 ± 0.3 | ||
| d18:1-N26:1 | 916.65 | 0.6 ± 0.1 | |||||||
| d18:1-N26:0 /d18:1-OHN25:1 | 918.67 | 66.5 ± 16.0 | 0.6 ± 0.0 | ||||||
| d18:0-N26:0 /d18:1-OHN25:0 | 920.69 | 76.6 ± 3.8 | 0.4 ± 0.0 | ||||||
| Total | 15.5 ± 2.3 | 3.9 ± 0.5 | 42.3 ± 8.6 | 879.3 ± 58.1 | 8310.3 ± 524.3 | 16.5 ± 1.3 | 86.6 ± 5.8 | 151.1 ± 9.5 |
Total lipids of each biological sample were extracted by using a modified Bligh and Dyer method as described in Materials and Methods. Negative-ion MALDI-MS analyses of sulfatide species in the lipid extracts were performed using 9-AA as matrix and quantitation by ratiometric comparison of the intensity with that of one or two internal standards was obtained as described in “Materials and Methods.” Data represent the means ± SEM of at least six separate animals.
Data are in pmol/mg of protein.
Data are in nmol/mg of protein.
MALDI-MS analyses showed that sulfatides were present in most tissues examined. As is well known, sulfatides are very enriched in the nervous system, particularly where myelin sheath is present, such as spinal cord and sciatic nerve (Table 1). We also demonstrated that abundant sulfatides are also present in kidney medulla, then in kidney cortex. Small amounts of sulfatides were identified in other peripheral organs (e.g., skeletal muscle and liver). Whether sulfatides are synthesized locally in these peripheral organs or transported from the nervous system or from the kidney is unknown at present but is now readily accessible using the developed method. It should be pointed out that we have determined the levels of sulfatides in mouse plasma and found that they are in very low abundance (Table 1), which is different from human plasma, where sulfatide content is much higher than that present in mouse plasma as previously reported (23). However, it is always advised to clean the potential contamination from plasma sulfatides to the tissue samples through perfusion prior to the analysis of any tissue samples.
It is intriguing that the composition of sulfatide species in the peripheral organs is very different from that present in the central nervous system where d18:1-N24:1 sulfatide species is always predominant (Fig. 6; Table 1). For example, hydroxy sulfatide species are very abundant in kidney (e.g., the ions at m/z 878.64 and 906.67 in Fig. 6B, C). In addition, sulfatide species containing sphinganine (d18:0) (e.g., the ion at m/z 892.66 in Fig. 6B, C) are also very enriched in comparison to the counterparts in the nervous system. These profiles certainly not only reflect the different biosynthesis loci but also likely correspond to specific biological functions in each tissue.
We also noted that there exists a caveat in the developed method. Sulfatide species can be identified by accurate mass, selective desorption/ionization under the experimental conditions, and the presence of m/z 97 fragment (corresponding to sulfate) in the product-ion spectra. However, the identities of the fatty acyl substituents or the sphingoid bases of sulfatide species, particularly those in low abundance, cannot be directly determined and must be inferred after assuming the sphingoid base is sphingosine.
SUMMARY
We exploited the unusual properties of 9-AA for the selective desorption/ionization of sulfatides over other anionic lipids present in lipid extracts of biological samples by MALDI-MS. Through this approach, we extended shotgun sphingolipidomics to quantitative analysis of very low abundance sulfatide molecular species present in crude lipid extracts of biological samples. A broad linear dynamic range for quantitation of sulfatide species and a detection limit at the high attomole level were achieved using this approach. Many potential factors that might influence quantitation of sulfatide species were examined and their effects are within the acceptable experimental errors under the conditions identified. The developed method is the most effective and high throughput approach for sulfatide analysis demonstrated to date. Accordingly, we anticipate that this methodology should be very useful for studying sulfatide metabolism, trafficking, and homeostasis at the health and disease states.
Footnotes
Abbreviations:
- 9-AA
- 9-aminoacridine
- DHB
- 2,5-dihydroxybenzoic acid
- PI
- phosphatidylinositol
- TOF
- time-of-flight
This work was supported by National Institutes of Health Grants P01 HL-57278 (R.W.G. and X.H.) and R01 HL-41250 (R.W.G.) as well as National Institutes of Health/National Institute on Aging Grant R01 AG-31675 (X.H.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies. R.W.G. and X.H. have financial relationships with LipoSpectrum LLC. R.W.G. also has a financial relationship with Platomics, Inc.
REFERENCES
- 1.Vos J. P., Lopes-Cardozo M., Gadella B. M. 1994. Metabolic and functional aspects of sulfogalactolipids. Biochim. Biophys. Acta. 1211: 125–149. [DOI] [PubMed] [Google Scholar]
- 2.Eckhardt M. 2008. The role and metabolism of sulfatide in the nervous system. Mol. Neurobiol. 37: 93–103. [DOI] [PubMed] [Google Scholar]
- 3.Kakinuma K., Yamaguchi T., Suzuki H., Nagai Y. 1982. Sulfatide activation of the oxygen radical generating system of leucocytes. FEBS Lett. 145: 16–20. [DOI] [PubMed] [Google Scholar]
- 4.Chiba T., Nagai Y., Kakinuma K. 1987. Cerebroside sulfuric ester (sulfatide) induces oxygen radical generation in guinea-pig leukocytes. Biochim. Biophys. Acta. 930: 10–18. [DOI] [PubMed] [Google Scholar]
- 5.Bengtsson T., Grenegard M., Olsson A., Sjogren F., Stendahl O., Zalavary S. 1996. Sulfatide-induced L-selectin activation generates intracellular oxygen radicals in human neutrophils: modulation by extracellular adenosine. Biochim. Biophys. Acta. 1313: 119–129. [DOI] [PubMed] [Google Scholar]
- 6.Constantin G., Laudanna C., Baron P., Berton G. 1994. Sulfatides trigger cytokine gene expression and secretion in human monocytes. FEBS Lett. 350: 66–70. [DOI] [PubMed] [Google Scholar]
- 7.Laudanna C., Constantin G., Baron P., Scarpini E., Scarlato G., Cabrini G., Dechecchi C., Rossi F., Cassatella M. A., Berton G. 1994. Sulfatides trigger increase of cytosolic free calcium and enhanced expression of tumor necrosis factor-alpha and interleukin-8 mRNA in human neutrophils. Evidence for a role of L-selectin as a signaling molecule. J. Biol. Chem. 269: 4021–4026. [PubMed] [Google Scholar]
- 8.Merten M., Thiagarajan P. 2001. Role for sulfatides in platelet aggregation. Circulation. 104: 2955–2960. [DOI] [PubMed] [Google Scholar]
- 9.Merten M., Beythien C., Gutensohn K., Kuhnl P., Meinertz T., Thiagarajan P. 2005. Sulfatides activate platelets through P-selectin and enhance platelet and platelet-leukocyte aggregation. Arterioscler. Thromb. Vasc. Biol. 25: 258–263. [DOI] [PubMed] [Google Scholar]
- 10.Han X., Holtzman D. M., McKeel D. W., Jr., Kelley J., Morris J. C. 2002. Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer's disease: potential role in disease pathogenesis. J. Neurochem. 82: 809–818. [DOI] [PubMed] [Google Scholar]
- 11.Cheng H., Xu J., McKeel D. W., Jr., Han X. 2003. Specificity and potential mechanism of sulfatide deficiency in Alzheimer's disease: an electrospray ionization mass spectrometric study. Cell. Mol. Biol. 49: 809–818. [PubMed] [Google Scholar]
- 12.von Figura K., Gieselmann V., Jaeken J. 2001. Metachromatic leukodystrophy: lysosomal disorders. The Metabolic and Molecular Bases of Inherited Diseases Sachdev H. S., Beaudet A. L., Sly W. S., Valle D., McGraw-Hill, New York: 3695–3724. [Google Scholar]
- 13.Sandhoff R., Hepbildikler S. T., Jennemann R., Geyer R., Gieselmann V., Proia R. L., Wiegandt H., Grone H. J. 2002. Kidney sulfatides in mouse models of inherited glycosphingolipid disorders: determination by nano-electrospray ionization tandem mass spectrometry. J. Biol. Chem. 277: 20386–20398. [DOI] [PubMed] [Google Scholar]
- 14.Molander-Melin M., Pernber Z., Franken S., Gieselmann V., Mansson J. E., Fredman P. 2004. Accumulation of sulfatide in neuronal and glial cells of arylsulfatase A deficient mice. J. Neurocytol. 33: 417–427. [DOI] [PubMed] [Google Scholar]
- 15.Eckhardt M., Hedayati K. K., Pitsch J., Lullmann-Rauch R., Beck H., Fewou S. N., Gieselmann V. 2007. Sulfatide storage in neurons causes hyperexcitability and axonal degeneration in a mouse model of metachromatic leukodystrophy. J. Neurosci. 27: 9009–9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han X., Fagan A. M., Cheng H., Morris J. C., Xiong C., Holtzman D. M. 2003. Cerebrospinal fluid sulfatide is decreased in subjects with incipient dementia. Ann. Neurol. 54: 115–119. [DOI] [PubMed] [Google Scholar]
- 17.Hu R., Li G., Kamijo Y., Aoyama T., Nakajima T., Inoue T., Node K., Kannagi R., Kyogashima M., Hara A. 2007. Serum sulfatides as a novel biomarker for cardiovascular disease in patients with end-stage renal failure. Glycoconj. J. 24: 565–571. [DOI] [PubMed] [Google Scholar]
- 18.Morichika H., Hamanaka Y., Tai T., Ishizuka I. 1996. Sulfatides as a predictive factor of lymph node metastasis in patients with colorectal adenocarcinoma. Cancer. 78: 43–47. [DOI] [PubMed] [Google Scholar]
- 19.Natowicz M. R., Prence E. M., Chaturvedi P., Newburg D. S. 1996. Urine sulfatides and the diagnosis of metachromatic leukodystrophy. Clin. Chem. 42: 232–238. [PubMed] [Google Scholar]
- 20.Hsu F-F., Bohrer A., Turk J. 1998. Electrospray ionization tandem mass spectrometric analysis of sulfatide. Determination of fragmentation patterns and characterization of molecular species expressed in brain and in pancreatic islets. Biochim. Biophys. Acta. 1392: 202–216. [DOI] [PubMed] [Google Scholar]
- 21.Hsu F-F., Turk J. 2004. Studies on sulfatides by quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes that include an unusual internal galactose residue loss and the classical charge-remote fragmentation. J. Am. Soc. Mass Spectrom. 15: 536–546. [DOI] [PubMed] [Google Scholar]
- 22.Sugiyama E., Hara A., Uemura K. 1999. A quantitative analysis of serum sulfatide by matrix-assisted laser desorption ionization time-of-flight mass spectrometry with delayed ion extraction. Anal. Biochem. 274: 90–97. [DOI] [PubMed] [Google Scholar]
- 23.Li G., Hu R., Kamijo Y., Nakajima T., Aoyama T., Inoue T., Node K., Kannagi R., Kyogashima M., Hara A. 2007. Establishment of a quantitative, qualitative, and high-throughput analysis of sulfatides from small amounts of sera by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Anal. Biochem. 362: 1–7. [DOI] [PubMed] [Google Scholar]
- 24.Merrill A. H., Jr., Sullards M. C., Allegood J. C., Kelly S., Wang E. 2005. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 36: 207–224. [DOI] [PubMed] [Google Scholar]
- 25.Han X., Gross R. W. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda K., Shimizu T., Taguchi R. 2008. Targeted analysis of ganglioside and sulfatide molecular species by LC/ESI-MS/MS with theoretically expanded multiple reaction monitoring. J. Lipid Res. 49: 2678–2689. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X., Cheng H., Yang K., Gross R. W., Han X. 2007. Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low abundance regime of cellular sphingolipids. Anal. Biochem. 371: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sjovall P., Lausmaa J., Johansson B. 2004. Mass spectrometric imaging of lipids in brain tissue. Anal. Chem. 76: 4271–4278. [DOI] [PubMed] [Google Scholar]
- 29.Kyogashima M., Tamiya-Koizumi K., Ehara T., Li G., Hu R., Hara A., Aoyama T., Kannagi R. 2006. Rapid demonstration of diversity of sulfatide molecular species from biological materials by MALDI-TOF MS. Glycobiology. 16: 719–728. [DOI] [PubMed] [Google Scholar]
- 30.Jackson S. N., Wang H. Y., Woods A. S. 2005. Direct profiling of lipid distribution in brain tissue using MALDI-TOFMS. Anal. Chem. 77: 4523–4527. [DOI] [PubMed] [Google Scholar]
- 31.Cha S., Yeung E. S. 2007. Colloidal graphite-assisted laser desorption/ionization mass spectrometry and MSn of small molecules. 1. Imaging of cerebrosides directly from rat brain tissue. Anal. Chem. 79: 2373–2385. [DOI] [PubMed] [Google Scholar]
- 32.Benabdellah F., Touboul D., Brunelle A., Laprevote O. 2009. In situ primary metabolites localization on a rat brain section by chemical mass spectrometry imaging. Anal. Chem. 81: 5557–5560. [DOI] [PubMed] [Google Scholar]
- 33.Sun G., Yang K., Zhao Z., Guan S., Han X., Gross R. W. 2007. Shotgun metabolomics approach for the analysis of negatively charged water-soluble cellular metabolites from mouse heart tissue. Anal. Chem. 79: 6629–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun G., Yang K., Zhao Z., Guan S., Han X., Gross R. W. 2008. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal. Chem. 80: 7576–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiller J., Suss R., Arnhold J., Fuchs B., Lessig J., Muller M., Petkovic M., Spalteholz H., Zschornig O., Arnold K. 2004. Matrix-assisted laser desorption and ionization time-of-flight (MALDI-TOF) mass spectrometry in lipid and phospholipid research. Prog. Lipid Res. 43: 449–488. [DOI] [PubMed] [Google Scholar]
- 36.Schiller J., Suss R., Fuchs B., Muller M., Zschornig O., Arnold K. 2007. MALDI-TOF MS in lipidomics. Front. Biosci. 12: 2568–2579. [DOI] [PubMed] [Google Scholar]
- 37.Christie W. W., Han X. 2010. Lipid analysis: isolation, separation, identification and lipidomic analysis. 4th edition The Oily Press, Bridgwater, England. [Google Scholar]
- 38.Yang K., Cheng H., Gross R. W., Han X. 2009. Automated lipid identification and quantification by multi-dimensional mass spectrometry-based shotgun lipidomics. Anal. Chem. 81: 4356–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng H., Jiang X., Han X. 2007. Alterations in lipid homeostasis of mouse dorsal root ganglia induced by apolipoprotein E deficiency: a shotgun lipidomics study. J. Neurochem. 101: 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X., Yang K., Gross R. W. 2008. Microfluidics-based electrospray ionization enhances intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom. 22: 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han X., Cheng H., Fryer J. D., Fagan A. M., Holtzman D. M. 2003. Novel role for apolipoprotein E in the central nervous system: modulation of sulfatide content. J. Biol. Chem. 278: 8043–8051. [DOI] [PubMed] [Google Scholar]
- 42.Woods A. S., Jackson S. N. 2006. Brain tissue lipidomics: direct probing using matrix-assisted laser desorption/ionization mass spectrometry. AAPS J. 8: E391–E395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han X., Cheng H. 2005. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J. Lipid Res. 46: 163–175. [DOI] [PubMed] [Google Scholar]
- 44.Fuchs B., Bischoff A., Suss R., Teuber K., Schurenberg M., Suckau D., Schiller J. 2009. Phosphatidylcholines and -ethanolamines can be easily mistaken in phospholipid mixtures: a negative ion MALDI-TOF MS study with 9-aminoacridine as matrix and egg yolk as selected example. Anal. Bioanal. Chem. 395: 2479–2487. [DOI] [PubMed] [Google Scholar]
- 45.Hsu F. F., Turk J. 2005. Analysis of sulfatides. The Encyclopedia of Mass Spectrometry. Caprioli R. M., editor Elsevier, New York: 473–492. [Google Scholar]
- 46.Jiang X., Yang K., Han X. 2009. Direct quantitation of psychosine from alkaline-treated lipid extracts with a semi-synthetic internal standard. J. Lipid Res. 50: 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]