Abstract
The biomedical literature holds our understanding of pharmacogenomics, but it is dispersed across many journals. In order to integrate our knowledge, connect important facts across publications and generate new hypotheses we must organize and encode the contents of the literature. By creating databases of structured pharmocogenomic knowledge, we can make the value of the literature much greater than the sum of the individual reports. We can, for example, generate candidate gene lists or interpret surprising hits in genome-wide association studies. Text mining automatically adds structure to the unstructured knowledge embedded in millions of publications, and recent years have seen a surge in work on biomedical text mining, some specific to pharmacogenomics literature. These methods enable extraction of specific types of information and can also provide answers to general, systemic queries. In this article, we describe the main tasks of text mining in the context of pharmacogenomics, summarize recent applications and anticipate the next phase of text mining applications.
Keywords: BioNLP, classification, curation, data mining, gene-drug relationships, information extraction, information retrieval, machine learning, natural language processing, NLP, pharmacogenetics, pharmacogenomics, text mining
After several decades of pharmacogenomics research, it is clear that the overall pharmacologic effects of medications are typically not monogenic traits, but are determined by the interactions among several genes encoding proteins involved in numerous pathways [1]. Polygenic determinants of drug response are often difficult to elucidate in clinical studies; however, recently functional genomics and high-throughput screening methods have been providing powerful new tools to reveal these interactions. To uncover the relationships between biological systems and drug response, pharmacogenomic researchers must assimilate knowledge from a multitude of disciplines, on levels ranging from genomic, molecular, cellular, tissue, organ and organismic.
Therefore, researchers need the ability to query the ‘bibliome’ (the collection of biomedical text) in order to answer their questions. One can imagine a network of biological entities (genes, proteins, drugs, diseases, symptoms and so on) that are connected by links indicating relationships hypothesized, established and/or discussed in the literature. Biologists need a deep view of the network around a specific drug or gene of interest, and also need to ask broad questions such as ‘what are all the genes known to metabolize drugs used to treat heart conditions?’
With the rapid growth of research and publications in all fields ranging from genomic to clinical, and the numbers of genome-wide studies and genes now characterized, it has become crucial to provide tools for scientists to organize and integrate these vast amounts of information [2]. Such tools to assist scientists must encode the known facts in a standardized structured format, to allow subsequent use, exploration, visualization and discovery. Therefore we must structure the unstructured textual information.
Structuring unstructured knowledge
There has been much effort in recent years focused on constructing databases that encapsulate findings published in the scientific literature in domain-specific knowledge bases. In the field of pharmacogenomics, the Pharmacogenomics knowledge base (PharmGKB) seeks to capture all information relating human genetic variation to drug response phenotypes [3,201]. PharmGKB is part of the NIH-sponsored Pharmacogenetics Research Network, a nationwide collaboration of hundreds of scientists in multidisciplinary research groups addressing research questions in pharmacogenomics [202]. PharmGKB has a team of curators who survey the literature regularly and annotate gene variants and gene–drug–disease relationships. They also summarize drug pathways and important pharmacogenomic genes. Their activities add structure to the pharmacogenomic literature.
Conversion of unstructured free-text information into a computable form, such as entries in a database, assists two tasks: first, providing information on specific entities of interest, such as a single protein or a single drug, and second, providing broad, systemic information such as that needed to extract evidence to support analysis of high-throughput assays, generate hypotheses or aid curators in the creation of databases. Enormous, unmanageable amounts of information in the scientific literature demand automated approaches [4]. To view publication trends of the last 50 years see Figure 1. Therefore we must integrate automatic methods, such as text mining.
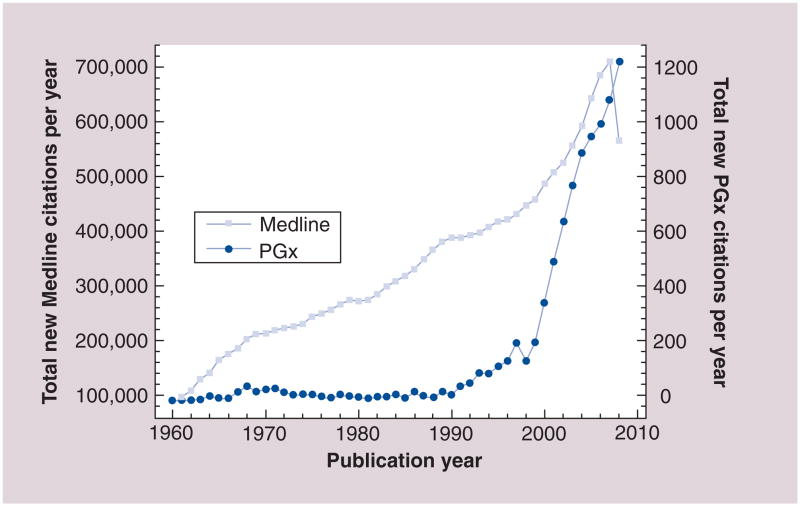
Figure 1. Annual growth of Medline and of pharmacogenomics-related publications.
The graph shows the annual growth in Medline, which is growing at a double-exponential rate [70], adding more than two citations per minute [136]. Also shown is the growth of PGx publications specifically, which has steeply risen in the last decade. PGx citations corresponds to articles returned by PubMed query ‘pharmacogenomics OR pharmacogenetics’; the two terms are considered interchangeable today.
PGx: Pharmacogenomics.
Extracting pharmacogenomic knowledge from text via text mining
Text mining is defined as deriving structured information from text, usually to fill some specific information need. Text mining allows aggregation of information culled from the entire corpus of published literature, and allows disparate information to be combined and presented to users, regardless of its original source. Some define text mining as ‘the discovery by computer of new, previously unknown information, by automatically extracting information from different written resources. A key element is the linking together of the extracted information together to form new facts or new hypotheses to be explored further by more conventional means of experimentation’ [203]. Others in the field claim that text mining is simply defined as mining, or extracting, the desired information from the text and thereby providing useful information of the sought after type directly from the text. In this article, we use the latter definition.
For example, how can text mining assist researchers to identify new gene variants associated with drug response? Using the candidate gene approach, text mining can assist by generating new biological hypotheses that will facilitate the choice of new candidate genes and variants to be tested. In the genome-wide screening approach, text mining can be used to comprehensively identify all of the connections and associations that we do know from decades of research. Thus, in prioritizing which predictions should be investigated as novel and potentially important ones we can distinguish known associations from novel ones. Text mining can assist scientists in answering questions like ‘what are all the known inhibitors or inducers of a gene?’ and ‘What are all the drugs tested for a certain disease?’
Recent work in biomedical text mining has drawn on techniques developed for natural language processing (NLP), and text mining can be thought of as a subset of NLP. NLP is defined as converting human language into computer-manipulatable formats [5]. This generally combines the fields of computer science with linguistics, and actually includes processing of both text and speech of human language. BioNLP or biomedical language processing is the field of research focusing on development of methods and tools to process biomedical texts. A simple example of how we might use NLP in recognizing drugs in text is by focusing on sentence fragments that are nouns or noun phrases, because drugs are never verbs. Thus, we first process sentences in the biomedical text, identify their grammatical parts of speech (such as verb, noun, adjective and preposition) automatically, and then use this to determine which words or phrases are drugs.
Text mining also uses techniques developed in the field of machine learning. A major focus of machine learning research is to automatically recognize complex patterns in large datasets. In the case of using machine learning in text mining, the ‘data’ is the text itself. An example of a complex pattern to recognize in text is a gene name; automatically detecting gene names is a task that we shall see (later) is quite nontrivial.
Much of the progress in BioNLP has been catalyzed by shared evaluation tasks between research groups, assessing their systems on the same dataset. The main such shared tasks are the Critical Assessment of Information Extraction systems in Biology (BioCreAtIvE) challenge, BioNLP ’09 Shared Task, KDD Cup challenge, and the genomics track of annual Text Retrieval Conferences (TREC), all of which have focused community efforts on timely challenges in the domain, such as gene name recognition and information retrieval methods to assist database curation efforts [6–9,204]. These evaluations provide evidence about the comparative performance of various tools. We will not draw conclusions about these comparative assessments in this article. Krallinger et al. provide an excellent online compendium of applications developed to provide access to information contained in the biomedical literature [2,205].
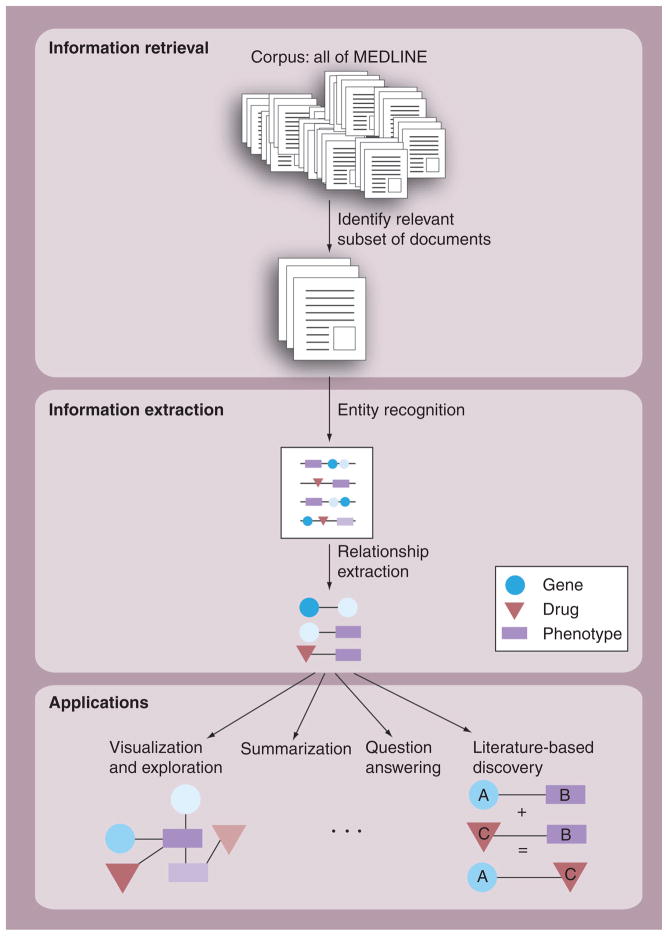
We divide text mining into two main steps: identification of documents that may contain the desired information, and then extraction of the information itself from this set of documents. Each step can subsequently be divided into several tasks. We review current methods for each, relevant to the field of pharmacogenomics. See Figure 2 for a visual overview of the main tasks of text mining.
Figure 2. Overview of text mining.
The figure shows typical flow of text mining, as outlined in this article. Information retrieval methods select a subset of relevant documents from the entire corpus. Subsequently, information extraction methods extract facts from the documents, typically by identifying entities and relationships of interest. These facts populate a structured database and can be used for a variety of applications, such as visualization and exploration, summarization, question answering systems and literature-based discovery.
Identification of relevant documents: information retrieval
Information retrieval is the process of identifying a subset of documents within a larger set that are relevant to a query of interest, such as ‘all documents discussing warfarin’. This process is often called information retrieval, document retrieval or document classification. When searching the World Wide Web, these documents are web pages and the goal is to retrieve web pages relevant to the user search. When searching the scientific literature, documents are journal publications and typically PubMed is the interface used to search the MEDLINE repository of over 19,000,000 publications. In a typical Web or PubMed search, a query may retrieve thousands of documents from the entire corpus, while only a small number of documents or ‘needles’ in this ‘haystack’ are truly relevant to the user. Information retrieval research has addressed methods to prioritize search results such that the most relevant documents are highly ranked.
Why perform information retrieval? Any user of PubMed or Google utilizes document retrieval techniques on a daily basis: when we simply query for ‘pharmacogenomics’, the search engine has already indexed the words or terms in all documents, and utilizes these indices in sophisticated ways to decide which documents to present, as it is unfeasible to read the entire corpus. In biomedical text mining, information retrieval is often performed as a step prior to information extraction, to aid in intelligently limiting the documents processed in the information extraction step to only the most relevant documents. This is done for a number of reasons: The researcher or curator is limited in time and thus in number of results they are able to read, and so we first enrich for most relevant documents to increase specificity before extracting text snippets from them that the user will have to read; the information extraction task, especially when using machine learning techniques, is computationally expensive and so it is unfeasible to process the entire corpus; visualization of a complete graph of interacting gene variants, drugs and diseases may be unfeasible if we do not first limit the ‘world’ we are looking at to a subset of entities of interest.
Typically the first step in text mining is to select the corpus of interest. To date, most pharmacogenomic information has appeared in scientific publications indexed by MEDLINE. However, other corpora (collections of documents) of interest may include patent literature, clinical patient records, US FDA-approved drug labels, drug adverse event reports in the Adverse Event Reporting System, web logs (blogs), websites or online health discussion forums. If we select MEDLINE as our corpus, we may want to limit our search to a subset of journals because MEDLINE contains 22,542 journals, many of which are not in English. For example, one might desire to limit to the English language, and to those journals relevant to pharmacogenomics. Most publications containing pharmacogenomic information are published in a set of approximately 20 key journals, as described by Lascar and Barnett [10] and from our experience at the PharmGKB [3]. However, important publications are also found in many other journals at a lower frequency, and so sophisticated methods to identify such publications automatically are critical.
Document classification methods determine whether a document has particular characteristics of interest, such as including a certain type of information or discussing a specific topic. Rather than requiring the user to specify the type of information explicitly, the user typically provides a set of documents that contain the characteristics of interest, a ‘positive training set’ and another set that does not, ‘negative training set’. These methods then automatically learn the characteristic ‘features’, to help determine positives from negatives using machine-learning techniques. Typical classification features used in the biomedical domain are terms used in abstracts and Medical Subject Headings (MeSH), which are manually assigned to publications by curators from a controlled terminology. One such classification system is the MScanner system, which uses a Naive Bayes classifier to search MEDLINE for articles most relevant to a given set of articles, by using a user-provided input set of PubMed IDs as a positive example set, indicative of the type of articles the user is searching for [11]. The authors describe the use of a corpus of pharmacogenomics-related articles curated by PharmGKB curators as input to extract other such articles to be reviewed, where the features used by the classifier were MeSH and journal of publication. Terms such as ‘Pharmacogenetics’ and ‘Cytochrome P-450 CYP2D6’ were found to be features that allowed for distinguishing papers on pharmacogenomics, from all other publications. Rubin et al. developed a similar system fine-tuned to pharmacogenomic literature, which experimented with a number of classifiers and used words in abstracts and MeSH as features [12]. Cohen et al. developed a voting perceptron-based citation classification system to assist production of systematic drug class evidence reviews by selecting the papers with the highest likelihood of containing high-quality evidence [13]. The authors used words from the title and abstract, MeSH, and MEDLINE publication types as classification features, and demonstrated the utility of the classifier in reduction of reviewer effort (as a function of number of articles that must be read), with examples of reduction as high as 50%.
A number of other algorithms have been developed for finding relevant literature. These have been developed as general-purpose tools for any biomedical domain, but can be applied to pharmacogenomics. GoPubMed performs a keyword-based search but then classifies the returned abstracts using Gene Ontology terms [14,206]. PubFocus prioritizes citations based on journal impact factor and number of times an article is cited [15]. The ReleMed system requires multiple words of a query to appear in proximity and uses sentence-level co-occurrence as a statistical surrogate for the existence of a relationship between the words of a query [16]. The system also calculates a relevance score for articles, which incorporates the proximity of search terms in the article. XPlorMed maps PubMed results to the eight main MeSH categories and extracts topic keywords and their co-occurrences to provide the user with an overview of the biomedical literature relevant to his query [17]. iHOP structures and links the biomedical literature based on genes and proteins; it maps a given gene or protein query name to its corresponding database identifier and retrieves a collection of sentences and allows interactive literature exploration through a network interface where these sentences and their corresponding publications are associated with edges in the network [18]. Pharmspresso, based on the Textpresso system, identifies articles that contain query keywords or categories (such as a drug category or polymorphism category) co-occurring within a sentence, from a corpus of full text pharmacogenomic articles [19,20]. See Winnenburg et al. for a thorough comparison of the features of many of these systems [21]. These document classification methods can be used to provide search results to a biomedical researcher, or as a filtering technology on an input flow of documents identified for database curation.
Once documents containing relevant information have been identified, the task remains to extract the information of interest from the text. This task is generally called information extraction.
Identification of information within the documents: information extraction
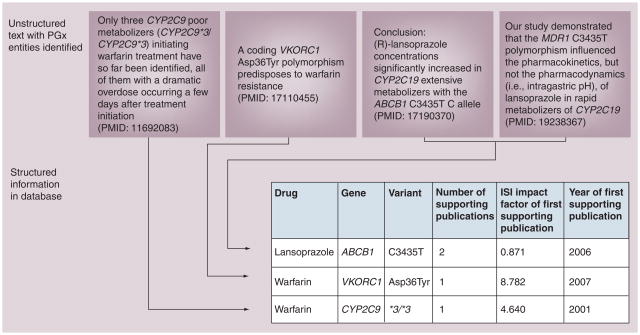
Information extraction systems all share the goal of extracting explicitly stated facts from unstructured text, often targeting a restricted set of assertion types such as ‘drug–gene–variant relationships’ or ‘metabolizing enzymes’. To illustrate information extraction targeting these two types of assertions, see Figures 3 & 4, respectively. Figure 3 also demonstrates the power of converting free text to computable knowledge: it allows us to deduce information such as ‘most well-established knowledge’ in pharmacogenomics (based on number of supporting publications), to highlight novel relationships recently discovered, and high-impact discoveries, among many other application examples.
Figure 3. Information extraction: structuring unstructured data using text mining to create a database of facts.
Sentences appearing in publications are processed, key pharmacogenomics entities are identified and facts are used to populate a structured database. Note that MDR1 is a synonym for gene name ABCB1, a gene normalization system identifies this and resolves the issue. Multiple publications can support the same fact (e.g., lansoprazole–ABCB1–C3435T relationship). The database shown on the bottom contains ‘computable’ information: data structured in a table can easily be used and analyzed using software. Examples of tasks enabled by the information extraction include: identification of high-confidence relationships by mandating a minimum number of supporting articles per fact; identification of high-impact discoveries by mandating high-impact factor of the journal that was the first to publish the finding; identification of novel relationships by restricting ‘year of first supporting publication’ to the present year.
PGx: Pharmacogenomics.
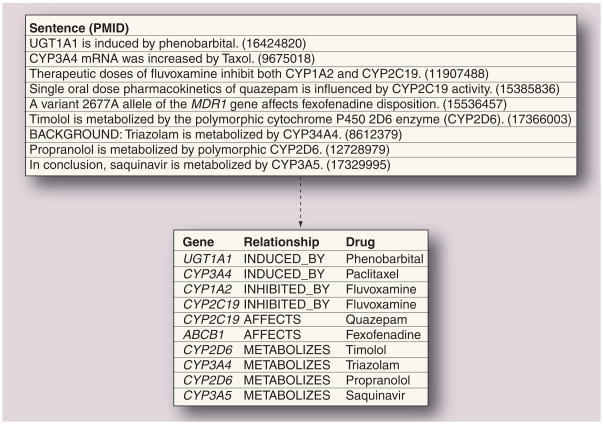
Figure 4. Example of relation extraction.
Relationships between genes and drugs are extracted from sentences and normalized using background knowledge such as lexicons and ontologies. Normalization mappings used: increased → induced, Taxol → paclitaxel, influence → affect, MDR1 → ABCB1. Relation extraction systems can be developed to extract a range of relationships, such as induction, inhibition, general affect and metabolism, or may be fine-tuned to specific relationships (such as to extract metabolizing enzymes and their metabolites). Background knowledge is also used to reconcile inverse relationships such as inhibits → inhibited by.
The broad goal of information extraction can be further subdivided into a number of subtasks, which we review.
Information extraction: identification of key entities using named entity recognition
In the context of pharmacogenomics, the key entities of interest are genes and gene variants, drugs and phenotypes. Specific entities, such as BRCA1 or codeine, can be referred to as ‘named entities’ (as opposed to unnamed entities such as gene or drug, and thus the task of identifying them in text has been called Named Entity Recognition (NER). Although entity recognition may seem trivial at first, it is possibly the most difficult task in biomedical text mining and is a prerequisite for almost all subsequent tasks and goals [22]. There has been significant progress in recent years in recognizing biomedical entities in text. We review the main approaches for NER, provide examples of difficulties in identifying genes, gene variants, drugs and phenotypes, and present examples of work that address these challenges.
Main approaches for biomedical NER
The approaches for biomedical NER fall into three main categories: lexicon-based, rule-based, and statistical or machine-learning based. Recent work has shown that combinations of these approaches often obtain the best performance.
Lexicon-based approaches
These utilize a lexicon, or dictionary of terms, to identify specific terms in the text. These lexica include the well-known nomenclatures of the entity type. For example, one might use all names, aliases and symbols of the HUGO gene nomenclature committee (HGNC) to identify gene names in the text [207]. Example systems that have applied the lexicon-based approach include those of Krauthammer et al., Hansich et al., and Tsuruoka and Tsujii, who developed systems for gene and protein recognition [23–25]. Dictionary-based approaches cannot identify new names added to the biomedical literature such as novel genes or drugs, or entries with word-order variations (integrin α 4 vs α 4 integrin). This has led researchers to consider rule-based systems, which can deal with a broader range of variations.
Rule-based NER approaches
These approaches manually or automatically construct rules or patterns to match them to the literature to identify entities of interest. An early method recognized protein names by beginning with identification of core terms (words with special characteristics such as capitalization, hyphens, brackets) and feature terms (like receptor and protein), and then concatenating the terms using handcrafted patterns, to extend the boundaries to adjacent nouns and adjectives [26]. For example, in order to extract the term ‘Ras guanine nucleotide exchange factor Sos’, they first identify the words Ras, factor and Sos as core and feature terms and then construct the entire phrase by application of the rule ‘Connect nonadjacent annotations if every word between them are either noun, adjective, or a numeral’. Tanabe and Wilbur [27] (AbGene) created a successful early rule-based system that was trained on 7000 hand-tagged example sentences from biomedical text. The system uses parts of speech and postprocessing rules to identify the context in which gene names are used, which greatly reduces false positives when compared with simple lexical matching used in lexicon-based methods. Rule-based approaches utilize manually created rules and patterns, which makes it difficult to apply them to new domains. Among the various disciplines in biomedicine, naming conventions vary significantly. This has led to the rising popularity of machine-learning based methods.
Machine-learning based approaches
Machine-learning based approaches automatically learn to recognize entities by using characteristics ‘features’ that distinguish between a set of examples ‘positive set’ and a set of counterexamples ‘negative set’. The developer of the system specifies which features to use; these features are the heart of the construction of a successful machine-learning application. Careful crafting of features enables the distinction between positive and negative examples. For example, one feature may be the presence of special characters (such as hyphens, digits and brackets) to distinguish gene names (COX-2, BRCA1) from nongene terms. Another simple feature may be adjacent words (e.g., the word ‘gene’ may often appear adjacent to gene names, such as in ‘the BRCA1 gene’, and this can be utilized in recognizing gene names). These machine-learning based approaches require annotated training sets or corpora as input, such as documents in which gene, drug or disease mentions in the text were manually annotated as such. These corpora are used as ‘gold-standards’ to evaluate results of the methods and are also used to compare performance between different methods. With the increased availability of such annotated sets, machine-learning approaches to NER have become mainstream research [6,28–31,208]. These approaches develop a statistical model for entity recognition. Three commonly used types of machine learning methods are Hidden Markov Models, Support Vector Machines (SVM), and Conditional Random Fields (CRFs). Markov models have used language characteristics (features) such as morphological patterns (such as prefix and suffix), bigrams (pairs of words that occur in tandem such as ‘BRCA1 gene’ or ‘warfarin dosage’), parts-of-speech (such as noun or adjective), presence of special characters (such as hyphens, digits, brackets), and intradocument name alias features (to determine whether one noun is the same entity as another noun) [30,32–34]. SVM methods often use information from surrounding words to provide input on the word of interest being classified [35]. CRFs have recently been shown to be successful at NER [36,37]. Interestingly, several methods have been developed that use a combination of models to achieve better performance; Huang et al. used a model that combines two SVM models and one CRF model such that the ‘vote’ of all three in recognizing an entity is taken into account when scoring predicted entities; this method achieved one of the best scores (ranked 3rd) in the BioCreAtIvE II gene mention recognition task [38]. All in all, it appears that the choice of features is at least as important as the choice of algorithm [31,39].
NER of pharmacogenomic entities
Identifying genes
In this genomics era, much work in biomedical NER has focused on identifying gene and protein names in the text. (Often NER applications do not distinguish between genes and proteins and will use the term genes to mean both genes and gene products, despite clear biological differences between the two). Genes are particularly difficult to recognize automatically owing to the frequent use of common English words as gene names, nonstandard nomenclature, varying use of hyphens and other separators, multiple nonunique synonyms per gene, and other such issues [40]. The BioCreAtIvE challenge included a community-wide effort to compare systems identifying genes in the text, and catalyzed good progress in this task [6,29–31]. Several research groups have developed freely available tools to tag genes found in text, such as BANNER, ABNER, and LingPipe [41,42,209]. Chang et al. did early work on extracting genes mentioned in the pharmacogenomic literature using machine-learning techniques [40]. Their method, called GAPSCORE, identifies gene names in text, scores words using a statistical model of gene names based on their context, morphology, and appearance, and then gives a score to the identified gene name signifying its probability of actually representing a gene.
Why are genes so difficult to identify? Genes often have a multitude of names; human genes have on average 5.5 different names [29], and they can appear in free text in multiple variations. For example, Cohen et al. note the example of BRCA1, which could be referred to with the spelling variants BRCA-1 or BRCA 1, or by any of its alternate symbols such as BRCC1 or RNF53, or their spelling variants, or by the full name ‘breast cancer susceptibility gene-1’, or by its official HGNC name ‘breast cancer 1, early onset’ [43]. Another problem stems from the fact that genes have frequently been given names, aliases, or symbols that are commonly used words in English, such as a, to, and, large, mice, mass, impact, minor, cord, task, wave, and aim. This makes the task of disambiguating whether the gene or the common word is being referred to quite difficult. In some cases, requiring these aliases to appear in capital letters reduces many false identifications of genes, however consider the sentence ‘AIM: To study prevalence of angiotensin-converting enzyme gene polymorphism and its correlation with angiotensin-converting enzyme level in Kyrgyz population suffering from chronic glomerulonephritis’. (PMID: 16078593). In this example, AIM (the symbol for three different genes: DNMT1, CD5L and CD69) is used as a common English word, chronic glomerulonephritis (the symbol for the cingulin gene) is an abbreviation for a disease, and only angiotensin-converting enzyme is actually a gene (angiotensin I converting enzyme 1) in this context. In addition, gene names may have character level variations (VKORC1 and VKORC-1), word level variations (SIN3 homolog B transcription regulator, SIN3 homolog B transcriptional regulator), and word-order variations (3-α hydroxysteroid dehydrogenase type IIb, type IIb 3-α hydroxysteroid dehydrogenase). Tuason et al. reported that up to 79% of failures in gene name recognition could be caused by character-level and word-level variations [44].
Identifying gene variants
Several applications specialize in identification of genetic variations (e.g., C[3435]T, 80G>A, rs9923231, His452Tyr or L-23Q). Examples include MuteXT, MarkerInfoFinder, MutationFinder and Pharmspresso [45–47,19]. These systems all identify mutations using manually created patterns such as X[1–9][0–9]*Y. In this pattern, X and Y belong to a set of 20 amino acids or four nucleotides, [1–9] signifies one number ranging from one through to nine inclusive, * signifies zero or more occurrences of the preceding bracketed entry [0–9]; the pattern would identify entities such as L23Q or P207L. Other patterns recognize variants such as 80G>A or rs28942082.
However, even this seemingly obvious pattern, or rule, is not fully accurate: It correctly identifies the variant E23K in the sentence ‘The objective of the study was to investigate whether diabetic patients carrying the E23K variant in KCNJ11 are at increased risk for secondary sulfonylurea failure.’ (PMID: 16595597). However, in another sentence it mistakenly identifies the gene E2F as a polymorphism: ‘We now show that the RB protein is found in a complex with the E2F transcription factor and that only the under phosphorylated form of RB is in the E2F complex’ (PMID: 1828392). It also mistakenly identifies the cell cycle checkpoint G2M as a gene in yet another sentence: ‘Several important functions of BRCA1 and BRCA2 have been disclosed, including regulation of the G2M checkpoint.’ (PMID: 11400119). One might require two digits to appear between the amino acids, however this rule would still incorrectly identify the cell line T98G and the gene name L23A, as noted by Caporaso et al. [47]. Despite some of these challenges and the existence of exceptions to the rules, the MutationFinder system achieves impressive near-perfect precision and high recall by using a combined set of rules, and is available for free download and use in a number of programming languages [47].
Identifying drugs
In comparison to gene names, drugs are easier to identify using standard dictionaries of drugs and their synonyms that include both generic and trade names. However names such as ‘Duration’, which is a brand name for phenylephrine, and the addition of new drugs and drug names make it difficult to maintain perfect accuracy. Identification of drugs in text is an area that has been far less addressed by the community than gene recognition, and one that is clearly necessary for text mining of the pharmacogenomics literature. For a recent review of drug name recognition and classification in biomedical texts, and for a review of literature mining in support of drug discovery see [48,49]. The early EDGAR system recognized drugs using the Unified Medical Language System (UMLS) metathesaurus, in a lexicon-based approach [50,51]. Kolárik et al. took a rule-based approach to extend drug identification to include drug effect terms, thereby identifying drugs more expressively described in the text than those names included in existing drug dictionaries [52]. For example, they identify drugs such as ‘cytochrome P-450 monooxygenase inhibitor’ and ‘aspirin like anti inflammatory drug’ and ‘selective high-affinity antagonist of human substance P/neurokinin 1 (NK1) receptor’. The authors first identify drugs with the help of lexicon-based NER using the DrugBank drug dictionary [53]. They then identify patterns used in conjunction with these drug mentions, to collect additional terms that are used to describe those same drugs. As a simple example, the phrase ‘Adinazolam is a benzodiazepine derivative’ matches the pattern [NP is a NP] where NP is a noun phrase, and would result in the identification of the drug ‘Adinazolam’ and the drug property term ‘benzodiazepine derivative’. The latter can be used to extend the drug dictionary. The authors compared drug identification using DrugBank’s drug lexicon [53] to that identified by their syntactic and semantic pattern-based sentence processing of MEDLINE. They showed that the latter approach captured a significant amount (29–53%) of valid new drug annotation terms not yet applied to drugs in DrugBank, thus enabling extension and update of such resources with novel descriptions of drugs. Segura-Bedmar et al. also developed a drug recognition system that combines lexicons and rules to detect possible candidates for drug names not detected by other systems [48]. Recently, Hettne et al. developed a rule-based method to identify drugs and small molecules, thus broadening the level of chemical identification to include metabolites and endogenous molecules [54].
Identifying phenotypes
In theory, phenotype identification is not harder than gene or drug identification. In practice, it is made harder by the lack of a convenient terminology or ontology. This lack is more obvious when one is interested in identifying not only diseases (which are pathologic phenotypes) but rather all phenotypes (both pathologic and physiological). Among the different existing vocabularies, MeSH, SNOMED-CT and ICD-9, which are part of the UMLS [210], contain rich sets of disease names and synonyms. MetaMap is a NER tool designed to recognize entities of these UMLS vocabularies and has been used to identify diseases [55]. To avoid the limitations of using a set vocabulary, Xu et al. take an approach similar to that used by Kolarik et al. in the context of drug recognition. Their method learns a disease vocabulary from the corpus text, using the context of known disease mentions as a reference to find new disease mentions [56]. The resulting vocabulary can subsequently be used for disease identification. Recently work has been carried out to extract side effects from the literature as well as from labels of FDA-approved drugs. Kuhn et al. developed the SIDER resource to connect drugs to their phenotypic effect by extracting side effects from drug labels, and were able to use the extracted information to estimate side effect frequency for over half of the drugs [57].
Several initiatives focus on the standardization and unified information management of phenotype information. Two such projects are the European project Genotype to Phenotype Databases: a Holistic Approach (GEN2PHEN), which aims to develop a knowledge web portal integrating information from the genotype to the phenotype, and PhenX.org [211] which is a web-based catalog of high priority measures for consideration and inclusion in genome-wide association studies.
Some initiatives for the normalization of phenotype descriptions are focusing on the process by which information is collected. For example, the project Data Schema and Harmonization Platform for Epidemiological Research (DataSHaPER) is a joint initiative that is constructing a suite of harmonization schemas for biobanks and major epidemiological studies [212]. In a similar way, the Experimental Factor Ontology (EFO) proposes a shared schema to harmonize annotations of gene expression experimentation (Malone et al. [58]). Others ontologies and controlled vocabularies are publicly shared on portals like the BioPortal [59] or the OBO Foundry [60] and used to harmonize phenotype descriptions over various biological databases.
A recent analysis of terms used in MEDLINE show that only 13% of the terms in UMLS (518,835 out of over 5.3 million) have ever appeared in MEDLINE [61]. This study shows that text can be used to uncover which phenotypes are actually discussed in the literature quantitatively, of the terms defined in a ‘top down’ approach when constructing UMLS using clinicians’ description of phenotypes. Text analysis also can point to new terms that should be included in UMLS owing to frequent usage in MEDLINE. Such work allows the emergence of resources for ‘phenotypic variables of interest’ driven by the research community, statisticians for example, rather than purely by clinicians and physicians (e.g., UMLS). It is important to note that statistical analysis of MEDLINE only captures language used in the scientific discourse, and may miss terminology used outside of this corpus, such as a new disease or drug name, or informal names that are not used in the corpus (scientific literature) but are common elsewhere (e.g., patient files). Future work will address these challenges, to bridge between scientific publications and clinical records.
Common challenges of NER
As shown previously, several types of problems confound the identification of these key entities. Ambiguous synonyms and abbreviations are two main issues. Hettne et al. note the example of ‘BAP’, which is a shared synonym between two chemicals ‘Benzo(a)pyrene’ and ‘Benzyladenine’ and also has 44 additional meanings as abbreviations (such as ‘Blood Agar Plate’, ‘British Association of Psychotherapists’) [54].
Identifying abbreviations
Abbreviations are very common in biomedical literature. For example, in the sentence ‘There was no statistical difference in DBP reduction or therapeutic response rate between telmisartan and lisinopril.’ (PMID: 18987649), DBP is the official HGNC gene name for ‘D site of albumin promoter binding protein’ but in this context it is the abbreviation of ‘diastolic blood pressure’. Acronym and abbreviation definitions are ambiguous, in fact almost 22% of abbreviations in one sample of biomedical text have several possible expansions, and there are an average of 4.61 possible definitions for abbreviations six or fewer characters long [62]. A number of systems devoted to resolving abbreviations in the text have been developed. Several methods rely on the proximity of full forms and their abbreviations, and use features such as the full form or abbreviation appearing within parenthesis. For example, ‘the vitamin D receptor (VDR) gene’ would be identified by pattern <full form> (<abbreviation>) and the ‘VDR (vitamin D receptor) gene’ would match the pattern <abbreviation> (<full form>). The system by Schwartz and Hearst is based on alignment of the characters of the abbreviation to the full form and subsequent scoring, and obtains an impressive precision of 96% with 82% recall for a set of 1000 abstracts [63]. Yu et al. also used pattern matching to discover the full forms of abbreviations and received similar precision [64]. Chang et al. used machine learning techniques, training a logistic regression classifier to score candidate full forms [62]. See Torii et al. [65] for a comparison study of the three publically available detection systems including Chang’s, Schwartz and Hearst’s and ALICE [62,63,66]. Also see Wren et al. for a review of four methods [67]: ARGH, the Stanford Biomedical Abbreviation Server, AcroMed and SaRAD [67,68,213–216]. A recent paper by Xu et al. describes MBA, a system that achieves similarly high performance [69]. It specializes in identifying nonacronym abbreviations such as ‘Fas’ used as an abbreviation for the gene ‘CD95’, using a statistical method in which they count the number of articles that contain both the candidate definition and the abbreviation and then use this in scoring each candidate definition/abbreviation pair.
Identifying unique identifiers: named entity normalization & disambiguation
Normalization, the process of mapping an entity mention in text to a unique identifier, is also a research area that has received significant focus, mostly related to gene normalization, owing to the high number of gene names or symbols that can refer to more than one gene. Morgan et al. report that on average each human synonym maps to more than one human gene identifier, a large source of ambiguity [29]. It has also been noted that if the organism is uncertain for a gene reference to the gene ‘p60’ in the text, the correct unique identifier can be one of over 800 distinct gene identifiers in the Entrez Gene database [70]. The BioCreAtIvE II challenge had an entire task devoted to gene normalization; (for a review see [29]), as did the BioNLP ’09 Shared Task [7]. GeNo is one recent high-performing state-of-the-art system for gene normalization, fully available to the public, which addresses the complete set of tasks necessary to perform gene mapping [71]. In reviewing the gene name disambiguation task, it is important to mention the efforts of the HGNC that aim at defining unambiguous and reference gene symbols that stand for a set of synonymous gene names [72]. Gene variant normalization can consist of mapping gene variant mentions to a unique identifier [73], which may be, for example, a dbSNP identifier [217] such as rs28942082, or a description that follows the Human Genome Variation Society (HGVS) nomenclature such as NT_011295.9:g.2489679G>T [74,218].
To conclude this section, NER can actually be thought of as a two-step process: recognizing the words that comprise the entity name in the text (entity recognition), and subsequently unique identification of the entity that those words refer to (entity normalization).
Having identified the entities of interest in the article, abstract, or sentence, in a consistent and normalized form, the next task is often to identify relationships between entities, which is an active field of research known as relationship extraction (RE).
Information extraction: identification of relationships
The aim of RE is to identify relationships between previously recognized entities. Related entities can be of similar kind such as two proteins, in the case of protein–protein interactions, or of distinct kind such as a drug and a gene or a gene and a disease, in the case of pharmacogenomic relationships. Most research on RE has focused on extracting binary relationships from the biomedical literature and has named extracted pieces of information as either relationships, facts or events. Extracted relationships can be very general, such as a nontyped relationship between two atomic entities (e.g., CYP2C9–codeine) or a more specific one such as a typed relationship between two composite entities (e.g., hypothetically affect CYP2C9 variant and codeine response. One challenge of RE is to structure and normalize relationships to enable their integrative use. Such normalization enables reconciliation of differences in natural language structure and integration of facts across boundaries of scientific disciplines. This in turn enables us to establish intricate pathways and networks that are often split across hundred of publications and too complex to commit to memory. Thus, the resulting relationships can be visualized in the form of a connected map of entities, providing the opportunity to assess our understanding of the complex domain and providing a computational resource valuable for knowledge discovery. Such maps have been used to represent protein–protein interaction, gene regulatory and gene–disease networks [75–77]. The benefit is clear: an accurate and comprehensive knowledge map would assist researchers, save their time, allow them to summarize literature content, and generate hypotheses to test in the laboratory. In pharmacogenomics, such a map would allow us to provide answers to questions such as ‘Which gene products metabolize drug X?’, ‘Which gene variant increases response to drug Y?’ or ‘What drug response phenotypes are affected by gene Z?’ We now review work that has been done in relationship extraction.
Main approaches for relationship extraction
As with the three main categories of biomedical NER, we distinguish here three main methods for extracting relationships between entities: co-occurrence based, rule based (or knowledge based), and machine learning based. It is important to note that, in practice, current work often combines these methods to obtain better results.
Co-occurrence-based methods
Co-occurence-based methods extract hypothetical relationships by analyzing the frequency of co-occurrence of two entities in a given corpus. The hypothesis behind this approach is that entities appearing frequently together in pieces of text, such as in the scope of a single sentence or abstract, are likely to be related. Chang et al., XPlorMed, FACTA and CoPub Mapper used abstract level co-occurrence, while methods such as AliBaba, EBIMed, iHOP, and Pharmspresso identify co-occurrence of drugs, genes and diseases at the sentence level [17–19,78–82]. Pharmspresso, based on the Textpresso engine, actually detects co-occurrence in the full text of pharmacogenomic-related articles, rather than the abstract only [19,20]. The ‘search tool for interactions of chemicals’ (STITCH) tool, connects chemicals by integrating information for over 68,000 chemicals, including 2200 drugs and 1.5 million genes [83,219]. The authors mine both MEDLINE and OMIM [84] for co-occurrence and use these as evidence (as well as other metrics such as chemical structure similarity) to predict relationships between chemicals. Many of the applications listed here provide user interfaces to visualize the extracted information. Traditional co-occurrence methods find relationships between pairs of entities, but do not describe their type, which can range from vague associations to very specific interaction such as one between a ligand and a target. Therefore co-occurrence has been expanded to extract typed relationships by searching for ‘tri-co-occurence’ [85]. Tri-co-occurrence refers to the co-occurrence of two named entities and one type of relationship in a single piece of text. The type of the relationship is constituted of one (or several) word(s)that describe(s) the quality of the relationship. For instance this word can clarify that the relationship is about transport or inhibition or localization. Empirical approaches based on the observation of multiple co-occurrences in large corpora are also used to identify sets of entities and of relationship types that are mentioned in the same context [86]. This can be used to derive similarity measure between entities and then compare different relationship types. Ultimately, the appearance of two (or three) entities in several abstracts or sentences does not guarantee the existence of a relationship between them, consequently co-occurrence based methods that do not employ additional filtering are prone to false positives.
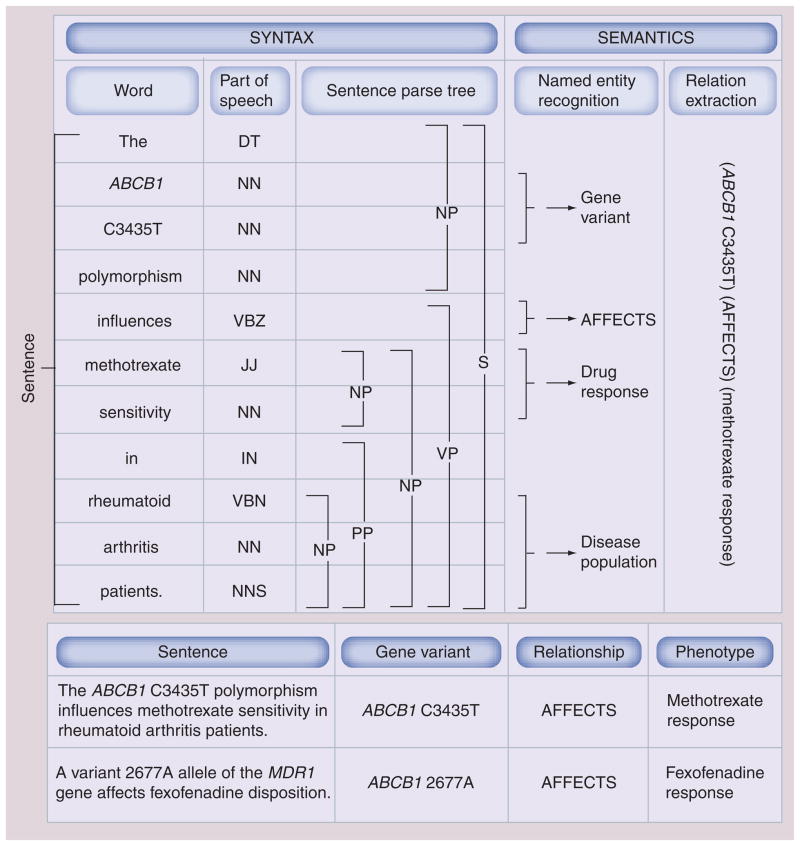
A number of methods have been developed to reduce the number of false positives that characterize co-occurrence based methods, and to identify the type of relationship between the entities. These methods take particularly into account the sentence structure, its ‘syntax’, and the sentence meaning, its ‘semantics’. Figure 5 shows the main levels of analysis used to ultimately extract a relationship from a sentence, including both syntactic and semantic analysis.
Figure 5. Levels of natural language processing analysis for a sentence.
The main levels of analysis are shown for the sentence ‘The ABCB1 C3435T polymorphism influences methotrexate sensitivity in rheumatoid arthritis patients.’ (PMID: 17181924) [137]. Processing uses syntax (sentence structure) and semantics (sentence meaning) to extract the relationship between gene variant and drug response. Sentence is tokenized into words, which are tagged with part of speech tags: DT, NNP, NN, VBZ, JJ, IN, NNS. Based on this sequence, parse tree is subsequently created for the sentence, which determines dependencies between words and groups words into phrases. Sentence parse tags: NP, PP, VP, S. Entities are recognized by combining the output of the syntactic analysis with external knowledge such as dictionaries of gene, drug and disease names in addition to categorization of relationship terms into classes (e.g., the class ‘AFFECTS’ would include the terms ‘affects’, ‘influences’, ‘has an effect on’; capitalization is used to indicate that this is a class name, not the textual term). Finally, relationship is extracted. Relationship term ‘influences’ found in raw text is normalized to <affects> class. Two rules are used: ‘Y sensitivity’ where Y is a drug, maps to Y <response>. Subsequently, the rule X <affects> Y <response> where X is a gene or protein or gene variant and Y is a drug is utilized. Using a similar process, the sentence ‘A variant 2677A allele of the MDR1 gene affects fexofenadine disposition.’ (PMID: 15536457) [138] can be processed to extract relationship between gene variant and drug response (‘MDR1’ maps to its synonym ABCB1, ‘disposition’ maps to <response>). Syntactic tagging is based on the output provided by the Stanford parser.
DT: Singular determiner; IN: Preposition; JJ: Adjective; NN: Singular or mass noun; NNP: Noun, singular proper; NNS: Plural noun; NP: Noun phrase; PP: Prepositional phrase; S: Sentence; VBZ: Verb, third person singular present; VP: Verb phrase.
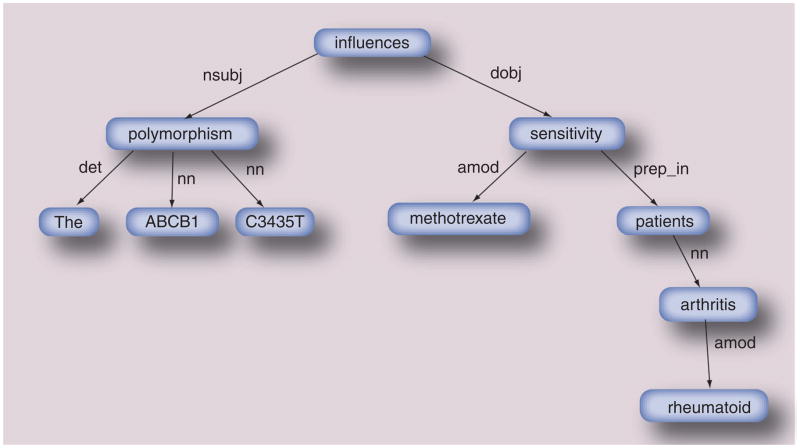
The syntax includes properties such as parts of speech and functional dependencies between constructs of words in a sentence. The syntax of a sentence can be represented in various computable formats including parse trees and dependency graphs. These are produced by NLP tools such as Gate or the Stanford Parser [87,220]. The latter is a statistical natural language parser. It uses a set of training sentences in which the grammatical functions of words were manually annotated by experts to record the most likely syntactical structure of a sentence. Figure 6 shows the dependency graph created for the sentence analyzed in Figure 5; the reader can compare the parse tree of Figure 5 to the dependency graph of Figure 6; both are created using the Stanford Parser.
Figure 6. Dependency graph.
Dependency graph is shown for the sentence analyzed in Figure 5. Figure 5 shows the parse tree for this sentence in a vertical view. The sentence: ‘The ABCB1 C3435T polymorphism influences methotrexate sensitivity in rheumatoid arthritis patients’. This representation enables the identification of a type of relationship (qualified by the verb ‘influences’) between a subject and an object.
amod: Adjective modifier; det: Determinant; dobj: Direct object; nn: Noun modifier; nsubj: Nominal subject; prep_in: Preposition in.
Data taken from [134].
Semantics try to capture the meaning of the text. Methods based on semantics can assist in entity recognition and relationship extraction using background knowledge that is frequently encoded in ontologies. In this setting, ontologies represent a shared interpretation of entities and their relations in a formal way that is represented within the computer. The major utility of ontologies is to provide support for synonym definition for normalization of named entities, and term hierarchies for generalization/specialization of named entities. The hierarchy of named entities can include objects (e.g., genes, drugs and diseases) as well as relationships between the objects (e.g., metabolizes, inhibits and induces). Ontologies are typically built for a focused area, such as pharmacogenomics, where certain specialized semantics may apply (e.g., ‘metabolism’ has a specific technical meaning in pharmacokinetics).
Uses of syntax and semantics are illustrated in the next sections on rule-based and machine-learning-based relationship extraction.
Rule-based methods
Rule-based methods use manually or automatically defined patterns to extract relationships of interest. For example, one might define the pattern <drug> <action> <gene> where <drug> and <gene> are recognized drug and gene, and <action> can be any of a list of verbs such as ‘inhibits’, ‘induces’, ‘is metabolized by’. Patterns can be applied either strictly or loosely. A strict application of patterns will not allow words between its elements, while a loose application requires the right succession of elements but allows the interposition of words between them. For example, using the previously mentioned pattern, a relationship between codeine and CYP2D6 can be extracted from the sentence ‘codeine is metabolized by CYP2D6’, and from ‘the codeine molecule is metabolized in the liver by the protein CYP2D6’ in the case of a loosely applied pattern. Tari et al. use a wildcard character (‘_’) to define loose patterns such as <drug> _<action>_ <gene> [88]. Loose patterns are prone to false positives since they allow the extraction of relationships not actually mentioned, such as (codeine, CYP2C9) from the sentence ‘codeine is metabolized by the liver where CYP2C9 is synthesized’.
Recent rule-based approaches use syntactic constraints to extract relationships, such as forcing entities to be the subject and object of a single verb. This approach has been successfully used to identify protein–protein interactions and protein transport and localization [75,89].
Ahlers et al. used vocabularies and semantic types defined in the UMLS associated with syntactic constraints to extract gene–disease and drug–disease relationships [90]. The EDGAR system extracts drugs, genes, and cell types and their relationships from sentences with syntactically complex structures, using a combination of syntactic and semantic processing techniques [50]. The EDGAR system uses a background knowledge representation that describes gene–drug–cell relationships. This representation is used both to constrain relationships extracted by the system (e.g., <drug>suppress<gene expression>) and to infer new relationships. Inferences are made on the basis of the EDGAR data model that states that information about a drug, gene or cell can be inferred from relationship extracted about other drugs, genes or cell lines. Importantly, the EDGAR system extracts relationships between three entities, while most systems to date have focused only on pairs.
Hakenberg et al. developed SNPshot, which contains information on phenotypic effects of genetic variants, focusing on effects on drug response [221]. They make available summarized information linking genes, variants, diseases, drug efficacy, adverse drug reactions, populations and allele frequencies, with cross-references to the literature, EntrezGene, PharmGKB, DrugBank and dbSNP [91]. SNPshot manages the impressive performance of 90–92% precision for recognition of the main entity types (gene, drug, diseases) and 76–84% for relationships involving these types.
We have recently published a method to extract normalized relationships of interest to the domain of pharmacogenomics [92]. Our manually created grammatical patterns and pharmacogenomic relationship ontology enable the use of both the syntax and the semantics of text to extract precise (70–87.7% precision) and unambiguous pharmacogenomic statements.
Machine-learning methods
Machine-learning methods are also used to address RE. These methods use a training set of text for which experts have manually annotated relationships; typically the reference set includes a set of valid relationships and invalid relationships that appear in the corpus text. When new text is submitted to the system, it uses the training set as a basis to identify new valid relationships. The constitution of such training set requires a large degree of human involvement and is consequently a bottleneck for machine learning approaches. Several important efforts have focused on constituting and sharing such corpora [28,93,94]. Craven et al. proposed various machine learning algorithms (statistical text classification and relationships learning) to extract relationships from text. One originality of their approach is to face the annotation bottleneck by using the content of existing databases to annotate automatically (and thus ‘weakly’ according to the authors) a training set [95]. Syntax has been used in machine-learning methods by adapting dependency graphs (Figure 6) to a format handled by graph kernel algorithms. This approach has been successfully applied to the extraction of protein–protein interactions and to relationships related to protein localization, binding and regulation [96,97].
Combinations of methods
Combinations of these three methods have been implemented for RE in pharmacogenomics. For example, Chang et al. identified drug–gene pairs using co-occurrence, but then classified the relationships into five different classes using machine learning classification methods [78]. Chilibot identifies co-occurrence of entities and syntactic structure of sentences [98]. Rules then classify the sentences into classes (e.g., stimulatory or inhibitory) based on the presence or absence of words describing relationships (such as ‘activate’, ‘induce’, ‘stimulate’, ‘suppress’ and ‘block’). The resulting network of relationships is then provided in a viewing tool, which displays the supporting evidence for each relationship. AliBaba uses two techniques in parallel: pattern matching (i.e., rule based) and co-occurrence filtering [81]. The patterns take into account syntax and entity classes, and the method assigns confidence scores for extracted relationships based on the quality of the match between the sentence and a pattern.
In some cases, relationship extraction methods will extract a relationship between entities X and Y that is not stated as a research conclusion, but rather as a hypothesis (such as in the sentence ‘We hypothesize that X metabolizes Y’). The level of certainty associated with the assertion can often be gleaned from the text, and a future challenge includes distinguishing hypothetical relationships from demonstrated ones. The location in the abstract or document (such as in introduction versus conclusion section) can be used as a clue. Positive results are most frequently published, and so we postulate that the majority of hypothetical relationships stated in abstracts are in fact proven by the publication and are subsequently demonstrated in the results section. However, this remains to be tested.
Challenges in relationship extraction
Negation
It is of importance when extracting a relationship from text to know if this relationship is actually affirmed or negated. Otherwise, a relationship of the form involved in (CYP2C9 polymorphism, pitavastatin metabolism) can be extracted from the sentence ‘The CYP2C9 polymorphism was not involved in the pitavastatin metabolism.’ (PMID: 12442637). The simplest methods consider the presence of words like no, not and so on. in the vicinity of recognized entities. Others go beyond syntactic analysis and use rules or machine learning to detect negation [99].
Contradiction
The detection of contradictory statements in text is of interest to identify debated or invalidated (and potentially interesting) knowledge. Identifying negation can obviously be used to find contradiction. In a similar manner relationship types can be defined as being contradictory to detect contradictions. For example if relationship types ‘inhibits’ and ‘stimulates’ are defined as being in contradiction, the occurrence of relationships inhibits and stimulates between two same entities X and Y (inhibits [X, Y] and stimulates [X, Y]) enables the identification of contradictions [100].
Context extraction
A challenge of current methods is the difficulty in handling the context that surrounds facts. For example, we may extract that X is related to Y, but in which species, at which temperature, under which pressure, what is the substrate? Extraction of this type of information could resolve seemingly contradictory statements.
In addition, in biomedical literature, when a new finding is claimed, it is often claimed as a probable relationship, because the claim is usually based on a set of observations that are not sufficient to state that the claim is always true. For example, ‘X may be related to Y’ or ‘According to our hypothesis, X is related to Y’ are typical statements. This type of ‘probable relationship’ statement is challenging to represent computationally; fuzzy logics can help to address this, but precise values on the probability of the relation must be assigned. This is an area for future research to address. Existing computational formalisms for language representation are currently limited in their ability to represent the flexibility of natural language.
Full text, figure captions & tables
Because of the accessibility of scientific abstracts, most biomedical text mining research has used abstracts to extract information from publications. Nevertheless, other sections of articles are of equal interest [101]. Cohen et al. evaluated differences between abstracts and full text [102]. They found marked distributional differences in entity mentions, such as significantly higher frequency of mutations mentioned in the bodies of articles, which did not mention the mutations in the abstracts at all. The authors also evaluated differential performance of text mining tools, reporting for example that commonly used gene taggers perform substantially better in abstracts than in article bodies. Future work will have to focus on overcoming the technical challenges of mining full text.
Recent studies have tried to go one step further by extracting relationships from figure captions and tables, widespread in biomedical articles and previously inaccessible to automated systems [103].
Relationships described over several sentences
Instead of reporting a relationship between two entities explicitly named, many sentences report a relationship between one entity and a reference (e.g., a pronoun) to an entity mentioned elsewhere in the text. This is the case of the sentence ‘This drug reduces the expression of BRCA1.’ Sophisticated algorithms can explore the vicinity of the sentence to find the explicit name that the reference points to [104]. This process is called anaphora resolution.
Relationships involving numerical values
In an interesting work by Wang et al., the authors proposed the extraction of relationships between one entity and one numeric value to capture the value of pharmacokinetic parameters such as the clearance of a drug [105]. The extraction of this information is of particular interest to automatically inform pharmacokinetic compartmental models.
Normalization of extracted relationships
The normalization of relationships between two atomic entities such as (CYP2D6, codeine) is usually addressed using simple lexicons. Rinaldi et al. proposed an approach to normalize typed relationships mentioned in the active form (X inhibits Y), in the passive form (Y is inhibited by X) or within a nominalization (inhibition of Y by X) [106]. In more complex cases, relationships can connect between two composite entities such as in the case of: is related to (CYP2C9 expression, codeine response) [107]. Owing to natural language redundancy these relationships are highly heterogeneous and necessitate normalization using both syntax and semantics [108].
Using the extracted structured information: applications
Visualization of extracted relationships
As Cohen and Hunter point out, the problem in biomedical search ‘is not the Google-task of finding a needle in a haystack – the problem is that the whole haystack is made of needles’ [109]. A query about CYP2C9 to the PubMed search engine returns over 2000 documents, a search of MEDLINE abstracts for sentences containing both ‘CYP2C9’ and any drug results in over 5000 individual sentences. How does one navigate those documents or the facts contained within them, without having to read them all? Therefore, the challenge is to organize the retrieved documents and extracted facts, such that the user can effectively navigate them. Several systems allow visual navigation of the network of interacting factoids embedded in the scientific literature. Notable ones relevant to pharmacogenomic interactions are iHOP, AliBaba and Chilibot [18,81,98]. The Cytoscape open source software platform is extensively used by researchers to visualize networks and integrate these relationships with other data in a flexible manner [110]. These visualization systems can, for example, assist scientists in researching a gene candidate when designing a pharmacogenomic clinical trial for one drug.
Identification of novel relationships through literature-based discovery
An exciting use of the information extraction techniques described previously is in linking the disjunct sets of literature to uncover ‘hidden’ links between biological entities. Literature-based discovery (LBD) systems automatically induce novel promising hypotheses by processing existing publications, and extracting indirect relationships. A few exciting results were obtained in the 1980s and 1990s, pioneered by Swanson [111]. He proposed a simple model of ‘A influences B’ and ‘B influences C’, therefore ‘A may influence C’; this model is commonly referred to as Swanson’s ABC model. Swanson was able to predict connections years before clinical trials established them. Weeber et al. used term co-occurrences in publication titles and abstracts and found potential new uses for thalidomide [112], and Srinivasan and Libbus [113] used weighted vectors of MeSH terms and UMLS semantic types to discover evidence of turmeric’s therapeutic effect on retinal diseases, Crohn’s disease and spinal cord injuries. More recently, LBD systems have been used to generate hypotheses that are then carried out in animal models for validation. Wren et al. suggested that chlorpromazine may reduce cardiac hypertrophy, and in fact validated their finding in a rodent model [114]. See Srinivasan and Libbus [113] for an overview and review of early LBD systems and the more recent Yetisgen-Yildiz and Pratt [115] for a review of the four main methods used today.
Literature-based discovery systems are not yet a standard tool used by scientists, but we predict that they will have to become so someday, in order to uncover connections that may be critical but are missed owing to the overload of dispersed published knowledge. Several online systems have been developed to aid researchers. Anni 2.0 is an online LBD tool that provides an ontology-based interface to the literature [116]; users can visually explore the literature and the proposed hypotheses. The authors reproduced the implicit relationships suggesting thalidomide uses as predicted by Weeber et al. [112]. Chilibot extracts relationships between genes, chemicals, and diseases, and visualizes these in a network view [98]. The authors identified novel hypothetical relationships, and provide the tool online for researchers to use. Many other systems described in this article can be used as LBD systems, such as iHOP [18]. Other systems have been specifically developed for the goal of LBD. The GeneWays project by Rzhetsky et al. has been used extensively to generate statistically significant predictions that can be tested experimentally, such as relationships between genes and cerebellar phenotypes [117–119]. LitLinker by Yetisgen-Yildiz and Pratt [68] implements an open-discovery approach, where a starting term ‘C’ is specified, but target term ‘A’ is left open. The authors evaluate their ability to capture novel and interesting relationships between diseases and chemicals, drugs, genes or molecular sequences. BITOLA by Hristovski et al. was used to identify candidate disease genes, and differs from LitLinker in its statistical processing as it uses weighted co-occurrence of MeSH terms rather than co-occurrence of terms in the document [120]. The authors later refined the BITOLA system by leveraging semantics extracted by other systems (BioMedLEE and SemRep) to provide more precise information about the ‘B’ entities that connect between the two entities (‘A’ and ‘C’) predicted to be related [121–123]. In this work they focused on predicting drugs that may treat diseases. There are other systems that use text mining as one of several sources of evidence for knowledge discovery. For example, we have reported that text-mined relationships can sometimes perform as well as manually curated ones in the context of candidate gene prediction when integrated with other data types [124]. In this section, however, we focused on those systems that use only text mining as the source of knowledge for discovery.
Summarization
Automatic summarization of information is an active field of research, and in biomedicine this has been applied to tasks ranging from gene overviews to clinical trial summarizations. Yang et al. created automatic summaries of genes; their system clusters genes studied in microarray experiments by MeSH, Gene Ontology and free text features, and then presents summaries for each gene ranked by using sentences extracted from abstracts in MEDLINE [125]. They show that informative sentences are ranked higher by their algorithm, see subsequent evaluation [126]. Such systems allow users to read an overview created from integrating textual information from multiple sources. Summarization can prove very useful for curators of databases, by decreasing the time spent aggregating information sources and allowing curators to focus on reading the information and synthesizing it and advancing it on to the next steps of their pipeline.
Fiszman et al. identified adverse drug events and drug interactions in MEDLINE citations using automatic summarization methods [127]. They presented the results to the user in a graph structure with links to the source text, thereby presenting an overview of the research literature. Pathway diagrams are another form of summarized information. Tari et al. developed a novel approach for automated pharmacokinetic pathway synthesis using facts from hand-curated knowledge bases, as well as from automated extraction via the mining of MEDLINE abstracts [128]. Their method uses a logic program solver to reason out the direction of the relationship between entities, a critical step in automated pathway generation.
Question answering
Question answering methods attempt to automatically provide an answer for questions asked in natural language, such as ‘What are all of the drugs known to induce CYP2C9?’ This can be thought of as ‘a special case of high accuracy information retrieval’ [39]. The goal is to provide very short, specific answers to questions with supporting evidence as context. TREC has had a conference track devoted to general question answering for several years, encouraging progress in this field. More research will be necessary to adapt these technologies for the biomedical field in particular. In 2006, the TREC Genomics Track had a task devoted to question answering in genomics specifically. The clinical domain saw active research earlier, but genomics has only recently been addressed. The TREC Genomics Roadmap includes question answering as one of its long-term goals, and we expect progress in this field in the coming years. For a review see [39,129]. Today, artificial intelligence researchers are still actively developing question answering systems. Results from this research will likely enhance knowledge discovery in pharmacogenomics (as in other domains) by enabling biologists to find the answers to their questions more quickly, as Google has enabled us to efficiently find relevant documents.
Future perspective
There is large demand and significant utility for the application of text mining to the study of pharmacogenomics. Research on the use of text mining applied specifically to the pharmacogenomics domain is gaining attention and has advanced greatly in the past few years, as evinced by the recent workshop devoted to this area at the Pacific Symposium on Biocomputation, which focused on the extraction of genotype–phenotype–drug relationships from text and involved several research groups now focusing on this exact area of research [130,222].
As personalized medicine and consumer genomics become more feasible for the population at large, even more focus will be given to this arena. Pharmacogenomic information appears not only in scientific literature, but also in patent documents and Investigational New Drug applications, and text mining will be employed internally at institutions such as the FDA to help manage the wealth of knowledge. Clinical records will increasingly contain pharmacogenomic clues, and NLP methods to analyze these records will prove critical. Several initiatives have aimed at encouraging such work, such as the i2b2 smoking challenge, First Shared Task for Challenges in NLP for Clinical Data and the repository of de-identified clinical reports made available by the University of Pittsburgh [131,223,224]. In order to develop and evaluate systems specific to text mining of pharmacogenomics literature, it is critical for the community to produce a large shared corpus of annotated pharmacogenomic relationships, and collaborative annotation may be the way to achieve such a corpus.
We predict that the coming years will focus on improving information extraction from full text and figures, and access to the full text of publications is critical for extraction of the complete report of interactions between gene variants and drug response. PubMed Central is a good start towards this goal. The time seems to be ripe for research that goes beyond the mere extraction of explicitly stated knowledge in documents, to linking text-mined and database data through formal reasoning to uncover implicit new knowledge. Close collaboration between pharmacogenomic researchers and computational developers of these systems will advance the field to produce tools useful to the researchers in a mutually beneficial way.
Some attempts have been made to ask authors to represent their publication in a structured digital abstract, using a controlled terminology that makes the essential information of that paper computable. However, recent experiments show that authors do not re-express their findings well using controlled terminologies [132]. Therefore the challenge of text mining will likely remain. Automatic structuring of unstructured abstracts is a possible avenue; we have shown this can be used as a step to assist in extraction of patient demographics from clinical trial reports [133,134].
We envision advances in visualization of high-throughput data. Reasoning on relationship types will allow the creation of a navigable interactive network of all pharmacogenomic relationships, with typed relationships between entities, and evidence connected to edges allowing easy links out to the original publications containing the reported knowledge. As described in our recent editorial [135], the current advances made and focus placed on the extraction of useful and semantically accurate information about pharmacogenomics will enable us to fine-tune our understanding of pharmacogenomics, generate new hypotheses, and uncover novel relationships from the accurate aggregation of all published observations.
Acknowledgments
The authors would like to thank Connie M Oshiro for comments on the manuscript and Nicholas P Tatonetti for useful discussions and assistance creating the figures. We also thank the three anonymous reviewers for their excellent comments.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
The authors acknowledge support from NIH LM07033 (Yael Garten), GM61374 (Yael Garten, Adrien Coulet and Russ B Altman), LM05652 (Russ B Altman) and the National Center for Biomedical Computing (NCBC) National Institute of Health roadmap initiative; NIH grant U54 HG004028 (Adrien Coulet). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286(5439):487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 2▪▪.Krallinger M, Valencia A, Hirschman L. Linking genes to literature: text mining, information extraction, and retrieval applications for biology. Genome Biol. 2008;9(Suppl 2):S8. doi: 10.1186/gb-2008-9-s2-s8. Excellent review of text mining in biology; describes methods and available tools. Has an excellent figure on text mining applications from the biology user’s perspective and provides an online compendium of biomedical language processing applications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein TE, Chang JT, Cho MK, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics research network and knowledge base. Pharmacogenomics J. 2001;1(3):167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 4▪.Baumgartner WA, Cohen KB, Fox LM, Acquaah-Mensah G, Hunter L. Manual curation is not sufficient for annotation of genomic databases. Bioinformatics. 2007;23(13):I41–I48. doi: 10.1093/bioinformatics/btm229. Article motivates biomedical text mining as it proves that manual curation will not allow us to keep up with the growth rate of new knowledge and publications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubitzky W, Azuaje F. Artificial Intelligence Methods and Tools for Systems Biology. Vol. 5. Springer; The Netherlands: 2005. pp. 147–173. [Google Scholar]
- 6.Hirschman L, Yeh A, Blaschke C, Valencia A. Overview of bioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005;6(Suppl 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsujii J. Proceedings of the Workshop on BioNLP: Shared Task: Shared Task. BioNLP ’09: Proceedings of the Workshop on BioNLP: Shared Task; 2009. [Google Scholar]
- 8.Yeh AS, Hirschman L, Morgan AA. Evaluation of text data mining for database curation: lessons learned from the KDD Challenge Cup. Bioinformatics. 2003;19(Suppl 1):I331–I339. doi: 10.1093/bioinformatics/btg1046. [DOI] [PubMed] [Google Scholar]
- 9.Hersh WR, Bhupatiraju RT, Ross L, Cohen AM, Kraemer D, Johnson P. TREC 2004 genomics track overview. Proceedings of annual Text Retrieval Conferences; 2004. [Google Scholar]
- 10.Lascar C, Barnett P. Defining and searching pharmacogenetics and pharmacogenomics to identify its core research journals. Science and Technology Libraries. 2005;26 (1):69–88. [Google Scholar]
- 11.Poulter GL, Rubin DL, Altman RB, Seoighe C. MScanner: a classifier for retrieving MEDLINE citations. BMC Bioinformatics. 2008;19(9):108. doi: 10.1186/1471-2105-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubin DL, Thorn CF, Klein TE, Altman RB. A statistical approach to scanning the biomedical literature for pharmacogenetics knowledge. J Am Med Inform Assoc. 2005;12(2):121–129. doi: 10.1197/jamia.M1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen AM, Hersh WR, Peterson K, Yen PY. Reducing workload in systematic review preparation using automated citation classification. J Am Med Inform Assoc. 2006;13(2):206–219. doi: 10.1197/jamia.M1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doms A, Schroeder M. GoPubMed: exploring PubMed with the gene ontology. Nucleic Acids Res. 2005;33(Web Server issue):W783–W786. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plikus MV, Zhang Z, Chuong CM. PubFocus: semantic MEDLINE/PubMed citations analytics through integration of controlled biomedical dictionaries and ranking algorithm. BMC Bioinformatics. 2006;7:424. doi: 10.1186/1471-2105-7-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siadaty MS, Shu J, Knaus WA. Relemed: sentence-level search engine with relevance score for the MEDLINE database of biomedical articles. BMC Med Inform Decis Mak. 2007;7:1. doi: 10.1186/1472-6947-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Iratxeta C, Bork P, Andrade MA. XplorMed: a tool for exploring MEDLINE abstracts. Trends Biochem Sci. 2001;26(9):573–575. doi: 10.1016/s0968-0004(01)01926-0. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann R, Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21(Suppl 2):II252–II258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 19.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2(11):E309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21▪.Winnenburg R, Wächter T, Plake C, Doms A, Schroeder M. Facts from text: can text mining help to scale-up high-quality manual curation of gene products with ontologies? Brief Bioinformatics. 2008;9(6):466–478. doi: 10.1093/bib/bbn043. Excellent review. Includes table comparing existing tools for information retrieval and information extraction across many attributes. [DOI] [PubMed] [Google Scholar]
- 22▪.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet. 2006;7(2):119–129. doi: 10.1038/nrg1768. Excellent review from 2006, details the rise of text mining to support biology and importance of biologists working with computational linguists to further this goal. [DOI] [PubMed] [Google Scholar]
- 23.Krauthammer M, Rzhetsky A, Morozov P, Friedman C. Using BLAST for identifying gene and protein names in journal articles. Gene. 2000;259(1–2):245–252. doi: 10.1016/s0378-1119(00)00431-5. [DOI] [PubMed] [Google Scholar]
- 24.Hanisch D, Fluck J, Mevissen HT, Zimmer R. Playing biology’s name game: identifying protein names in scientific text. Pac Symp Biocomput. 2003:403–414. [PubMed] [Google Scholar]
- 25.Tsuruoka Y, Tsujii J. Improving the performance of dictionary-based approaches in protein name recognition. J Biomed Inform. 2004;37(6):461–470. doi: 10.1016/j.jbi.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda K, Tamura A, Tsunoda T, Takagi T. Toward information extraction: identifying protein names from biological papers. Pac Symp Biocomput. 1998:707–718. [PubMed] [Google Scholar]
- 27.Tanabe L, Wilbur WJ. Tagging gene and protein names in biomedical text. Bioinformatics. 2002;18(8):1124–1132. doi: 10.1093/bioinformatics/18.8.1124. [DOI] [PubMed] [Google Scholar]
- 28.Kim JD, Ohta T, Tateisi Y, Tsujii J. GENIA corpus–semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(Suppl 1):I180–I182. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- 29.Morgan AA, Lu Z, Wang X, et al. Overview of BioCreative II gene normalization. Genome Biol. 2008;9(Suppl 2):S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith L, Tanabe LK, Ando RJ, et al. Overview of BioCreative II gene mention recognition. Genome Biol. 2008;9(Suppl 2):S2. doi: 10.1186/gb-2008-9-s2-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh A, Morgan A, Colosimo M, Hirschman L. BioCreAtIvE task 1A: gene mention finding evaluation. BMC Bioinformatics. 2005;6(Suppl 1):S2. doi: 10.1186/1471-2105-6-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collier N, Nobata C, Tsujii J. Extracting the names of genes and gene products with a hidden Markov model. Proceedings of the 18th conference on Computational linguistics. 2000;1:201–207. [Google Scholar]
- 33.Zhou G, Zhang J, Su J, Shen D, Tan C. Recognizing names in biomedical texts: a machine learning approach. Bioinformatics. 2004;20(7):1178–1190. doi: 10.1093/bioinformatics/bth060. [DOI] [PubMed] [Google Scholar]
- 34.Smith LH, Rindflesch TC, Wilbur WJ. The importance of the lexicon in tagging biological text. Natural Language Engineering. 2005;1:1. [Google Scholar]
- 35.Kazama J, Makino T, Ohta Y, Tsujii J. Tuning support vector machines for biomedical named entity recognition. Proceedings of the ACL-02 Workshop on Natural language Processing in the Biomedical Domain; 2002. [Google Scholar]
- 36.McDonald R, Pereira F. Identifying gene and protein mentions in text using conditional random fields. BMC Bioinformatics. 2005;6(Suppl 1):S6. doi: 10.1186/1471-2105-6-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu CN, Chang YM, Kuo CJ, Lin YS, Huang HS, Chung IF. Integrating high dimensional bi-directional parsing models for gene mention tagging. Bioinformatics. 2008;24(13):I286–I294. doi: 10.1093/bioinformatics/btn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, Lin Y, Lin K, et al. High-Recall Gene Mention Recognition by Unification of Multiple Backward Parsing Models. Enhancing Recall by Unifying Backward Models. 2007 [Google Scholar]
- 39.Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB. Frontiers of biomedical text mining: current progress. Brief Bioinformatics. 2007;8(5):358–375. doi: 10.1093/bib/bbm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang JT, Schütze H, Altman RB. GAPSCORE: finding gene and protein names one word at a time. Bioinformatics. 2004;20(2):216–225. doi: 10.1093/bioinformatics/btg393. [DOI] [PubMed] [Google Scholar]
- 41.Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pac Symp Biocomput. 2008:652–663. [PubMed] [Google Scholar]
- 42.Settles B. ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics. 2005;21(14):3191–3192. doi: 10.1093/bioinformatics/bti475. [DOI] [PubMed] [Google Scholar]
- 43.Cohen KB, Hunter L. Getting started in text mining. PLoS Comput Biol. 2008;4(1):E20. doi: 10.1371/journal.pcbi.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuason O, Chen L, Liu H, Blake JA, Friedman C. Biological nomenclatures: a source of lexical knowledge and ambiguity. Pac Symp Biocomput. 2004:238–249. doi: 10.1142/9789812704856_0023. [DOI] [PubMed] [Google Scholar]
- 45.Horn F, Lau AL, Cohen FE. Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors. Bioinformatics. 2004;20(4):557–568. doi: 10.1093/bioinformatics/btg449. [DOI] [PubMed] [Google Scholar]
- 46.Xuan W, Wang P, Watson SJ, Meng F. MEDLINE search engine for finding genetic markers with biological significance. Bioinformatics. 2007;23(18):2477–2484. doi: 10.1093/bioinformatics/btm375. [DOI] [PubMed] [Google Scholar]
- 47.Caporaso JG, Baumgartner WA, Randolph DA, Cohen KB, Hunter L. MutationFinder: a high-performance system for extracting point mutation mentions from text. Bioinformatics. 2007;23(14):1862–1865. doi: 10.1093/bioinformatics/btm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segura-Bedmar I, Martínez P, Segura-Bedmar M. Drug name recognition and classification in biomedical texts. A case study outlining approaches underpinning automated systems. Drug Discov Today. 2008;13(17–18):816–823. doi: 10.1016/j.drudis.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 49▪.Agarwal P, Searls DB. Literature mining in support of drug discovery. Brief Bioinformatics. 2008;9(6):479–492. doi: 10.1093/bib/bbn035. Interesting review including a GlaxoSmithKline case study showing how text mining is used to support their drug discovery pipeline. [DOI] [PubMed] [Google Scholar]
- 50.Rindflesch TC, Tanabe L, Weinstein JN, Hunter L. EDGAR: extraction of drugs, genes and relations from the biomedical literature. Pac Symp Biocomput. 2000:517–528. doi: 10.1142/9789814447331_0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolárik C, Hofmann-Apitius M, Zimmermann M, Fluck J. Identification of new drug classification terms in textual resources. Bioinformatics. 2007;23(13):I264–I272. doi: 10.1093/bioinformatics/btm196. [DOI] [PubMed] [Google Scholar]
- 53▪.Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–D906. doi: 10.1093/nar/gkm958. Manuscript describes DrugBank, a rich annotated source of detailed drug information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hettne KM, Stierum RH, Schuemie MJ, et al. A dictionary to identify small molecules and drugs in free text. Bioinformatics. 2009;25(22):2983–2991. doi: 10.1093/bioinformatics/btp535. [DOI] [PubMed] [Google Scholar]
- 55▪.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. Useful recent overview of MetaMap, an important resource in structuring biomedical text, particularly for phenotype recognition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu R, Supekar K, Morgan A, Das A, Garber A. Unsupervised method for automatic construction of a disease dictionary from a large free text collection. AMIA Annu Symp Proc. 2008:820–824. [PMC free article] [PubMed] [Google Scholar]
- 57▪.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343. doi: 10.1038/msb.2009.98. Useful resource connecting 888 drugs to their side effects, freely available online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malone J, Holloway E, Adamusiak T, et al. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics. 2010;26(8):1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noy NF, Shah NH, Whetzel PL, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37(Web Server issue):W170–W173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu R, Musen M, Nigam S. A comprehensive analysis of five million UMLS metathesaurus terms using eighteen million MEDLINE citations. AMIA Annu Symp Proc. 2010 (In Press) [PMC free article] [PubMed] [Google Scholar]
- 62.Chang JT, Schütze H, Altman RB. Creating an online dictionary of abbreviations from MEDLINE. J Am Med Inform Assoc. 2002;9(6):612–620. doi: 10.1197/jamia.M1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwartz AS, Hearst MA. A simple algorithm for identifying abbreviation definitions in biomedical text. Pac Symp Biocomput. 2003:451–462. [PubMed] [Google Scholar]
- 64.Yu H, Hripcsak G, Friedman C. Mapping abbreviations to full forms in biomedical articles. J Am Med Inform Assoc. 2002;9(3):262–272. doi: 10.1197/jamia.M0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torii M, Hu ZZ, Song M, Wu CH, Liu H. A comparison study on algorithms of detecting long forms for short forms in biomedical text. BMC Bioinformatics. 2007;8(Suppl 9):S5. doi: 10.1186/1471-2105-8-S9-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ao H, Takagi T. ALICE: an algorithm to extract abbreviations from MEDLINE. J Am Med Inform Assoc. 2005;12(5):576–586. doi: 10.1197/jamia.M1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wren JD, Chang JT, Pustejovsky J, Adar E, Garner HR, Altman RB. Biomedical term mapping databases. Nucleic Acids Res. 2005;33(Database issue):D289–D293. doi: 10.1093/nar/gki137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yetisgen-Yildiz M, Pratt W. Using statistical and knowledge-based approaches for literature-based discovery. J Biomed Inform. 2006;39(6):600–611. doi: 10.1016/j.jbi.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Xu Y, Wang Z, Lei Y, Zhao Y, Xue Y. MBA: a literature mining system for extracting biomedical abbreviations. BMC Bioinformatics. 2009;10:14. doi: 10.1186/1471-2105-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hunter L, Cohen KB. Biomedical language processing: what’s beyond PubMed? Mol Cell. 2006;21(5):589–594. doi: 10.1016/j.molcel.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wermter J, Tomanek K, Hahn U. High-performance gene name normalization with GeNo. Bioinformatics. 2009;25(6):815–821. doi: 10.1093/bioinformatics/btp071. [DOI] [PubMed] [Google Scholar]
- 72.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36(Database issue):D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coulet A, Smail-Tabbone M, Benlian P, Napoli A, Devignes M. SNP-converter: An ontology-based solution to reconcile heterogeneous SNP descriptions for pharmacogenomic studies. In: Leser U, Naumann F, Eckman B, editors. Data Integration in the Life Sciences. Springer; Berlin, Germany: 2006. pp. 82–93. [Google Scholar]
- 74.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15(1):7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 75.Fundel K, Küffner R, Zimmer R. RelEx – relation extraction using dependency parse trees. Bioinformatics. 2007;23(3):365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 76.Friedman C, Kra P, Yu H, Krauthammer M, Rzhetsky A. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics. 2001;17(Suppl 1):S74–S82. doi: 10.1093/bioinformatics/17.suppl_1.s74. [DOI] [PubMed] [Google Scholar]
- 77.Rindflesch TC, Libbus B, Hristovski D, Aronson AR, Kilicoglu H. Semantic relations asserting the etiology of genetic diseases. AMIA Annu Symp Proc. 2003:554–558. [PMC free article] [PubMed] [Google Scholar]
- 78.Chang JT, Altman RB. Extracting and characterizing gene-drug relationships from the literature. Pharmacogenetics. 2004;14(9):577–586. doi: 10.1097/00008571-200409000-00002. [DOI] [PubMed] [Google Scholar]
- 79.Tsuruoka Y, Tsujii J, Ananiadou S. FACTA: a text search engine for finding associated biomedical concepts. Bioinformatics. 2008;24(21):2559–2560. doi: 10.1093/bioinformatics/btn469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alako BT, Veldhoven A, van Baal S, et al. CoPub Mapper: mining MEDLINE based on search term co-publication. BMC Bioinformatics. 2005;6:51. doi: 10.1186/1471-2105-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81▪.Plake C, Schiemann T, Pankalla M, Hakenberg J, Leser U. AliBaba: PubMed as a graph. Bioinformatics. 2006;22(19):2444–2445. doi: 10.1093/bioinformatics/btl408. Authors provide an easy-to-use interactive application for browsing biological networks extracted on-the-fly from results of PubMed queries. Edges of the graph display the supporting evidence (sentence from PubMed abstract) linking two entities. [DOI] [PubMed] [Google Scholar]
- 82.Rebholz-Schuhmann D, Kirsch H, Arregui M, Gaudan S, Riethoven M, Stoehr P. EBIMed–text crunching to gather facts for proteins from MEDLINE. Bioinformatics. 2007;23(2):E37–E244. doi: 10.1093/bioinformatics/btl302. [DOI] [PubMed] [Google Scholar]
- 83▪.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36(Database issue):D684–D688. doi: 10.1093/nar/gkm795. Useful interactive online resource for exploring interactions between proteins and over 68,000 small molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80(4):588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blaschke C, Andrade MA, Ouzounis C, Valencia A. Automatic extraction of biological information from scientific text: protein-protein interactions. Proc Int Conf Intell Syst Mol Biol. 1999:60–67. [PubMed] [Google Scholar]
- 86.Cohen T, Widdows D. Empirical distributional semantics: methods and biomedical applications. J Biomed Inform. 2009;42(2):390–405. doi: 10.1016/j.jbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klein D, Manning CD. Accurate unlexicalized parsing. ACL ’03: Proceedings of the 41st Annual Meeting on Association for Computational Linguistics. 2003;1:423–430. [Google Scholar]
- 88.Tari L, Hakenberg J, Gonzalez G, Baral C. Querying parse tree database of MEDLINE text to synthesize user-specific biomolecular networks. Pac Symp Biocomput. 2009:87–98. [PubMed] [Google Scholar]
- 89.Hunter L, Lu Z, Firby J, et al. OpenDMAP: an open source, ontology-driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-type-specific gene expression. BMC Bioinformatics. 2008;9:78. doi: 10.1186/1471-2105-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ahlers CB, Fiszman M, Demner-Fushman D, Lang FM, Rindflesch TC. Extracting semantic predications from MEDLINE citations for pharmacogenomics. Pac Symp Biocomput. 2007:209–220. [PubMed] [Google Scholar]
- 91.Hakenberg J, Voronov D, Nguyen VH, et al. Taking a SNPshot of PubMed – a repository of genetic variants and their drug response phenotypes. GPD-Rxn Workshop: Genotype-Phenotype-Drug Relationship Extraction from Text at Pac. Symp. Biocomput; 2010. [Google Scholar]
- 92.Coulet A, Shah N, Garten Y, Musen M, Altman R. Using text to build semantic networks for pharmacogenomics. J Biomed Inform. 2010 doi: 10.1016/j.jbi.2010.08.005. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thompson P, Iqbal SA, McNaught J, Ananiadou S. Construction of an annotated corpus to support biomedical information extraction. BMC Bioinformatics. 2009;10:349. doi: 10.1186/1471-2105-10-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cano C, Monaghan T, Blanco A, Wall DP, Peshkin L. Collaborative text-annotation resource for disease-centered relation extraction from biomedical text. J Biomed Inform. 2009;42(5):967–977. doi: 10.1016/j.jbi.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Craven M, Kumlien J. Constructing biological knowledge bases by extracting information from text sources. Proc Int Conf Intell Syst Mol Biol. 1999:77–86. [PubMed] [Google Scholar]
- 96.Airola A, Pyysalo S, Björne J, Pahikkala T, Ginter F, Salakoski T. All-paths graph kernel for protein-protein interaction extraction with evaluation of cross-corpus learning. BMC Bioinformatics. 2008;9(Suppl 11):S2. doi: 10.1186/1471-2105-9-S11-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buyko E, Faessler E, Wermter J, Hahn U. Event extraction from trimmed dependency graphs. Proceedings of the Workshop on BioNLP: Shared Task; 2009. pp. 19–27. [Google Scholar]
- 98.Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics. 2004;5:147. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goldin IM, Chapman WW. Learning to Detect Negation with ‘Not’ in Medical Texts. Workshop at: 26th ACM SIGIR Conference; 28 July–1 August; Toronto, Canada. 2003. [Google Scholar]
- 100.de Marneffe MC, Rafferty AN, Manning CD. Finding contradictions in text. Proceedings of ACL-08:HLT, Annual Meeting of the Association for Computational Linguistics; 15–20 June; OH, USA. 2008. pp. 1039–1047. [Google Scholar]
- 101.Shah PK, Perez-Iratxeta C, Bork P, Andrade MA. Information extraction from full text scientific articles: where are the keywords? BMC Bioinformatics. 2003;4:20. doi: 10.1186/1471-2105-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cohen KB, Johnson H, Verspoor K, Roeder C, Hunter L. Tool performance and semantic type distribution for genotype-phenotype-drug relationship. Pac Symp Biocomput. 2010 [Google Scholar]
- 103▪.Hearst MA, Divoli A, Guturu H, et al. BioText search engine: beyond abstract search. Bioinformatics. 2007;23(16):2196–2197. doi: 10.1093/bioinformatics/btm301. Authors describe BioText, an application that mines and searches within figures, captions and tables, as well as titles and abstracts. Important advance in mining the scientific literature. [DOI] [PubMed] [Google Scholar]
- 104.Segura-Bedmar I, Crespo M, de Pablo-Sánchez C, Martínez P. Resolving anaphoras for the extraction of drug-drug interactions in pharmacological documents. BMC Bioinformatics. 2010;11(Suppl 2):S1. doi: 10.1186/1471-2105-11-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Z, Kim S, Quinney SK, et al. Literature mining on pharmacokinetics numerical data: a feasibility study. J Biomed Inform. 2009;42(4):726–735. doi: 10.1016/j.jbi.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rinaldi F, Schneider G, Kaljurand K, Hess M, Romacker M. An environment for relation mining over richly annotated corpora: the case of GENIA. BMC Bioinformatics. 2006;7(Suppl 3):S3. doi: 10.1186/1471-2105-7-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ramakrishnan C, Mendes PN, Wang S, Sheth AP. Unsupervised Discovery of Compound Entities for Relationship Extraction. Proceedings of EKAW Knowledge Engineering: Practice and Patterns; 29 September–2 October 2; Acitrezza, Italy. 2008. pp. 146–155. [Google Scholar]
- 108.Coulet A, Altman RB, Musen MA, Shah NH. Integrating heterogeneous relationships extracted from natural language sentences. Proceedings of the Bio-ontologies SIG, ISBM; 9–10 July; MA, USA. 2010. [Google Scholar]
- 109.Cohen KB, Hunter L. Artificial Intelligence Methods and Tools for Systems Biology. Vol. 5. Springer; 2005. Natural Language Processing and Systems Biology; pp. 147–173. [DOI] [Google Scholar]
- 110.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Swanson DR. Complementary structures in disjoint science literatures. SIGIR ’91: Proceedings of the 14th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; 1991. [Google Scholar]
- 112.Weeber M, Vos R, Klein H, De Jong-Van Den Berg LT, Aronson AR, Molema G. Generating hypotheses by discovering implicit associations in the literature: a case report of a search for new potential therapeutic uses for thalidomide. J Am Med Inform Assoc. 2003;10(3):252–259. doi: 10.1197/jamia.M1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Srinivasan P, Libbus B. Mining MEDLINE for implicit links between dietary substances and diseases. Bioinformatics. 2004;20(Suppl 1):I290–I296. doi: 10.1093/bioinformatics/bth914. [DOI] [PubMed] [Google Scholar]
- 114.Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004;20(3):389–398. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- 115.Yetisgen-Yildiz M, Pratt W. A new evaluation methodology for literature-based discovery systems. J Biomed Inform. 2009;42(4):633–643. doi: 10.1016/j.jbi.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 116.Jelier R, Schuemie MJ, Veldhoven A, Dorssers LC, Jenster G, Kors JA. Anni 2.0: a multipurpose text-mining tool for the life sciences. Genome Biol. 2008;9(6):R96. doi: 10.1186/gb-2008-9-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rzhetsky A, Iossifov I, Koike T, et al. GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J Biomed Inform. 2004;37(1):43–53. doi: 10.1016/j.jbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 118.Rzhetsky A, Iossifov I, Loh JM, White KP. Microparadigms: chains of collective reasoning in publications about molecular interactions. Proc Natl Acad Sci USA. 2006;103(13):4940–4945. doi: 10.1073/pnas.0600591103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iossifov I, Rodriguez-Esteban R, Mayzus I, Millen KJ, Rzhetsky A. Looking at cerebellar malformations through text-mined interactomes of mice and humans. PLoS Comput Biol. 2009;5(11):e1000559. doi: 10.1371/journal.pcbi.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hristovski D, Peterlin B, Mitchell JA, Humphrey SM. Using literature-based discovery to identify disease candidate genes. Int J Med Inform. 2005;74(2–4):289–298. doi: 10.1016/j.ijmedinf.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 121.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 122.Lussier Y, Borlawsky T, Rappaport D, Liu Y, Friedman C. PhenoGO: assigning phenotypic context to gene ontology annotations with natural language processing. Pac Symp Biocomput. 2006:64–75. [PMC free article] [PubMed] [Google Scholar]
- 123.Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Exploiting semantic relations for literature-based discovery. AMIA Annu Symp Proc. 2006:349–353. [PMC free article] [PubMed] [Google Scholar]
- 124.Garten Y, Tatonetti NP, Altman RB. Improving the prediction of pharmacogenes using text-derived drug-gene relationships. Pac Symp Biocomput. 2010:305–314. doi: 10.1142/9789814295291_0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang J, Cohen AM, Hersh W. Automatic summarization of mouse gene information by clustering and sentence extraction from MEDLINE abstracts. AMIA Annu Symp Proc. 2007;11:831–835. [PMC free article] [PubMed] [Google Scholar]
- 126.Yang J, Cohen A, Hersh W. Evaluation of a gene information summarization system by users during the analysis process of microarray datasets. BMC Bioinformatics. 2009;10(Suppl 2):S5. doi: 10.1186/1471-2105-10-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fiszman M, Rindflesch TC, Kilicoglu H. Summarizing drug information in MEDLINE citations. AMIA Annu Symp Proc. 2006:254–258. [PMC free article] [PubMed] [Google Scholar]
- 128.Tari L, Anwar S, Liang S, Hakenberg J, Baral C. Synthesis of pharmacokinetic pathways through knowledge acquisition and automated reasoning. Pac Symp Biocomput. 2010:465–476. doi: 10.1142/9789814295291_0048. [DOI] [PubMed] [Google Scholar]
- 129.Zweigenbaum P. Question answering in biomedicine. Proc Workshop on Natural Language Processing for Question Answering, EACL; 2003. [Google Scholar]
- 130.Coulet A, Shah N, Hunter L, Barral C, Altman RB. Extraction of genotype-phenotype-drug relationships from text: from entity recognition to bioinformatics application. Pac Symp Biocomput. 2010:485–487. doi: 10.1142/9789814295291_0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heinze DT, Morsch ML, Potter BC, Sheffer RE. Medical i2b2 NLP smoking challenge: the A-Life system architecture and methodology. J Am Med Inform Assoc. 2008;15(1):40–43. doi: 10.1197/jamia.M2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ceol A, Chatr-Aryamontri A, Licata L, Cesareni G. Linking entries in protein interaction database to structured text: the FEBS Letters experiment. FEBS Lett. 2008;582(8):1171–1177. doi: 10.1016/j.febslet.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 133.Xu R, Supekar K, Huang Y, Das A, Garber A. Combining text classification and Hidden Markov Modeling techniques for categorizing sentences in randomized clinical trial abstracts. AMIA Annu Symp Proc. 2006:824–828. [PMC free article] [PubMed] [Google Scholar]
- 134.Xu R, Garten Y, Supekar KS, Das AK, Altman RB, Garber AM. Extracting subject demographic information from abstracts of randomized clinical trial reports. Stud Health Technol Inform. 2007;129(Pt 1):550–554. [PubMed] [Google Scholar]
- 135.Garten Y, Altman RB. Teaching computers to read the pharmacogenomics literature … so you don’t have to. Pharmacogenomics. 2010;11(4):515–518. doi: 10.2217/pgs.10.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hull D, Pettifer SR, Kell DB. Defrosting the digital library: bibliographic tools for the next generation web. PLoS Comput Biol. 2008;4(10) doi: 10.1371/journal.pcbi.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Takatori R, Takahashi KA, Tokunaga D, et al. ABCB1 C3435T polymorphism influences methotrexate sensitivity in rheumatoid arthritis patients. Clin Exp Rheumatol. 2006;24(5):546–554. [PubMed] [Google Scholar]
- 138.Yi SY, Hong KS, Lim HS, et al. A variant 2677A allele of the MDR1 gene affects fexofenadine disposition. Clin Pharmacol Ther. 2004;76(5):418–427. doi: 10.1016/j.clpt.2004.08.002. [DOI] [PubMed] [Google Scholar]
Websites
- 201.Collect, encode, and disseminate knowledge about the impact of human genetic variations on drug response. www.pharmgkb.org.
- 202.National institute of general medical science. www.nigms.nih.gov/Initiatives/PGRN.
- 203.What Is Text Mining? http://people.ischool.berkeley.edu/~hearst/text-mining.html.
- 204.Trec retrieval conference; http://trec.nist.gov. [Google Scholar]
- 205▪.Bio-NLP resources database. http://zope.bioinfo.cnio.es/bionlp_tools. Excellent online compendium of available BioNLP applications.
- 206.Gene ontology project, a major bioinformatics initiative. www.geneontology.org.
- 207.HUGO gene nomenclature committee. www.genenames.org.
- 208.Biomedical information extraction project. http://bioie.ldc.upenn.edu.
- 209.Natural language processing software for text analytics, text data mining and search. www.alias-i.com.
- 210.National libraries of medicine. www.nlm.nih.gov/research/umls.
- 211.PhenX. Consensus measures for phenotypes and exposures. www.PhenX.org.
- 212.Data schema and harmonization platform for epidemiological research. www.datashaper.org.
- 213.Biomedical acronym database. http://lethargy.swmed.edu/ARGH/argh.asp.
- 214.Biomedical term mapping database. http://bionlp.stanford.edu/abbreviation.
- 215.Biomedical term mapping database. http://medstract.med.tufts.edu/acro1.1/index.htm.
- 216.Simple and robust abbreviation dictionary. doi: 10.1093/bioinformatics/btg439. www.hpl.hp.com/research/idl/projects/abbrev.html. [DOI] [PubMed]
- 217.Database of SNPs. www.ncbi.nlm.nih.gov/sites/entrez?db=snp.
- 218.Human genome variation society. www.hgvs.org.
- 219.Resource to explore known and predicted interactions of chemicals and proteins. http://stitch.embl.de.
- 220.Open source software capable of solving almost any text processing problem. http://gate.ac.uk.
- 221.A repository of genetic variants linked to phenotypic effects on drug response. http://bioai4core.fulton.asu.edu/snpshot.
- 222.GPD-Rxn workshop. genotype-phenotype-drug relationship extraction from text. http://psb.stanford.edu/psb10/gpdrxn-workshop.html.
- 223.Informatics for integrating biology at the bedside, NPL research dataset. www.i2b2.org/NLP/DataSets/Main.php.
- 224.University of Pittsburgh NLP repository. www.dbmi.pitt.edu/blulab/nlprepository.html.