Abstract
Human paraoxonase 1 (PON1) is a high-density lipoprotein (HDL)-associated serum enzyme that exhibits a broad substrate specificity. In addition to protecting against exposure to some organophosphorus (OP) pesticides by hydrolyzing their toxic oxon metabolites, PON1 is important in protecting against vascular disease by metabolizing oxidized lipids. Recently, PON1 has also been shown to play a role in inactivating the quorum sensing factor N-(3-oxododecanoyl)-L-homoserine lactone (3OC12-HSL) of Pseudomonas aeruginosa. Native, untagged engineered recombinant human PON1 (rHuPON1) expressed in E. coli and purified by conventional column chromatographic purification is stable, active, and capable of protecting PON1 knockout mice (PON1-/-) from exposure to high levels of the OP compound diazoxon. The bacterially-derived rHuPON1 can be produced in large quantities and lacks the glycosylation of eukaryotic systems that can produce immunogenic complications when inappropriately glycosylated recombinant proteins are used as therapeutics. Previous studies have shown that the determination of PON1 status, which reveals both PON1192 functional genotype and serum enzyme activity level, is required for a meaningful evaluation of PON1’s role in risk of disease or exposure. We have developed a new two-substrate assay/analysis protocol that provides PON1 status without use of toxic OP substrates, allowing for use of this protocol in non-specialized laboratories. Factors were also determined for inter-converting rates of hydrolysis of different substrates. PON1 status also plays an important role in revealing changes in HDL-associated PON1 activities in male patients with Parkinson disease (PD). Immunolocalization studies of PONs 1, 2 and 3 in nearly all mouse tissues suggests that the functions of PONs 1 and 3 extend beyond the plasma and the HDL particle.
Keywords: paraoxonase 1, Parkinson disease, organophosphate, therapy for OP poisoning, chlorpyrifos/chlorpyrifos oxon, diazinon/diazoxon
1.1 Genetics of PON1
1.1.1 Early studies
In 1953, Aldridge divided esterases into two categories, those that catalytically hydrolyzed organophosphate substrates (A-esterases) and those that were inhibited by organophosphates (B-esterases) [1]. Plasma paraoxonase 1 (PON1) is an A-esterase. Studies in the 1960s and 1970s demonstrated that PON1 activity was polymorphically distributed in human populations and the frequency of the low activity phenotype varied among populations of different ethnic origins. These studies are summarized in an excellent review by Geldmacher-von Mallinckrodt and Diepgen [2].
1.1.2 Enzyme assays used to establish Q192R phenotype
The early assays used to assess PON1 phenotype varied from assays that measured the ability of serum samples to protect cholinesterase from inhibition to spectrophotometric assays run under varying conditions of pH, salt concentration and presence or absence of EDTA (subsequently shown to be an inhibitor of Ca2+-dependent PON1). These early assay protocols were reviewed by Ortigoza-Ferado et al. [3]. A major improvement in PON1 phenotype resolution was achieved with a two-substrate assay/analysis pioneered in La Du’s laboratory [4]. Plotting rates of paraoxon (PO) vs. phenyl acetate hydrolysis clearly resolved low metabolizers from high metabolizers but did not resolve heterozygotes from homozygous high metabolizers. This was achieved using the substrate pair diazoxon (DZO) and PO. Plots of rates of DZO hydrolysis vs. PO hydrolysis provided a clear resolution of all three phenotypes (Fig. 1), which by the time of development of the assay had been shown to be PON1Q192 homozygotes, PON1Q/R192 heterozygotes and PON1R192 homozygotes [5]. It had been shown earlier that it was the amino acid at position 192 that determined catalytic efficiency of PON1 for hydrolysis of some substrates [6-8]. The information gleaned from the two-substrate analysis has been referred to as an individual’s PON1 status since it provides both the functional PON1192 genotype as well as the level of plasma PON1 [9].
Figure 1.
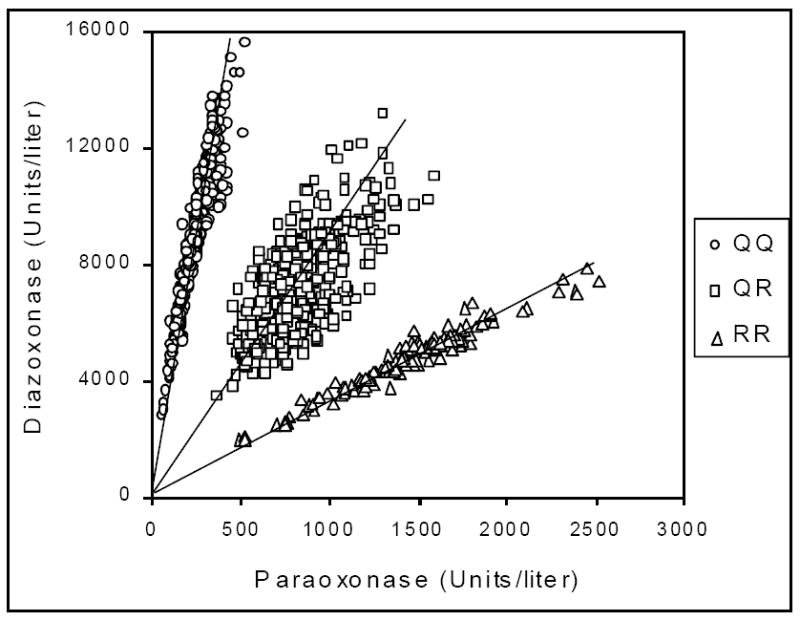
Determination of the functional genomics of plasma PON1 using citrate stored plasma. Plotting the rates of hydrolysis of diazoxon vs. paraoxon for plasma (or serum) samples from a population divides the population into 3 distinct groups, individuals functionally homozygous for PON1Q192, heterozygotes (PON1Q/R192) and individuals homozygous for PON1R192. While PCR analysis of the position 192 polymorphism may reveal that an individual possesses one copy of each allele, this analysis will pick up alleles in heterozygotes that are inactivated by any number of mutations, i.e., the analysis provides the functional status of an individual’s PON1 genomics. Reproduced from [77] with permission.
More recently, we have developed a two substrate assay/analysis protocol that clearly resolves the three PON1 phenotypes (functional genotypes) without the use of the highly toxic OP substrates [10-11]. Plots of rates of phenyl acetate hydrolysis at high salt vs. rates of 4-(chloromethylphenyl) acetate (CMPA) at low salt provide a resolution of phenotypes and functional genotypes comparable to the DZO vs. PO plot. Tables to convert rates of hydrolysis of one substrate to rates of hydrolysis of other substrates were included in the two recent publications [10-11]. Rates of in vivo detoxication at different oxon levels can also be calculated from the in vitro rates and the kinetic parameters of the two PON1192 alloforms. Since the PON1Q192R polymorphism does not affect the rate of hydrolysis of phenyl acetate at low salt concentration, measurement of this rate provides a surrogate measure of plasma PON1 protein level [12].
1.1.3 DNA assays used to establish PON1 genotypes
The elucidation of the initial human PON1 cDNA sequences revealed the two common coding region polymorphisms that have since been studied extensively [13]. Since both the L55M and Q192R polymorphisms included restriction sites for one allele, it was easy to genotype individuals for these two polymorphisms [6]. Single nucleotide polymorphisms (SNPs) were also found in the promoter and 3’ untranslated regions of the PON1 gene. Five of the promoter region polymorphisms have been characterized by three research groups [14-16]. The functionality of the 3’ untranslated region polymorphisms has yet to be studied. Of the promoter region polymorphisms, the C-108T polymorphism appears to have the most significant effect on the regulation of PON1 expression [14, 17]. On average, the PON1C-108 allele expresses approximately twice the level as the PON1T-108 allele [14]. It is interesting that this polymorphism occurs in an Sp1 transcription factor binding site [17]. Sequencing the entire PON1 gene from 47 individuals revealed nearly 200 additional SNPs in the PON1 gene (Fig. 2). There is a relatively low level of linkage disequilibrium across the PON1 gene with non-recombinant blocks of only several thousand bases observed; thus haplotype analysis is not highly informative [18-19].
Figure 2.
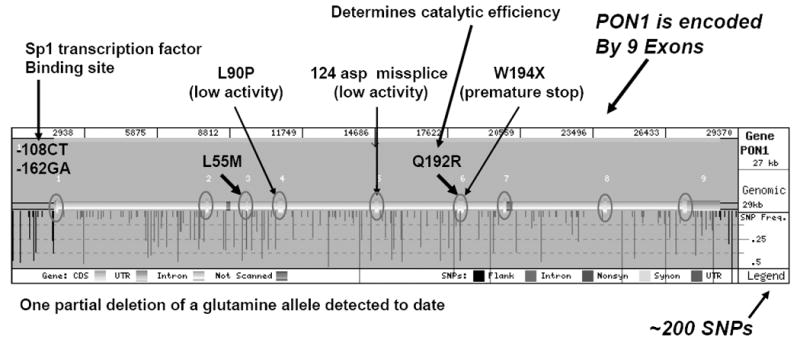
The human PON1 gene with known polymorphisms and their frequencies. The 5’ end of the gene is on the left. (SeattleSNPs, http://pga.gs.washington.edu/)
1.1.4 PON1 genetic variability and risk of OP exposure
1.1.4.1 Rodent models
Early experiments by Main [20] demonstrated that the injection of partially purified rabbit PON1 provided some protection against exposure to PO. This observation was followed up by our laboratory, initially in rats [21] and more extensively in a mouse model system with wild-type mice and genetically modified mice. Injection of purified rabbit PON1 provided some protection against dermal exposure of wild-type mice to PO, but much better protection against exposure to chlorpyrifos oxon [22]. The injected rabbit PON1 also protected against chlorpyrifos exposure [23]. These experiments prompted us to propose the use of PON1 as a therapeutic for OP poisoning [23].
Experiments with PON1-/- mice illustrated the importance of catalytic efficiency of hydrolysis for protecting against OP exposure [24]. Injection of the purified PON1Q192 or PON1R192 alloform into the PON1-/- mice restored resistance to CPO and DZO and provided information on the ability of each alloform to protect against OP exposure. In the case of CPO, the PON1R192 alloform protected better than the PON1Q192 alloform, whereas both alloforms protected equally as well against DZO. Surprisingly, neither alloform protected against PO exposure. Determination of the catalytic efficiency of OP hydrolysis by the two PON1192 alloforms provided a clear explanation for these results. PON1R192 had a higher catalytic efficiency than PON1Q192 for hydrolyzing CPO, whereas both alloforms had nearly equivalent catalytic efficiencies for hydrolyzing DZO. Even though human PON1R192 has approximately 9-times the catalytic efficiency of PON1Q192 for the hydrolysis of PO, it was not high enough to provide protection against exposure. The early in vitro assays that revealed the PON1 Q192R polymorphism was responsible for the large difference in rates of PON hydrolysis between the PON1192 alloforms, leading to the incorrect assumption that high PO hydrolytic activity would protect against PO and parathion (PS) exposure [2].
Experiments with transgenic mouse lines that expressed either recombinant human PON1R192 (rHuPON1R192) or PON1Q192 (rHuPON1Q192) were in complete accord with the results of the injection experiments [25]. Mice expressing tgHuPON1R192 were more resistant to CPO exposure than mice expressing rHuPON1Q192. At high CPO concentrations, the rHuPON1Q192 mice were as sensitive as the PON1-/- mice. This is a significant finding, since approximately one-half of individuals of Northern European origin are homozygous for PON1Q192.
1.1.4.2 Development and OP sensitivity
It was shown early on that the level of plasma arylesterase, since shown to be PON1, in newborns was low at birth and increased over time [26-27]. A study that tracked PON1 activity over time in individual children showed that plasma PON1 levels reach a plateau between 6 and 24 months of age [28]. However, a more recent study reported that PON1 levels can continue to increase beyond 5 years of age [29]. To examine the significance of variable plasma PON1 levels during development, a dose response (sq) to CPO was carried out with PON1-/- and wild-type mice at postnatal day 4. The wild-type mice were already 2.5-times more resistant to CPO than the PON1-/- mice (Cole et al. unpublished data).
1.1.4.3 Human epidemiological studies
Relatively few epidemiological studies in humans relating PON1 status or levels to OP sensitivity have been published. Some studies, e.g., [30] have examined only SNP data which provided no measure of plasma level of PON1, the most important factor in determining OP resistance. As noted above, it is important to determine PON1 status for individuals to obtain an accurate estimate of their ability to detoxify DZO and CPO, the only two OPs for which PON1 status appears to physiologically relevant. Genotyping alone will not provide this information.
A recent study by Hofmann et al. [31] of pesticide handlers in Washington State reported that individuals homozygous for PON1Q192 in the lowest tertile of plasma PON1 levels were the most sensitive to butyrylcholinesterase inhibition during the spraying season, as would be expected. A determination of PON1 status in farm worker mothers and their babies (cord bloods) predicted a range of sensitivity to DZO of 65-fold from the baby with the lowest PON1 status to the mother with the highest PON1 status. Due to the contribution of both PON1 levels and the PON1192 polymorphism for CPO detoxication, the range was even greater, i.e., ~131- to 164-fold [12]. Other studies have also examined PON1 levels and OP susceptibility [32]. Some of these investigators have interpreted the lower Vmax of the PON1R192 homozygous phenotype as a risk for higher sensitivity to diazoxon [32-33]. This is not the case. While the Vmax of PON1R192 is lower than that of PON1Q192, the affinity of PON1R192 is higher than that of PON1Q192 resulting in a slightly higher catalytic efficiency (Vmax/Km) of the PON1R192 alloform [8, 24, 34].
1.1.4.4 Ethics of testing for PON1 status among agricultural workers
The study of the Washington State agricultural workers by Hofmann et al. [31] raised the important ethical question of how to inform a worker that they would be predicted to be highly sensitive to DZO/diazinon (DZ) and CPO/chlorpyrifos (CPF) exposure based on the functional PON1 status assays. One approach to this issue would be to provide anonymous testing through the workers’ union with results provided to the workers along with educational materials in the native language of the workers, written at a level understandable to the worker. It might also be necessary in cases of illiterate workers to provide a verbal explanation and interpretation of the test results by an appropriate spokesperson. Since low PON1 status is also a risk factor for carotid artery disease [35] as well as other vascular disease [36] (see next section) and at-risk individuals can implement protective measures against vascular disease, knowledge of one’s PON1 status may have other health benefits.
2.1 PON1 genetic variability and risk of disease
2.1.1 Diseases associated with PON1 variability
In recent years, there have been approximately 500 papers published related to the association or lack of association of PON1 with specific diseases (PubMed searches). The largest number of studies has been carried out on vascular disease. Other diseases studied for their possible association with genetic variability in PON1 include Parkinson disease (PD), amyotrophic lateral sclerosis (ALS), kidney disease, eye diseases, systemic lupus erythematosus, abdominal aortic aneurysms, asthma, chronic idiopathic pancreatitis, diabetes and associated kidney disease, Chron’s disease, sarcoidosis, erectile dysfunction, systemic vasculitis, glomerulonephritis, breast cancer, prostate cancer, brain tumors, multiple myeloma, dementia and multiple chemical sensitivity, There have been too many studies to cover in this brief review. The reader is referred to recent reviews [37-38]. However some important comments are relevant to all of these studies as well as studies that examine PON1 activity variability as a risk of exposure, infection or other physiological function.
Many of the reported studies have examined PON1 SNPs and ignored plasma PON1 levels, the most important determinant of the rates of metabolism of endogenous or exogenous toxins. Not surprisingly, the results from these studies have provided conflicting information. A number of papers have reviewed these studies [19, 36, 39-42]. Studies that examined either PON1 levels or PON1 status (plasma PON1 activity levels and Q192R genotype) have shown that low PON1 levels are a risk factor for vascular disease [35-36, 43-44]. It stands to reason that higher levels of a protein involved in metabolizing oxidized lipids or toxic OP compounds would be more protective and the level of protection should be related to the activity level of the protein. Conversely, low PON1 levels would be a risk factor for disease or exposure. In some cases, such as the metabolism of CPO, the PON1192 genotype is also important [24], but in no case would the activity level of PON1 be unimportant [19]. The recently-developed high throughput protocol for determining PON1 status without the use of the highly toxic OP substrates PO and DZO should make it convenient for epidemiologists to do a proper study of the relationship of PON1 genetic variability (PON1 status) to CVD and other diseases [11, 45-46].
If new coding region polymorphisms that generate mutant PON1s are discovered, they will most likely be found through the current next-generation DNA sequencing efforts that examine entire exomes, or focused sequencing of PON1 exons (or complete genes) in cases where risk of CVD or other disease maps to the PON1 gene. Promoter mutations that affect expression level would be reflected in the PON1 status analysis as low plasma activity levels. Among individuals that genotype as heterozygotes for position 192, defects in one allele can be detected by comparing Q192R genotyping with the PON1 status analysis [47]. Samples that are discrepant between DNA analyses and PON1 status analyses or which have very low PON1 levels are candidates for defects in one PON1192 allele. For example, if the genotyping indicates a PON1192 heterozygote and the functional PON1 status analysis indicates a position PON1192Q or PON1192R homozygote, this would be presumptive evidence for a defective PON1192 allele that can be verified by DNA sequencing [47].
3.1 PON1 status as a possible indicator of defects in the modulation of oxidative stress
Several recent studies have suggested linkage between Parkinson disease (PD) and genetic variability in the PON region of chromosome 7 [48-51], while others have not [52-55]. Based on our knowledge of the importance of PON1 status in risk of disease or exposure, we felt that it would be important to carry out a PON1 status analysis on a sizeable cohort of patients with PD. We expected to find lower PON1 levels in PD patients compared with control subjects as we had seen in our earlier study of PON1 status and carotid artery disease (CAAD) [35]. We did not find such differences; however, we did find differences in ratios of rates of substrate hydrolysis in two dimensional plots of activity rates with different substrates. Plots of arylesterase activity vs. paraoxonase activity (POase) showed differences in males with PD compared with control subjects, but not in females with PD (Furlong et al. unpublished data). A likely explanation of these data is that the environment of the HDL particle in which PON1 is localized is affected by defects in the modulation of oxidative stress that in turn are reflected in subtle differences in rates of hydrolysis of different substrates. This explanation is reasonable since many of the mutations that lead to PD are related to defects in mitochondrial function [56-58].
4.1 PONs and quorum sensing
Ozer et al. reported that all three PONs hydrolyzed the quorum sensing factor of Pseudomonas aeruginosa N-3-oxododecanoyl homoserine lactone (3OC12-HSL) [59]. Exposure experiments with PON1-/- mice were inconclusive in demonstrating a protective effect for PON1 due to the abilities of PON2 and PON3 to inactivate 3OC12-HSL [59]. Experiments with PON2-/- mice were more convincing with respect to demonstrating a protective role of PON2 against P. aeruginosa infection [60]. The ability of PON1 to protect against the lethality of P. aeruginosa infection was demonstrated by expressing human PON1 in Drosophila [61]. These flies were also resistant to chlorpyrifos exposure. These observations led the authors to speculate about the roles for PON1 in human-pathogen interactions, including a role for PON1 as a regulator for normal bacterial florae, and a link between infection/inflammation and cardiovascular disease. The potential for therapy was also noted. Thus, in addition to the role of PON1 in protecting against OP exposure and the importance of all three PONs in modulating oxidative stress, all three PONs appear to function in innate immunity via their abilities to inactivate bacterial quorum sensing factors.
5.1 PON1 as a potential therapeutic
Our early studies [22-23] following those of Main [20] indicated that it would be possible to use PON1 purified from human plasma to treat cases of CPF/CPO and DZ/DZO exposure, with the PON1R192 alloform being the best choice of alloforms, since it hydrolyzes both CPO and DZO efficiently [24]. However, it will be necessary to engineer recombinant human PON1 for higher catalytic efficiency for use in treating exposure to OPs hydrolyzed with low catalytic efficiency [24, 62-63].
To test this hypothesis, we expressed three recombinant human PON1 alloforms in an E. coli expression system, rHuPON1R192, rHuPON1Q192 and the engineered variant rHuPON1K192. The replacement of Q/R-192 with lysine was suggested from our earlier experiments where we observed high catalytic efficiency of chlorpyrifos oxon with rabbit PON1, which has lysine at position 192 [64] and experiments with recombinant GSTHuPON1 constructs that showed increased OP hydrolase activity for PO, CPO and DZO when lysine was present at position 192 [65]. We were able to express and purify untagged rHuPON1 variants in the E. coli expression system using ion exchange and hydrophobic interaction chromatography [62]. The rHuPON1K192 variant was about twice as efficient as the rHuPON1R192 protein for hydrolyzing CPO, PO and DZO.
The requirements for a protein to be useful as a therapeutic are that it must be non-toxic and non-immunogenic when injected into an animal or human, it should persist in the system for some time, it should protect against or be useful in treating OP exposures and should have a useful shelf-life. Injection intraperitoneally (ip) of the purified rHuPON1K192 (1.2 U) into PON1-/- mice showed that the rHuPON1K192 was non-toxic and persisted in plasma beyond two days following injection. A second experiment where 3.91 U of rHuPON1K192 were injected intraperitoneally (ip) into the PON1-/- mice followed by a dermal challenge of 1 mg/kg DZO demonstrated that the PON1 still in the plasma 48 h post-injection was able to protect against this exposure, demonstrating that the rHuPON1K192 could be used prophylactically to prevent ChE inhibition by an OP exposure. These mice showed no ill effects from the injected PON1, and anti-PON1 antibodies were not detectable by ELISA 4 months postexposure. The mice survived more than 12 months post-exposure. To test the ability of rHuPON1 to be used as a therapeutic post OP exposure, two PON1-/- mice were exposed to 2 and 3 LD50 dermal doses of DZO followed 10 min later by ip and intramuscular (im) injections of a total of 3.91 U of rHuPON1K192. The injected rHuPON1K192 protected the mice against these lethal exposures of OP. Since these doses would have killed a PON1-/- mouse, the control group, that also received DZO, was only given 1.5 mg/kg. The two mice that received the high-dose exposures and the injected rHuPON1K192 post-exposure not only survived, but showed less severe symptoms of intoxication when compared to the control group which received the lower DZO exposure. They did not demonstrate any symptoms related to the injection of the rHuPON1K192. Thus, these experiments demonstrated that 1) it was possible to engineer human PON1 for higher catalytic efficiency on the native rHuPON1 scaffold in an E. coli expression system, 2) the injected rHuPON1 persisted for more than 2 days post exposure, 3) the injected rHuPON1 protected against high levels of OP exposure, 4) the injected rHuPON1 was not immunogenic in a mouse that lacked PON1, 5) the lack of glycosylation did not change the half-life of the injected PON1 significantly, nor the activity of the rHuPON1K192 and 6) the purified rHuPON1K192 was stable for more than two months at 4° C. These experiments also indicate that it should be possible to generate rHuPON1 for use in treating individuals whose vascular disease may result from very low PON1 levels [35, 66-67] and individuals who may be susceptible to infection by Pseudomonas aeruginosa due to low PON1 levels [61, 67]. PON1 may also be useful for treating other diseases that result from low PON1 levels.
6.1 Immunolocalization of the PON family of enzymes
Circulating PON1 is synthesized in the liver [13, 68] and the secreted PON1 circulates in plasma tightly bound to HDL [68], using its unprocessed hydrophobic signal sequence as an anchor into the lipoprotein particles [13, 69]. Some authors have reported PON1 protein expression in kidney and aorta [70-71]. Immunolocalization of PON1 has been described in nearly all the tissues of mice [72]. Specifically, PON1 was found in nearly all the studied epithelia, such as chondrocytes, enterocytes, eye lens and retinal layers, skin epidermis, stomach, tongue and trachea. Since PON1 metabolizes toxic agents such as oxidized lipids, it would be logical to find the protein where its function is needed. In this vein, PON1 was also found in the muscle fibers of both skeletal and cardiac muscle, areas where free radicals are produced as a consequence of energy metabolism. PON1 also plays a protective role against lipid peroxidation, so it is not surprising to find this protein in adipocytes, and acini from exocrine pancreas, submandibular gland and sebaceous gland. Consistent with this concept, PON1 is also expressed in cells where oxidative stress occurs and lack of detoxication could result in significant disease in liver, kidney proximal tubules, fiber tracts of the encephalon and the spinal cord. Finally, inability to modulate oxidative stress could contribute to infertility, emphasizing the importance of finding PON1 in ovary follicular fluid, seminiferous tubules and spermatozoa.
Similar results were reported in human tissues by the Swedish Human Protein Atlas Database (http://proteinatlas.org/tissue_profile.php?antibody_id=1610). Recently, PON1 has also been localized in macrophages, endothelial cells and smooth muscle cells of human aorta with or without atherosclerosis (Marsillach, unpublished data), and in human lens tissues [73]. These findings open a debate about where PON1 is synthesized and its possible functions.
Sorenson et al. [69] were the first to demonstrate that phospholipids competitively remove PON1 from HDL, suggesting that PON1 may migrate between HDL and cell membranes. Some years later, Deakin et al. [74] described the transfer of PON1 from membrane to HDL, supporting the idea of the transfer process. More recently, Efrat et al. [75] demonstrated that PON1 is able to bind to macrophages and be internalized in them. The authors speculated that the binding could be via the union of HDL to macrophage scavenger receptor B1 (SR-B1) and the anchor of PON1 to cell membrane phospholipids. These observations suggest that although synthesized by the liver and tightly bound to HDL, PON1 may also have an important role outside the HDL complex.
These findings also raise the possibility of local PON1 synthesis in these cells. In the mouse study [71], authors did not find co-localization of PON1 with apo A-I in tissues. This could result from local synthesis of PON1 or a transfer of PON1 from HDL without a co-transfer of apo A-I. However, as yet, no PON1 gene expression has been reported for other tissues. Additional studies should be carried out to address these questions. The finding of PON1 protein in so many tissues [72-73] [http://proteinatlas.org/tissue_profile.php?antibody_id=1610] [Marsillach et al. unpublished data] not only suggests the transfer of PON1 from the liver to the other tissues, but may also reveal an important role for this enzyme in preventing oxidative stress and inflammation in other tissues, making it an important candidate to be studied in the development of other diseases. PON2 and PON3 are the other members of the paraoxonase family and PON gene cluster. The three share common activities of modulating oxidative stress and inactivating quorum sensing factors. Like PON1, PON3 is also found in HDL while PON2 seems to be a ubiquitously expressed intracellular enzyme, not found in circulation. Unlike the gene expression results obtained for PON2 in humans [76], PON2 appears to be poorly expressed in mouse tissues studied by Marsillach et al. [72] who reported a very light staining of PON2 in some tissues where PON1 was found, and only strong staining in skeletal and cardiac muscle endomysium, ovary follicular fluid and ducts from submandibular glands. PON3 protein expression paralleled that of PON1. The antibodies used in these experiments were raised against peptides from specific sequence of each mature PON, so they lack of cross-reactivity between the 3 enzymes.
Summary
The many studies carried out on the PON family of enzymes have provided convincing evidence for the protective role of PON1 in modulating exposures to CPO/CPF and DZO/DZ, but not in modulating exposure to PS/PO or nerve agents. To protect against the latter OPs, PON1 will need to be engineered for increased catalytic efficiency. All three PONs appear to be important in modulating oxidative stress and protecting against vascular disease and perhaps other diseases that result from deficiencies in modulating oxidative stress. The recent findings that PONs hydrolyze microbial quorum sensing factors and that PON1 can protect against the lethality of infections from Pseudomonas aeruginosa show that they are also important components of the innate immunity system. Space does not permit a discussion of the roles of the PONs in the pharmacokinetics of drug metabolism, however, the papers published to-date suggest that this will be a fertile area for future investigation. Again, it is important to note that investigations of the role of PON1 in risk of disease of exposures should at a minimum determine PON1 status for the study subjects. Knowing only the PON1192 genotype or SNP haplotype provides wholly inadequate information for an epidemiological study. Convenient high-throughput assays have been developed for determining PON1 status. All of the nearly 200 SNPs in the PON1 gene could be characterized and one would not be able to accurately predict PON1 activity levels for an individual, the most important factor in resistance to exposure or disease [19].
Acknowledgments
This work was supported by the grants from the NIH (ES04696, ES09883; ES07033; ES09601, HL67406, R01 NS065070, P50 NS062684) and the Department of Veterans Affairs (Merit Review Award). JM was supported by a Beatriu de Pinós postdoctoral fellowship (2008 BP A 00166) from Comissionat per a Universitats i Recerca del Departament d’Innovació, Universitats i Empresa, Catalunya Spain.
Abbreviations
- CPF
chlorpyrifos
- CPO
chlorpyrifos oxon
- DZ
diazoxon
- DZO
diazoxon
- PO
paraoxon
- PD
Parkinson disease
- PON1
paraoxonase 1
- PON2
paraoxonase 2
- PON3
paraoxonase 3
- PCR
polymerase chain reaction
- tgHuPON1R192
transgenic human PON1R192
- tgHuPON1Q192
transgenic human PON1Q192
- PS
parathion
- CVD
cardiovascular disease
- CAAD
coronary artery disease
- POase
paraoxonase activity
- 3OC12-HSL
N-(3-oxododecanoyl)-L-homoserine lactone
- sq
subcutaneous
- ip
intraperitoneally
- im
intramuscularly
- SNP
single nucleotide polymorphism
- OP
organophosphorus compound
- EDTA
ethylenediaminetetraacetate
- rHuPON1K192, rHuPON1R192, rHuPON1K192
recombinant human PON1192 variants
- HDL
high density lipoprotein
- SR-B1
macrophage scavenger receptor B1
Footnotes
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Aldridge WN. Serum esterases. I. Two types of esterase (A and B) hydrolysing p-nitrophenyl acetate, propionate and butyrate, and a method for their determination. Biochem J. 1953;53:110–7. doi: 10.1042/bj0530110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geldmacher-von Mallinckrodt M, D TL. The Human Serum Paraoxonase-Polymorphism and Specificity. Toxicological and Environmental Chemistry. 1988;18:79–196. [Google Scholar]
- 3.Ortigoza-Ferado J, et al. Paraoxon hydrolysis in human serum mediated by a genetically variable arylesterase and albumin. Am J Hum Genet. 1984;36:295–305. [PMC free article] [PubMed] [Google Scholar]
- 4.Eckerson HW, Wyte CM, La Du BN. The human serum paraoxonase/arylesterase polymorphism. Am J Hum Genet. 1983;35:1126–38. [PMC free article] [PubMed] [Google Scholar]
- 5.Richter RJ, Furlong CE. Determination of paraoxonase (PON1) status requires more than genotyping. Pharmacogenetics. 1999;9:745–53. [PubMed] [Google Scholar]
- 6.Humbert R, et al. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet. 1993;3:73–6. doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- 7.Adkins S, et al. Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: glutamine or arginine at position 191, for the respective A or B allozymes. Am J Hum Genet. 1993;52:598–608. [PMC free article] [PubMed] [Google Scholar]
- 8.Davies HG, et al. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat Genet. 1996;14:334–6. doi: 10.1038/ng1196-334. [DOI] [PubMed] [Google Scholar]
- 9.Kaliste-Korhonen E, et al. Phosphotriesterase decreases paraoxon toxicity in mice. Toxicol Appl Pharmacol. 1993;121:275–8. doi: 10.1006/taap.1993.1154. [DOI] [PubMed] [Google Scholar]
- 10.Richter RJ, J GP, Furlong Clement E. Determination of Paraoxonase 1 Status Without the Use of Toxic Organophosphate Substrates. Circ Cardiovasc Genet. 2008;1:147–152. doi: 10.1161/CIRCGENETICS.108.811638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richter RJ, Jarvik GP, Furlong CE. Paraoxonase 1 (PON1) status and substrate hydrolysis. Toxicol Appl Pharmacol. 2009;235:1–9. doi: 10.1016/j.taap.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furlong CE, et al. PON1 status of farmworker mothers and children as a predictor of organophosphate sensitivity. Pharmacogenet Genomics. 2006;16:183–90. doi: 10.1097/01.fpc.0000189796.21770.d3. [DOI] [PubMed] [Google Scholar]
- 13.Hassett C, et al. Characterization of cDNA clones encoding rabbit and human serum paraoxonase: the mature protein retains its signal sequence. Biochemistry. 1991;30:10141–9. doi: 10.1021/bi00106a010. [DOI] [PubMed] [Google Scholar]
- 14.Brophy VH, et al. Effects of 5’ regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. Am J Hum Genet. 2001;68:1428–36. doi: 10.1086/320600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suehiro T, et al. A polymorphism upstream from the human paraoxonase (PON1) gene and its association with PON1 expression. Atherosclerosis. 2000;150:295–8. doi: 10.1016/s0021-9150(99)00379-2. [DOI] [PubMed] [Google Scholar]
- 16.Leviev I, James RW. Promoter polymorphisms of human paraoxonase PON1 gene and serum paraoxonase activities and concentrations. Arterioscler Thromb Vasc Biol. 2000;20:516–21. doi: 10.1161/01.atv.20.2.516. [DOI] [PubMed] [Google Scholar]
- 17.Deakin S, et al. Paraoxonase-1 promoter haplotypes and serum paraoxonase: a predominant role for polymorphic position - 107, implicating the Sp1 transcription factor. Biochem J. 2003;372:643–9. doi: 10.1042/BJ20021670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvik GP, et al. Paraoxonase activity, but not haplotype utilizing the linkage disequilibrium structure, predicts vascular disease. Arterioscler Thromb Vasc Biol. 2003;23:1465–71. doi: 10.1161/01.ATV.0000081635.96290.D3. [DOI] [PubMed] [Google Scholar]
- 19.Furlong CE, R R, Li W-F, Brophy VH, Carlson C, Meider M, Nickerson D, Costa LG, Ranchalis J, Lusis AJ, Shih DM, Tward A, Jarvik GP. The functional consequences of polymorphisms in the human PON1 gene. In: M M, Mackness B, Aviram M, Paragh G, editors. The Paraoxonases: Their Role in Disease Development and Xenobiotic Metabolism. Springer; Dordrecht, The Netherlands: 2008. [Google Scholar]
- 20.Main AR. The Role of A-esterase in the acute toxicity of paraoxon, TEEP and parathion. Can J Biochem Physiol. 1956;34:197–216. [PubMed] [Google Scholar]
- 21.Costa LG, M B, Murphy SD, Omenn GS, Richter RJ, Motulsky AG, Furlong CE. Serum paraoxonase and its influence on paraoxon and chlorpyrifos-oxon toxicity in rats. Toxicol Appl Pharmacol. 1990;103:66–76. doi: 10.1016/0041-008x(90)90263-t. [DOI] [PubMed] [Google Scholar]
- 22.Li WF, Costa LG, Furlong CE. Serum paraoxonase status: a major factor in determining resistance to organophosphates. J Toxicol Environ Health. 1993;40:337–46. doi: 10.1080/15287399309531798. [DOI] [PubMed] [Google Scholar]
- 23.Chae MY, et al. Utilization of copper as a paramagnetic probe for the binuclear metal center of phosphotriesterase. Arch Biochem Biophys. 1995;316:765–72. doi: 10.1006/abbi.1995.1102. [DOI] [PubMed] [Google Scholar]
- 24.Li WF, et al. Catalytic efficiency determines the in-vivo efficacy of PON1 for detoxifying organophosphorus compounds. Pharmacogenetics. 2000;10:767–79. doi: 10.1097/00008571-200012000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Cole TB, et al. Toxicity of chlorpyrifos and chlorpyrifos oxon in a transgenic mouse model of the human paraoxonase (PON1) Q192R polymorphism. Pharmacogenet Genomics. 2005;15:589–98. doi: 10.1097/01.fpc.0000167327.08034.d2. [DOI] [PubMed] [Google Scholar]
- 26.Augustinsson KB, B M. Age variation in plasma arylesterase activity in children. Clin Chim Acta. 1963;8:568–573. doi: 10.1016/0009-8981(63)90106-2. [DOI] [PubMed] [Google Scholar]
- 27.Ecobichon DJ, S D. Perinatal development of human blood esterases. Clin Pharmacol Ther. 1973;14:41–47. doi: 10.1002/cpt197314141. [DOI] [PubMed] [Google Scholar]
- 28.Cole TB, et al. Expression of human paraoxonase (PON1) during development. Pharmacogenetics. 2003;13:357–64. doi: 10.1097/00008571-200306000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Huen K, et al. Developmental Changes in PON1 Enzyme Activity in Young Children and Effects of PON1 Polymorphisms. Environmental Health Perspectives. 2009;117:1632–1638. doi: 10.1289/ehp.0900870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee BW, et al. Association between human paraoxonase gene polymorphism and chronic symptoms in pesticide-exposed workers. J Occup Environ Med. 2003;45:118–22. doi: 10.1097/01.jom.0000052953.59271.e1. [DOI] [PubMed] [Google Scholar]
- 31.Hofmann JN, et al. Serum cholinesterase inhibition in relation to paraoxonase- 1 (PON1) status among organophosphate-exposed agricultural pesticide handlers. Environ Health Perspect. 2009;117:1402–8. doi: 10.1289/ehp.0900682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackness B, et al. Paraoxonase and susceptibility to organophosphorus poisoning in farmers dipping sheep. Pharmacogenetics. 2003;13:81–8. doi: 10.1097/00008571-200302000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Cherry N, et al. Paraoxonase (PON1) polymorphisms in farmers attributing ill health to sheep dip. Lancet. 2002;359:763–4. doi: 10.1016/s0140-6736(02)07847-9. [DOI] [PubMed] [Google Scholar]
- 34.Jansen KL, et al. Paraoxonase 1 (PON1) modulates the toxicity of mixed organophosphorus compounds. Toxicol Appl Pharmacol. 2009;236:142–53. doi: 10.1016/j.taap.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarvik GP, et al. Paraoxonase (PON1) phenotype is a better predictor of vascular disease than is PON1(192) or PON1(55) genotype. Arterioscler Thromb Vasc Biol. 2000;20:2441–7. doi: 10.1161/01.atv.20.11.2441. [DOI] [PubMed] [Google Scholar]
- 36.Mackness B, et al. Paraoxonase status in coronary heart disease: are activity and concentration more important than genotype? Arterioscler Thromb Vasc Biol. 2001;21:1451–7. doi: 10.1161/hq0901.094247. [DOI] [PubMed] [Google Scholar]
- 37.Li HL, Liu DP, Liang CC. Paraoxonase gene polymorphisms, oxidative stress, and diseases. J Mol Med. 2003;81:766–79. doi: 10.1007/s00109-003-0481-4. [DOI] [PubMed] [Google Scholar]
- 38.Camps J, Marsillach J, Joven J. The paraoxonases: role in human diseases and methodological difficulties in measurement. Crit Rev Clin Lab Sci. 2009;46:83–106. doi: 10.1080/10408360802610878. [DOI] [PubMed] [Google Scholar]
- 39.Lawlor DA, et al. The association of the PON1 Q192R polymorphism with coronary heart disease: findings from the British Women’s Heart and Health cohort study and a meta-analysis. BMC Genet. 2004;5:17. doi: 10.1186/1471-2156-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheeler JG, et al. Four paraoxonase gene polymorphisms in 11212 cases of coronary heart disease and 12786 controls: meta-analysis of 43 studies. Lancet. 2004;363:689–95. doi: 10.1016/S0140-6736(04)15642-0. [DOI] [PubMed] [Google Scholar]
- 41.Deakin SP, James RW. Genetic and environmental factors modulating serum concentrations and activities of the antioxidant enzyme paraoxonase-1. Clin Sci (Lond) 2004;107:435–47. doi: 10.1042/CS20040187. [DOI] [PubMed] [Google Scholar]
- 42.La Du BN. Future studies of low-activity PON1 phenotype subjects may reveal how PON1 protects against cardiovascular disease. Arterioscler Thromb Vasc Biol. 2003;23:1317–8. doi: 10.1161/01.ATV.0000082600.42562.7F. [DOI] [PubMed] [Google Scholar]
- 43.Graner M, et al. Association of paraoxonase-1 activity and concentration with angiographic severity and extent of coronary artery disease. J Am Coll Cardiol. 2006;47:2429–35. doi: 10.1016/j.jacc.2006.01.074. [DOI] [PubMed] [Google Scholar]
- 44.Gur M, et al. Paraoxonase and arylesterase activities in coronary artery disease. Eur J Clin Invest. 2006;36:779–87. doi: 10.1111/j.1365-2362.2006.01727.x. [DOI] [PubMed] [Google Scholar]
- 45.Richter Rebecca J, J GP, Furlong Clement E. Determination of Paraoxonase 1 Status Without the Use of Toxic Organophosphate Substrates. Circ Cardiovasc Genet. 2008;1:147–152. doi: 10.1161/CIRCGENETICS.108.811638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loscalzo J. Paraoxonase and Coronary Heart Disease Risk: Language Misleads, Linkage Misinforms, Function Clarifies. Circ Cardiovasc Genet. 2008;1:79–80. doi: 10.1161/CIRCGENETICS.108.837179. [DOI] [PubMed] [Google Scholar]
- 47.Jarvik GP, et al. Novel paraoxonase (PON1) nonsense and missense mutations predicted by functional genomic assay of PON1 status. Pharmacogenetics. 2003;13:291–5. doi: 10.1097/00008571-200305000-00009. [DOI] [PubMed] [Google Scholar]
- 48.Duric G, et al. Polymorphisms in the genes of cytochrome oxidase P450 2D6 (CYP2D6), paraoxonase 1 (PON1) and apolipoprotein E (APOE) as risk factors for Parkinson’s disease. Vojnosanit Pregl. 2007;64:25–30. doi: 10.2298/vsp0701025d. [DOI] [PubMed] [Google Scholar]
- 49.Akhmedova SN, Yakimovsky AK, Schwartz EI. Paraoxonase 1 Met--Leu 54 polymorphism is associated with Parkinson’s disease. J Neurol Sci. 2001;184:179–82. doi: 10.1016/s0022-510x(01)00439-7. [DOI] [PubMed] [Google Scholar]
- 50.Carmine A, et al. Further evidence for an association of the paraoxonase 1 (PON1) Met-54 allele with Parkinson’s disease. Mov Disord. 2002;17:764–6. doi: 10.1002/mds.10172. [DOI] [PubMed] [Google Scholar]
- 51.Kondo I, Yamamoto M. Genetic polymorphism of paraoxonase 1 (PON1) and susceptibility to Parkinson’s disease. Brain Res. 1998;806:271–3. doi: 10.1016/s0006-8993(98)00586-1. [DOI] [PubMed] [Google Scholar]
- 52.Kelada SN, et al. Paraoxonase 1 promoter and coding region polymorphisms in Parkinson’s disease. J Neurol Neurosurg Psychiatry. 2003;74:546–7. doi: 10.1136/jnnp.74.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clarimon J, et al. Paraoxonase 1 (PON1) gene polymorphisms and Parkinson’s disease in a Finnish population. Neurosci Lett. 2004;367:168–70. doi: 10.1016/j.neulet.2004.05.108. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Liu Z. No association between paraoxonase 1 (PON1) gene polymorphisms and susceptibility to Parkinson’s disease in a Chinese population. Mov Disord. 2000;15:1265–7. doi: 10.1002/1531-8257(200011)15:6<1265::aid-mds1034>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 55.Fong CS, Cheng CW, Wu RM. Pesticides exposure and genetic polymorphism of paraoxonase in the susceptibility of Parkinson’s disease. Acta Neurol Taiwan. 2005;14:55–60. [PubMed] [Google Scholar]
- 56.Mandemakers W, Morais VA, De Strooper B. A cell biological perspective on mitochondrial dysfunction in Parkinson disease and other neurodegenerative diseases. J Cell Sci. 2007;120:1707–16. doi: 10.1242/jcs.03443. [DOI] [PubMed] [Google Scholar]
- 57.Pankratz N, Foroud T. Genetics of Parkinson disease. Genet Med. 2007;9:801–11. doi: 10.1097/gim.0b013e31815bf97c. [DOI] [PubMed] [Google Scholar]
- 58.Lim KL, Ng CH. Genetic models of Parkinson disease. Biochim Biophys Acta. 2009;1792:604–15. doi: 10.1016/j.bbadis.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 59.Ozer EA, et al. Human and murine paraoxonase 1 are host modulators of Pseudomonas aeruginosa quorum-sensing. FEMS Microbiol Lett. 2005;253:29–37. doi: 10.1016/j.femsle.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 60.Stoltz DA, et al. Paraoxonase-2 deficiency enhances Pseudomonas aeruginosa quorum sensing in murine tracheal epithelia. Am J Physiol Lung Cell Mol Physiol. 2007;292:L852–60. doi: 10.1152/ajplung.00370.2006. [DOI] [PubMed] [Google Scholar]
- 61.Stoltz DA, et al. Drosophila are protected from Pseudomonas aeruginosa lethality by transgenic expression of paraoxonase-1. J Clin Invest. 2008;118:3123–31. doi: 10.1172/JCI35147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stevens RC, et al. Engineered recombinant human paraoxonase 1 (rHuPON1) purified from Escherichia coli protects against organophosphate poisoning. Proc Natl Acad Sci U S A. 2008;105:12780–4. doi: 10.1073/pnas.0805865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lenz DE, et al. Stoichiometric and catalytic scavengers as protection against nerve agent toxicity: a mini review. Toxicology. 2007;233:31–9. doi: 10.1016/j.tox.2006.11.066. [DOI] [PubMed] [Google Scholar]
- 64.Furlong CE, et al. Spectrophotometric assays for the enzymatic hydrolysis of the active metabolites of chlorpyrifos and parathion by plasma paraoxonase/arylesterase. Anal Biochem. 1989;180:242–7. doi: 10.1016/0003-2697(89)90424-7. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki SM, S R, Richter RJ, Cole TB, Park S, Otto TC, Cerasoli DM, Lenz DE, Furlong CE, editors. Paraoxonases in Inflammation, Infection and Toxicology. Humana Press; 2010. Engineering Human PON1 in an E. coli Expression System. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shih DM, Lusis AJ. The roles of PON1 and PON2 in cardiovascular disease and innate immunity. Curr Opin Lipidol. 2009;20:288–92. doi: 10.1097/MOL.0b013e32832ca1ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackness MI, Durrington PN, Mackness B. The role of paraoxonase 1 activity in cardiovascular disease: potential for therapeutic intervention. Am J Cardiovasc Drugs. 2004;4:211–7. doi: 10.2165/00129784-200404040-00002. [DOI] [PubMed] [Google Scholar]
- 68.Blatter MC, et al. Identification of a distinct human high-density lipoprotein subspecies defined by a lipoprotein-associated protein, K-45. Identity of K-45 with paraoxonase. Eur J Biochem. 1993;211:871–9. doi: 10.1111/j.1432-1033.1993.tb17620.x. [DOI] [PubMed] [Google Scholar]
- 69.Sorenson RC, et al. Human serum Paraoxonase/Arylesterase’s retained hydrophobic N-terminal leader sequence associates with HDLs by binding phospholipids : apolipoprotein A-I stabilizes activity. Arterioscler Thromb Vasc Biol. 1999;19:2214–25. doi: 10.1161/01.atv.19.9.2214. [DOI] [PubMed] [Google Scholar]
- 70.Rodrigo L, et al. Immunohistochemical evidence for the expression and induction of paraoxonase in rat liver, kidney, lung and brain tissue. Implications for its physiological role. Chem Biol Interact. 2001;137:123–37. doi: 10.1016/s0009-2797(01)00225-3. [DOI] [PubMed] [Google Scholar]
- 71.Mackness B, et al. Increased immunolocalization of paraoxonase, clusterin, and apolipoprotein A-I in the human artery wall with the progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 1997;17:1233–8. doi: 10.1161/01.atv.17.7.1233. [DOI] [PubMed] [Google Scholar]
- 72.Marsillach J, et al. Immunohistochemical analysis of paraoxonases-1, 2, and 3 expression in normal mouse tissues. Free Radic Biol Med. 2008;45:146–57. doi: 10.1016/j.freeradbiomed.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 73.Hashim Z, et al. Expression and activity of paraoxonase 1 in human cataractous lens tissue. Free Radic Biol Med. 2009;46:1089–95. doi: 10.1016/j.freeradbiomed.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 74.Deakin S, et al. Enzymatically active paraoxonase-1 is located at the external membrane of producing cells and released by a high affinity, saturable, desorption mechanism. J Biol Chem. 2002;277:4301–8. doi: 10.1074/jbc.M107440200. [DOI] [PubMed] [Google Scholar]
- 75.Efrat M, Aviram M. Macrophage paraoxonase 1 (PON1) binding sites. Biochem Biophys Res Commun. 2008;376:105–10. doi: 10.1016/j.bbrc.2008.08.106. [DOI] [PubMed] [Google Scholar]
- 76.Ng CJ, et al. The paraoxonase gene family and atherosclerosis. Free Radic Biol Med. 2005;38:153–63. doi: 10.1016/j.freeradbiomed.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 77.Richter RJ, J R, Jarvik GP, Costa LG, Furlong CE. Determination of paraoxonase 1 (PON1) status and genotypes at specific polymorphic sites. Current Protocols in Toxicology. 2004;2004:4.12.1–4.12.19. doi: 10.1002/0471140856.tx0412s19. [DOI] [PubMed] [Google Scholar]


