Abstract
We performed alignment of apolipoprotein A-I (apoA-I) sequences from 31 species of animals. We found there is specific conservation of salt bridge-forming residues in the first 30 residues of apoA-I and general conservation of a variety of residue types in the central domain, helix 2/3 to helix 7/8. In the lipid-associating domain, helix 7 and helix 10 are the most and least conserved helixes, respectively. Furthermore, eight residues are completely conserved: P66, R83, P121, E191, and P220, and three of seven Tyr residues in human apoA-I, Y18, Y115, and Y192, are conserved. Residue Y18 appears to be important for assembly of HDL. E191-Y192 represents the only completely conserved pair of adjacent residues in apoA-I; Y192 is a preferred target for site-specific oxidative modification within atheroma, and molecular dynamic simulations suggest that the conserved pair E191-Y192 is in a solvent-exposed loop-helix-loop. Molecular dynamics testing of human apoA-I showed that M112 and M148 interact with Y115, a microenvironment unique to human apoA-I. Finally, conservation of Arg residues in the α11/3 helical wheel position 7 supports several possibilities: interactions with adjacent phospholipid molecules and/or oxidized lipids and/or binding of antioxidant enzymes through cation-π orbital interactions. We conclude that sequence alignment of apoA-I provides unique insights into apoA-I structure-function relationship.
Keywords: antioxidants, apolipoproteins, lipoproteins, molecular modeling
HDL represents a heterogeneous population of particles in which apolipoprotein A-I (apo A-I) is the major protein; apoA-I-containing lipoproteins in the form of HDL are inversely correlated with the risk of coronary artery disease and atherosclerosis (1). Conformation of apoA-I is highly flexible as apoA-I exists in different states: lipid-free, lipid-poor, and discoidal or spheroidal lipoproteins of different sizes (1). Furthermore, HDL, a supramolecular assembly of lipid and protein, is a soft form of condensed matter easily deformable by thermal fluctuations (2), so that direct experimental methods for studying HDL structure-function relationship have had only limited success.
HDL has been suggested to serve as a platform for the assembly of certain protein components with specific functions (3–5) that form the basis for functional heterogeneity of HDL. Shotgun proteomics has shown that many of these proteins correlate with antioxidant, anti-inflammatory, and antiatherogenic properties of HDL (4, 5). In particular, paraoxonase-1 (PON-1) is found to be increased in HDL from patients with coronary artery disease (4). In mice lacking both apoE and PON-1, there is an increased accumulation of oxidized phospholipids in the plasma and an increase in atherosclerosis compared with that in apoE-deficient mice (6). PON-1 has been shown to inhibit cholesterol biosynthesis (7), uptake of oxidized LDL by macrophages (8), and the inflammatory response (9).
Two possible mechanisms for the segregation of HDL proteins among HDL subspecies have been suggested: i) specific protein–protein interactions on the HDL surface, or ii) attraction of certain proteins to a particular particle biophysical characteristic, such as lipid surface pressure or surface curvature (5).
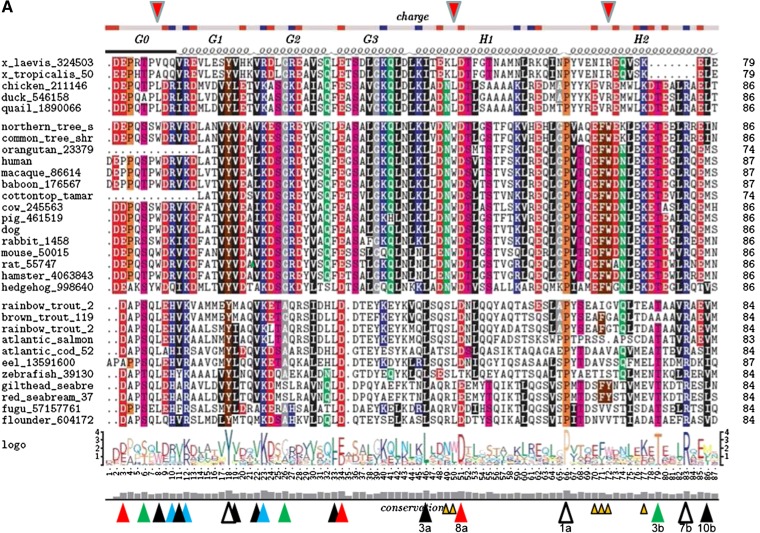
The mature sequence of human apo A-I has a length of 243 amino acid residues and is encoded by exons 3 and 4 of the apolipoprotein gene in chromosome 11. The common lipid-associating motif in apoA-I is the amphipathic α helix (10, 11). Exon 3 encodes residues 1–43 in human apoA-I. This domain, commonly referred to as the globular domain (12), contains an N-terminal segment of 8–10 residues (10 in human apoA-I), followed by three 11 mer amino acid tandem repeats, designated G1, G2, and G3, respectively (see Fig. 3).
Fig. 3.
Sequence alignment of 31 ApoA-I orthologs. The top bar shows the consensus position of charged residues. The second row shows the position of tandem 11/22 mer repetitive domains G0 to H10. Helix 5 (H5) in all 31 sequences is contained within a box. The diagram under the alignment of 31 sequences shows a traditional 4.2-bit conservation scale WebLogo representation of the alignment. The bottom histogram shows the degree of conservation of each sequence position.
Barker and Dayhoff (13), Fitch (14), and McLachlan (15) independently observed that apoA-I contains multiple repeats of 22 amino acids (22 mer), each of which consists of two 11 mer tandem arrays. DNA sequencing work has confirmed the existence of a 22 mer periodicity in apoA-I (16). The portion of apoA-I that follows the globular domain, residues 44–241 in human apoA-I, referred to as the lipid-associating domain (12), contains eight 22 mer and two 11 mer tandem repeats, each of which has the helical wheel signature of an amphipathic α helix. The fundamental unit of repeat in exon 4 is not an 11 mer but a 22 mer repeat made up of two 11 mer repeats designated a and b; most 11 mer repeats are more similar to the 11- mer repeat one unit removed (e.g., helix 1a vs. helix 2a) than to the adjacent 11 mer unit (e.g., helix 1a vs. helix 1b) (14, 16, 17). This is indicated by the fact that almost all Pro residues appear in the first column when the alignment is made using 22 amino acids as the basic unit of repeat, i.e., at position 1 in repeat a. It has therefore been suggested that the different apolipoprotein genes arose from a common ancestral gene by gene duplications of 11 codons (18).
This laboratory proposed in 1999 an atomic resolution double-belt model for discoidal HDL that consists of two copies of the lipid-associating domain of apoA-I arranged as continuous antiparallel amphipathic helixes around a bilayer disc containing 160 palmitoyloleoyl phosphatidylcholine (POPC) molecules that forms a 106-Å-diameter particle (19). The general features of the double-belt model, particularly the LL5/5 registry of the two chains (19), have been confirmed by multiple laboratories using physical chemical methods (20–35).
RATIONALE FOR THIS REVIEW
It has been approximately 10 years since the last fundamental review articles pertaining to phylogenetic comparisons of apoA-I sequences were published (36, 37). Each of these reviews compared the sequences of apoA-I from 11 species that were only partially identical, for a total of 14 species: 10 species of mammals, 2 of birds, and 2 of fish.
Based upon similarities among teleosts (a major infraclass of ray-finned fish) and human apoA-I sequences and the absence of an apoA-I ortholog in lamprey eels (a cyclostomatous, jawless fish), it is generally accepted that the repeat patterns of the apoA-I protein family arose between the teleost fish–mammal split, approximately 400 million years ago (38), and the cyclostomatous fish origin, approximately 430 million years ago. Sequences of apoA-I from sharks and other cartilaginous fish (animals that arose approximately 420 million years ago) have, as yet, not been published.
In this review, we performed detailed sequence alignment and conservation analyses using a database of unique apoA-I amino acid sequences from 31 species, consisting of 11 species of fish, 2 species of amphibians, 3 species of birds, and 18 species of mammals. When analyzed for correlations with current concepts of apoA-I and HDL structure and function, these analyses provide useful insights into the structure-function relationship of apoA-I.
ANALYTIC TOOLS
ApoA-I sequence alignments
All apoA-I sequences were retrieved from the GenBank protein database (39), and their corresponding version numbers are used throughout the text. The mature protein sequence of human apoA-I (version number 2A01_A) was used as a seed in two iterations of the position-specific iterated BLAST (PSI-BLAST) (40) algorithm used to search through all available taxonomic groups. Nonredundant protein sequence databases were searched, and general parameters were set using values for the expected threshold (41) of 10 and a word size of three; the scoring parameters used were BLOSUM62 protein substitution matrix (42) with costs of gap opening and extension of 11 and 1, respectively; no filtering or masking of the low-complexity regions were used; and the statistical significance of the PSI-BLAST threshold for the second iteration was 0.005. The search resulted in the selection of more than 100 sequences from fish, amphibians, birds, and mammal species. After removing protein sequences that were identical (although they possessed unique version numbers) and closely related paralogs (43), 31 orthologous sequences remained, and these were used for subsequent analysis.
Sequences were aligned using ClustalX version 1.83 software (44) (packaged through MultiCLUSTAL (45) script), performing 98 cycles of alignments and using six different parameters, as follows: different multiple protein substitution matrices; costs for gap openings between 0 and 20 in steps of 4; pairwise gap extensions between 0 and 0.5 with 0.1 intervals; and multiple gap extensions between 0.02 and 0.1 with an interval of 0.02. The highest scoring alignment, according to the MultiCLUSTAL estimate, was chosen for further analysis. Phylogenetic analyses were performed using a molecular evolutionary genetics analysis software suite (46).
Sequence alignment logos with a small sample correction applied (47) were created using the Web interface to WebLogo software (48). Subfamily logos (49), as implemented in TeXshade (50), a LaTeX macro software (Addison-Wesley, Reading MA) for setting and shading multiple sequence alignments, were used together with a WebLogo interface. Helical wheels were produced using the WHEEL set of software (51).
Molecular dynamics simulations
One PL-rich particle containing 160 POPC, 24 unesterified cholesterol (UC), and 2 apoA-I molecules was created by generating an all-atom particle containing 2 apoA-I molecules in a 100% α11/3 helical wheel conformation wrapped in an antiparallel double belt around a POPC/UC bilayer and then subjected to molecular dynamics (MD) simulated annealing (MDSA) 16 times by the methods described previously (52).
The fraction of frames in which each residue was determined to be α helical was measured over the last 10 ns of the ensemble of 16 MDSA 160:24:2 simulations by using the visual molecular dynamics (VMD) implementation of the secondary structure determination software, STRIDE (52a). Fractions for each MDSA were averaged and the SEM was calculated for each residue.
General phylogenetic analysis
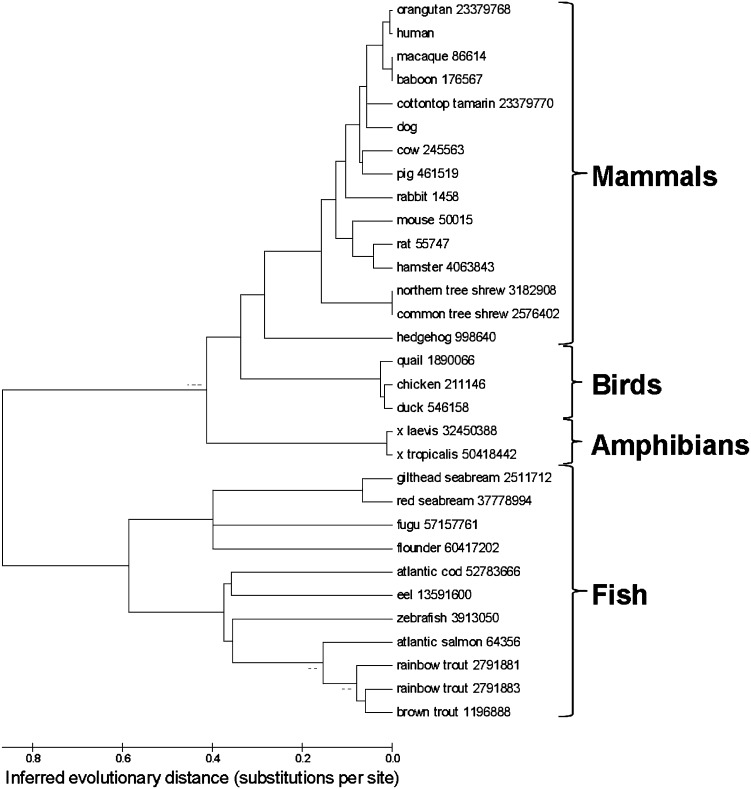
Figure 1 shows an unrooted phylogenetic tree of apoA-I sequence orthologs from 31 species of fish, amphibians, birds, and mammals (for sequence alignments of individual orthologs, see Fig. 3). Unrooted phylogenetic trees visualize the level of mutual similarity among homologous sequences without direct referral to chronological relationships among members of the set. Note the separation of the two largest branches of the sequences, fish from nonfish.
Fig. 1.
An unrooted phylogenetic tree shows apoA-I sequence orthologs from 31 species of fish, amphibians, birds, and mammals. Version numbers used are shown next to species names.
CORRELATION OF ALIGNMENT RESULTS WITH CURRENT STRUCTURAL AND FUNCTIONAL PARADIGMS
WHEEL motif of apoA-I
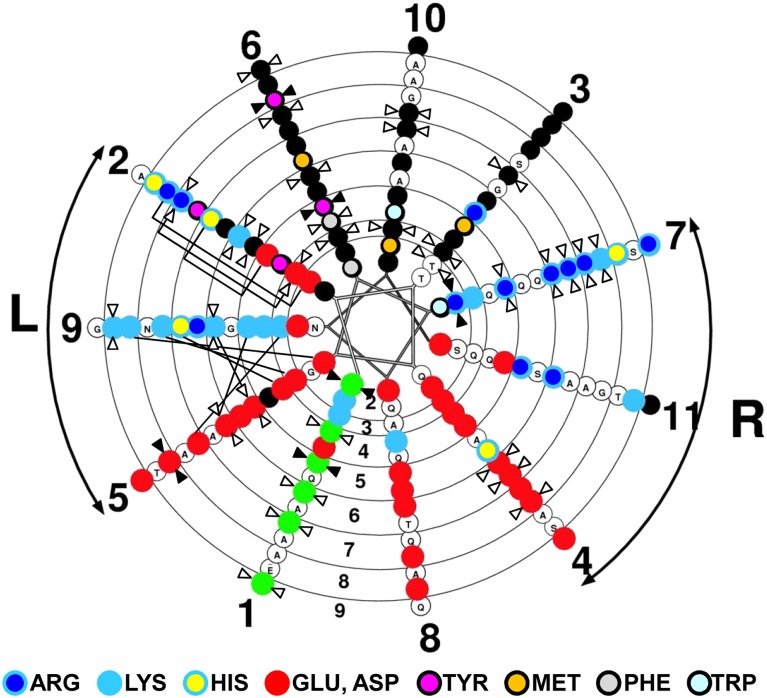
In our double-belt model for discoidal HDL (19), apoA-I monomers form a curved, flat amphipathic α helical ring with 11/3 (approximately 3.67) residues per turn, termed an α11/3 helix, which has since been reported in other lipid-associating proteins such as α-synuclein (53, 54), so that the hydrophobic surface faces inward toward the lipid disc. Figure 2 shows a continuous α11/3 helical wheel diagram of tandem helixes 2–9 of apoA-I. Assigning the six Pro residues to α11/3 helical wheel position 1 (wp1), wp6 is occupied entirely; wp10 and wp3 are occupied mostly by hydrophobic residues; wp9 and wp7 are occupied mostly by positively charged residues; and wp5, wp8, and wp4 are occupied mostly by negatively charged residues, the class A amphipathic α helix pattern (12). Only helical wp2 is equally divided between positively and negatively charged residues. We have suggested that the antiparallel helical rings are held together by 20 interhelical salt bridges (19, 55), 6 bridges between solvent-shielded α11/3 helical wp2-wp2 and 14 bridges between the more-solvent-exposed α11/3 helical wp5–wp9 and wp9–wp5 (Fig. 2, connector lines).
Fig. 2.
Conservation of helical repeats and wheel positions (for helixes 2–9) in apoA-I. Helical wheel analysis is shown of tandem amphipathic α helices 1–10 (residues 44–241) of human apoA-I, oriented with the N-terminal up, of tandem helices 2–9 of apoA-I plotted with a pitch of 11/3 residues/turn using the WHEEL program. Basic residues are represented by sky-blue circles with dark blue centers (Arg), sky-blue centers (Lys), and yellow centers (His); acidic residues are represented by solid red circles; hydrophobic residues are represented by solid black circles with magenta centers (Tyr), gold centers (Met), gray centers (Phe), and light-blue centers (Trp); and Pro is represented by solid green circles. Left and right docking interfaces are shown by arcs labeled L and R, respectively. Completely conserved residues are indicated by closed double arrowheads and largely conserved residues by open double arrowheads. Solvent-accessible salt bridges in human apoA-I are denoted by straight lines between the involved basic and acidic residues in wp9 and wp5, respectively, while the solvent-inaccessible salt bridges are shown by brackets linking the involved basic and acidic residues in wp2.
Completely conserved residues
Figure 3 shows alignments and conservation of sequences of 31 individual orthologs. There are eight sequence positions that are conserved in all 31 of the orthologs (Fig. 3, open arrowheads).
Of the 10 Pro residues in human apoA-I, 3 residues are conserved in all 31 of the orthologs (P66, P121, and P220). Of the 7 Tyr residues in human apoA-I, 3 residues are conserved in all 31 of the orthologs (Y18, Y115, and Y192). The remaining two fully conserved residues are basic (R83) and acidic (E191).
While Pro residues almost certainly have a predominantly structural role in apoA-I, that is less likely to be true for Tyr. Thus, full conservation of 43% of the Tyr residues in apoA-I is highly significant and suggests a major functional role for this amino acid residue. Two of these conserved Tyr residues are located in the lipid-associating domain, and one is located in the N-terminal domain.
Residue Y18 in helix G2 is predicted by MD simulations in the lipid-associated dimeric form of apoA-I and by hydrogen exchange-mass spectrometry in the monomeric lipid-free form (56) to be in a highly helical domain (Fig. 4). Residue Y18 has been shown by Tanaka et al. (57) to be important for assembly of HDL complexes through the interactions of apoA-I with phospholipid, “the α helix around Tyr18 [conceals] a potential lipid-binding region in the N-terminal domain, which [is] exposed by the disruption of the helix-bundle structure.” Results of that study indicate that Y18 in the α helical segment of the N terminus of apoA-I modulates lipid interactions of the lipid-free structure in concert with the C-terminal domain (58).
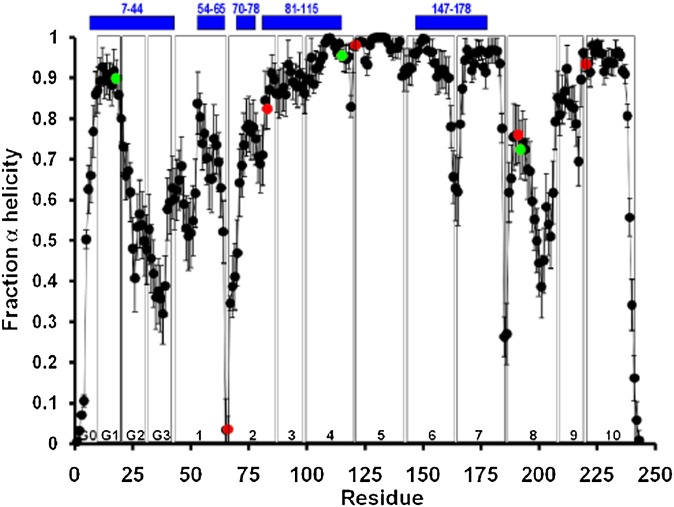
Fig. 4.
Locations of the eight completely conserved residues relative to the predicted α helicity for dimeric lipid-associated and monomeric lipid-free apoA-I are shown (56). Average local (per residue) changes in lipid-associated fractional α helicity (means ± 1 SEM) during the last 10 ns of the MDSA protocol are plotted for 16 160:24:2 particle replicas. Green data points represent positions of the three completely conserved Tyr residues, Y18, Y115, and Y192. Red data points represent positions of the other five completely conserved residues, P66, R83, P121, E191, and P220. The positions of five helical domains predicted for monomeric lipid-free apoA-I by hydrogen exchange-mass spectrometry (56) are represented as blue bars at the top of the figure. Vertical boxes in the figure denote the positions of each of the 10 helical and N-terminal G* repeats.
Residue Y115 in helix 4b, wp3 (Fig. 2), also predicted to be in a highly helical domain in the R2-2 lipid-associated form of apoA-I (Fig. 4), is completely conserved from fish to mammals. Furthermore, Y115 and R116 (helix 4b, wp4) are conserved as a pair in all but 3 of the 31 species and form, after the E191-Y192 pair, the second most conserved sequence pair in apoA-I (Fig. 3B). Furthermore, the completely conserved residues R83 and Y115 bracket the longest helical domain, residues 81–115 (Fig. 4), predicted for monomeric lipid-free apoA-I by hydrogen exchange-mass spectrometry (56). Thus, R83, Y115, and R166 could play a major role in the folding of monomeric apoA-I, perhaps required for proper membrane insertion and HDL assembly.
Residues Y192 (helix 8a, wp6) and E191 (helix 8a, wp5), predicted to be in an apparent loop-helix-loop domain in the lipid-associated form of apoA-I (Fig. 4), form a 100% conserved sequence motif, suggesting strongly that this site in apoA-I has an important biological function. It is therefore of interest that Shao et al. (59) suggest that Y192 acts as a preferred atherogenic oxidation target in apoA-I.
The final fully conserved residue, R83 (helix 2b, wp7), at the edge of a predicted helix-nonhelix junction in both lipid-associated dimers and lipid-free monomers (Fig. 4), has no known or suggested function. Perhaps it is important in HDL particle assembly, or it might be important in interactions with antioxidant proteins, such as PON-1 (see Discussion).
As shown in Fig. 2, the 22 mer/11 mer tandem repeats are often punctuated by Pro residues in the α11/3 wp1a position. In humans, three Pro residues are also located in the 10 residues of the N-terminal G0 domain of mature apoA-I. As shown in Fig. 3, these three residues are not well conserved between fish and mammals. In human apoA-I, Pro occurs at wp1 in helixes 2a, 4a, 5a, 6a, 7a, 9a, and 10a; in addition to helix 9, the other 11 mer repeat in the lipid-associating domain is helix 3, classified as helix 3b (see Fig. 6). These seven Pro residues are well conserved from fish to mammals; P66 (helix 2), P121 (helix 5), and P220 (helix 10) are conserved in 100% of the species shown in Fig. 3. The most variable Pro in the lipid-associating domain is at wp1 of helix 8a; in humans and many mammals, this Pro, located at human sequence position 187, is replaced by a flexible G185-G186-A187 loop and in several mammals by G184-G185-G186-A187. This variability suggests that structural flexibility is important for the helix7–helix 8 junction. As noted later, in MD simulations of lipid-associated apoA-I (52), helix 8 is the least stable of the last 10 helical repeats in human apoA-I (Fig. 4).
Fig. 6.
Comparison of differences between mammalian and fish sequences for each of the tandem amphipathic helical repeats 2–9 in the lipid-associating domain (residues 65–220 for humans) is shown. Top and bottom scales indicate conservation in mammals and fish, respectively. If the residue is completely conserved across all species, no residue is indicated on the graph (e.g., see residue 66). If there are differences between the species, the height of the letter indicates conservation of given residues within mammals and within fish (e.g., residue 77 is 100% Trp in mammals but is a mixture of residue types in fish). Black diamonds, 100% conserved residues; yellow diamonds, comments; red diamonds, conserved residue clusters across both fish and mammals.
Conservation of residues in the N-terminal (globular) domain
G0 domain.
Based upon its degree of conservation, the N-terminal G0 domain appears to be unusually important in apoA-I structure-function relationships. All apoA-I orthologs, except for those of eel, begin their N termini (G0-1) with Asp. Mammals, amphibians, and birds begin G0 with two N-terminal acidic residues, while fish (except eel) begin G0 with a single Asp residue. As noted earlier, G0 domains in all orthologs (except hedgehog) contain 1–3 Pro residues; in all but two orthologs, the third residue is Pro. Sequence position G0-6 relative to that of human apoA-I in all but one ortholog is either Ser or Thr, perhaps representing a site of phosphorylation. Sequence position G0-8 is almost always a hydrophobic residue (either Leu or Trp), G0-9 is generally an acidic residue, and G0-10 is generally basic, His in fish and Arg in mammals.
G1 domain.
Residue position G1-1 (residue 11) is almost always hydrophobic, while G1-2 (residue 12) is always basic, and G1-3 (residue 13) is always acidic (usually Asp) in nonfish. Residue G1-8 (Y18), as noted earlier, is conserved in all 31 orthologs. Finally, G1-9 (residue 19) is always hydrophobic.
G2 domain.
Residue position G2-1 (residue 22) is always hydrophobic (generally Val) or, in two cases, Ala, while residue position G2-2 (residue 23) is always basic and almost always Lys. G2-5 (residue 26) is neutral and small, generally Gly, while residue position G2-6 (residue 27) is always basic in nonfish and all but one fish.
G3 domain.
Residue position G3-1 (residue 33) is always hydrophobic, approximately equally Leu and Phe. Residue position G3-2 (residue 34) is always acidic, Asp in fish, and, except for hedgehog, always Glu in mammals, amphibians, and birds; while G3-4 (residue 36) is completely conserved in all nonfish but is acidic in most fish.
Based upon MD simulations, we propose that the two N-terminal domains of apoA-I are “sticky” (M. K. Jones, et al., unpublished observations): that is, they tend to be locked together, sort of like molecular Velcro, in the discoidal double-belt model. Figure 5 illustrates one example of this spatial arrangement in a 160:24:2 particle simulated by MDSA. An illustration of this spatial arrangement in the average ensemble of four MDSAs was published previously (52). This “stickiness” is derived at least partially through a complex series of salt bridges, until, at a critical threshold of particle size, the interacting domains separate to expose extensive hydrocarbon regions of the phospholipid bilayer to solvent, putatively resulting in particle growth by fusion. Relevant to the discussion of the G0, G1, and G2 domains above, we have noted the presence of a variable cluster of nine interhelical salt bridges among the G0, G1, and G2 domains: D1-R10, D1-K12, E2-R10, E2-K12, D1-K23, D1-R27, D9-R10, K12-D13, and R10-D13. The first four salt bridge pairs are those most commonly seen. Importantly, all seven of the charged residues that form these interhelical salt bridges are generally well conserved.
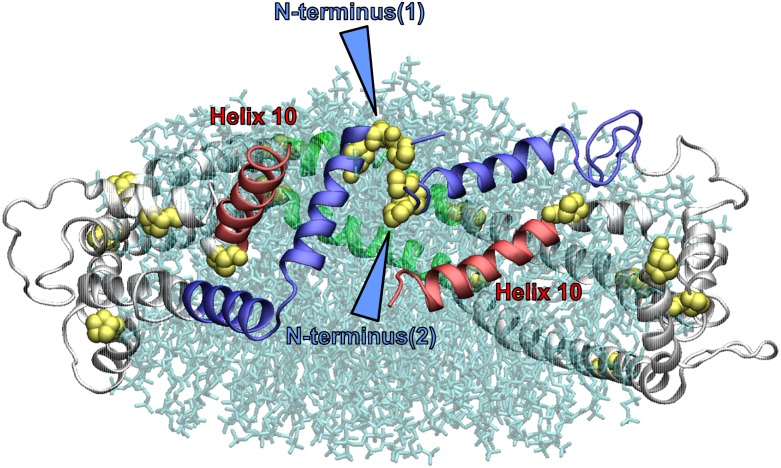
Fig. 5.
A molecular representation of an example of the spatial arrangement of the double belt resulting from the interlocking of the two N-terminal (“sticky”) domains of a 160:24:2 (R2-2) particle after MDSA. The protein is in a ribbons mode. The N-terminal domains (residues 1–43) are blue, the helix 10 domains are red, and the helix 5 domains are green. Blue arrowheads indicate the association of the Pro-rich N termini. Lipid molecules are in transparent licorice mode.
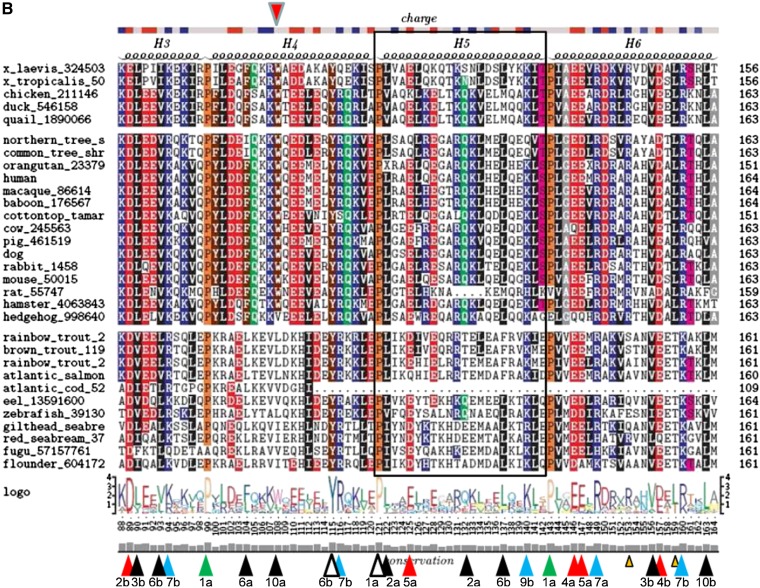
Well-conserved residues in the lipid-associating domain (helixes 1ndash10)
A total of 13 (Fig. 3) hydrophobic residues are well conserved in the lipid-associating domain. These conserved hydrophobic residues are located in hydrophobic wheel positions 2, 3, 6, and 10; five residues are at wp6, four are at wp10, four are at wp3, and one is at wp2 (Fig. 2).
There are 10 well-conserved basic residues (Fig. 3) in the lipid-associating domain. Six residues are located in wp7 (Fig. 2, right docking interface), and of these, it has been suggested that R149 and R160 in helix 6a and 6b (wp7) play a role as recognition sites for the binding of the LCAT enzyme (60). Two well-conserved basic residues are located in wp9 (a superficial salt bridge position), and two are in wp2 (a solvent-shielded salt bridge position).
Eight acidic residues (Fig. 3, red arrowheads) are well conserved in the lipid-associating domain, three in the wp4 position, three in the wp5 (superficial salt bridge) position, and one in the solvent-shielded salt bridge in position wp2 (Fig. 2), and one in helix 1 at position 8a (we do not consider helix 1 to be an a11/3 helix). Other than Pro, there is only one sequence position in the lipid-associating domain where neutral residues (Fig. 3, green arrowheads) are well conserved: T78 (helix 2b, wp3). This Thr residue is conserved in all species, except one, where it has been mutated to an Ala residue.
Met residues are generally not well conserved in apoA-I; M86 is conserved in most mammals but not in other animal classes.
Generally, the conserved residues of the lipid-associating domain are concentrated in its central domain, from the last half of helix 2 to the first portion of helix 8 (Figs. 3 and 4). This domain is both implicated in LCAT activation and the location of the solvent-shielded interhelical salt bridges (Fig. 2).
Additional conserved motifs in the lipid-associating domain
The motif in helix 1a, N49-W50 (wp6 and wp7) is completely conserved in mammals but not at all in fish, birds, or amphibians (Fig. 3, small gold arrowheads,). Similarly, the motif in helix 2a, E70-F71-W72 (wp5, wp6 and wp7), is conserved in mammals but not in the other classes. Residues K77 and L159 in helix 2b (wp1) and helix 6b (wp6), respectively, are conserved in mammals, birds, and amphibians; in fish, K77 and L159 are replaced with an acidic residue and Thr, respectively. Finally, R153 in helix 6a (wp11) is completely conserved in mammals, birds, and amphibians but not in fish. These differences, especially those between fish and mammals, suggest certain significant local structure-function relationship differences in the apoA-I sequences between the two classes. For example, R153 has been suggested to be important for LCAT association with lipid-bound apoA-I (60), and the lack of conservation in fish suggests the possibility of divergent evolutionary changes in LCAT structure between fish and land animals.
Location of conserved residues on the alpha11/3 helical wheel plot
Conserved residues, completely conserved residues, and well-conserved residues are shown in Fig. 2. The most conserved wheel positions are wp6, wp7, and wp1. Three of the seven conserved residues in the always-hydrophobic wp6 are aromatics, and two, the Tyr residues, are completely conserved. Five of the six conserved residues in wp7 are Arg, and one of these is completely conserved. All of the conserved residues in wp1 are Pro. The next most conserved wheel position is wp2, with four conserved residues. The remainder of the wheel positions have either three (wp5, wp10, wp3, and wp4) or two (wp9) conserved residues, except for wp8, which has none.
The pattern of conservation of amino acid residues shown in Fig. 2 strongly supports our model for apoA-I in which the lipid-associating domain forms an α11/3 helix (3 helical turns in 11 residues), rather than the classic Pauling-Corey-Branson α helix of 5 turns in 18 residues (61). Parenthetically, in the initial Pauling-Corey-Branson publication (61), 3 turns per 11 residues was mentioned as one possible solution to the protein helix problem.
Comparison of similarities and differences between apoA-I sequences of fish and mammals in the lipid-associating domain
Figure 6 shows a comparison of the differences between mammalian and fish sequences for each of the tandem amphipathic helical repeats 2–9 in the lipid-associating domain (residues 65–220 for humans). In addition to the completely conserved residues mentioned earlier, there is one charged motif of six residues in helix 7 that stands out as being well conserved in both fish and mammals (Fig. 6; see also Fig. 3C), namely, R177-L178-E179-E180-L181-K/R182 helix 7b (wp2–wp7, respectively). Residue R177 forms a solvent-shielded salt bridge with E89 in all PL-rich reconstituted particle sizes (R2-2, R2-1, and R2-0) subjected to MDSA simulations (52), and both are well conserved. A number of publications have suggested that the central domain of the apoA-I double-belt, helixes 3–7, is critical for LCAT activation; perhaps the conserved 177–182 motif is involved.
In addition to similarities, there are a number of dramatic differences between the two classes (Fig. 6). In helix 2a, residue 72 (wp7, is usually a basic residue in most helical repeats) is always a Trp in mammals but is quite variable in fish (see Fig. 2). At the junction of helix 2a and 2b, the E76-K77-E78 cluster (wp11-wp1-wp2, respectively) is extremely well conserved in mammals and poorly conserved in fish. In helix 4a, Y100, L101, and F104 (wp2, wp3, wp6, respectively) and helix 4b (wp2) are well conserved in mammals, and, while these residues are conserved in fish, they are conserved as different residues. In helix 4a mammals, N102 and W108 (wp3 and wp10) are also well conserved, but in fish, residue 102 is not well conserved, and residue 108, while hydrophobic, is not aromatic.
In helixes 5a and 5b, A130 and K133 (wp10 and wp2, respectively) are well conserved in mammals but are basic and acidic, respectively, in fish. On the basis of MD simulations, we have suggested that these two residues are critical to the formation of an amphipathic presentation tunnel located between the antiparallel helix 5-helix 5 pair of the double belt, for migration of hydrophobic acyl chains and amphipathic UC from the bilayer to the phospholipase A2-like and esterification active sites of LCAT, respectively (62). This model is similar to one proposed by Martin et al. (26) based upon structure studies suggesting a looped belt located between residues 133 and 146, centered at residue 129. The major differences in the two models are: i) our presentation tunnel is centered at residue 133 rather than 139, and ii) the looped-belt model requires a change in the registration of the antiparallel helix 5-helix 5 central domain. Because MD simulations suggest that K133 is a key residue in stabilizing the presentation tunnel (62), it is entirely possible that any mutation at this position that incorporates a spin label or a fluorescent molecule would significantly change the dynamics of the presentation tunnel structure.
In helix 6b, residue 155 (wp2, a solvent-shielded salt bridge) is well conserved as His in mammals and completely conserved as Asp in fish. In helix 6b, residue 159 (wp6, generally a hydrophobic residue) is completely conserved as Leu in mammals and as Thr in fish. Finally, in helix 7a, residue 166 (wp2) is conserved as Tyr in all but one mammal but is uniformly a nonaromatic hydrophobic residue in fish.
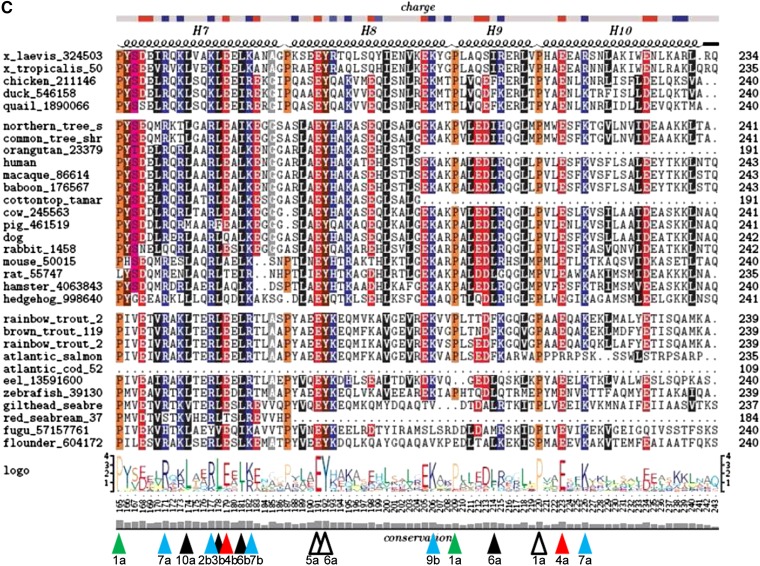
Finally, none of the 11 species of fish has even a single Trp residue; Trp first appears in apoA-I at position W108 in land animals. Four Trp residues appear in humans, W7 (G0), W50 (H1), W72 (H2, wp7), and W108 (H4, wp10). The first three Trp residues (Fig. 3A) are completely conserved in mammals but are present in no other classes of animals. The fourth Trp, W108, is conserved in all mammals but hedgehog and in all other land animals (Fig. 3B).
Comparison of consensus sequences of tandem amphipathic helical repeats in lipid-associating domains of apoA-I between fish and mammals
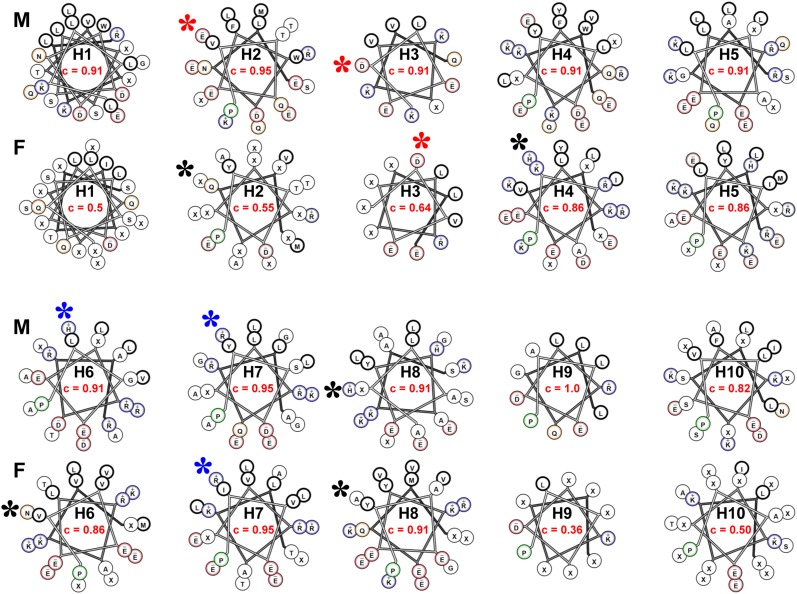
Figure 7 shows helical wheel diagrams of consensus sequences of lipid-associating domain helical repeats 1–10 of fish and mammals. Sequence positions with less than 50% identity are denoted by X. In the figure, the fraction of residues in each helical wheel that is at least 50% conserved is shown. With that as a measure of helical repeat conservation, helix 9 is the most conserved, and helix 10 is the least conserved consensus tandem repeat in mammals. Using a cutoff higher than 50% identity, helix 10 is also the least conserved helical repeat in mammals (data not shown). In fish, helix 7 is the most conserved, and helix 9 is the least conserved. As helix 7 is well conserved and helix 10 is poorly conserved in both mammals and fish, these appear to represent the most conserved and least conserved consensus helixes, respectively, across the phylogenetic tree.
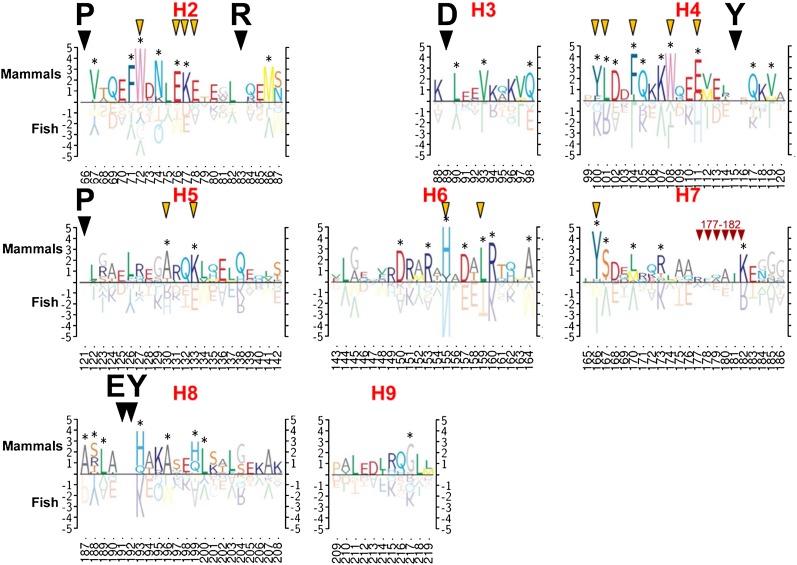
Fig. 7.
Helical wheel diagrams of consensus sequences of lipid-associating domain helical repeats 1–10 of fish (F) and mammals (M) are shown. Based upon our previous analysis (51), the helical wheel for helix 1 (residues 44–65) is displayed as an idealized α18/5 helix, while the remainder are displayed as α11/3 helixes. The helix number is shown in the center of each wheel (e.g., H1, H2, etc.). The fraction of residues in each helical wheel that is at least 50% conserved (c) is shown in red. The position of the residues involved in the putative solvent-inaccessible salt bridges in the human sequence are shown as stars: red stars denote acidic residues that are conserved, blue stars denote basic residues that are conserved, and black stars denote residues that are not conserved.
In fish, central helixes 4–8 are significantly more conserved than terminal helixes 1–3 and 9–10. Except for helix 10, this type of dichotomy between central and terminal helixes is not discernible in mammals, perhaps because of the greater phylogenic distances between different species of fish compared with that of mammals.
The six residues involved in forming the three pairs of solvent-inaccessible salt bridges in human apoA-I are E78 (helix 2)-R188 (helix 8), D89 (helix 3)-R177 (helix 7), and E111 (helix 4)-H155 (helix 6). Five of the putative solvent-inaccessible salt bridge residues appear in mammal consensus wheels; the lack of conservation of the sixth residue, R188, in mammals is compatible with our observation that the most terminal pair of solvent-inaccessible salt bridges, E78 (helix 2)-R188 (helix 8), unlike the two more central ones, are easily broken (and are thus unstable) during temperature-jump MD simulations (52, 55). In the fish consensus wheels, only the putative solvent-inaccessible salt bridge residues D89 (helix 3)-R177 (helix 7) are conserved; the other four putative solvent-inaccessible salt bridge residues are not conserved.
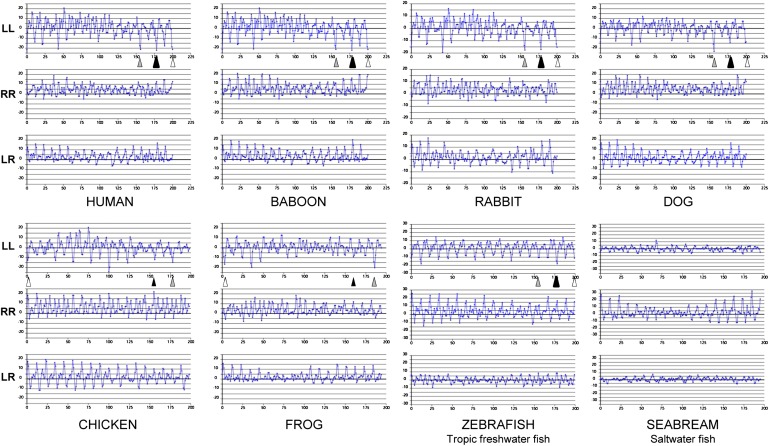
The conservation of one of the three pairs of putative solvent-inaccessible salt bridge residues in fish suggests that the LL5/5 registration of apoA-I dimers might persist in fish, a possibility briefly discussed in our original detailed double-belt model for discoidal HDL (19). To further test the phylogenetic persistence of the LL5/5 registration of apoA-I dimers from fish to mammals, we performed analyses, using ALIGN software (19), of apoA-I sequences for four mammals (human, baboon, rabbit, and dog), one bird (chicken), one amphibian (frog), and two fish species (zebrafish and seabream); results are shown in Fig. 8. While there are differences in seven of the eight species in the relatively weighted salt bridge scores of the three registrations emphasized previously, LL5/4, LL5/5, and LL5/6, each of the seven, including zebrafish, a tropical freshwater fish, has a pronounced 11/22 mer periodicity. However, seabream, a saltwater fish, shows no obvious 11/22 mer periodicity at all along the LL docking interface, suggesting that interhelical salt bridge formation between left docking interfaces of apoA-I monomers may be a recent evolutionary adaptation that occurred since freshwater fish split off from saltwater fish.
Fig. 8.
ALIGN analyses (19, 51) are shown of apoA-I sequences for four mammals (human, baboon, rabbit, and dog), one bird (chicken), one amphibian (frog), and two fish (the tropical freshwater zebrafish and the saltwater fish seabream). Positions of the LL5/4, LL5/5, and LL5/6 registries are indicated by gray, black, and open arrowheads, respectively.
Structure and function roles of completely conserved Pro and Tyr residues
The detailed sequence alignment and conservation analyses of apoA-I amino acid described here provide a phylogenetic window into the structure, assembly, function, and dynamics of apoA-I. These results are useful as stand-alone information but are more valuable if correlated with current concepts of apoA-I structure and function. The general overall conservation of Pro residue punctuations between most of the 11/22 mer tandem helical repeats is not surprising as Pro almost certainly plays a mostly structural role in apoA-I.
The complete conservation of three of seven Tyr residues in apoA-I, however, is functionally more significant. It has been suggested that certain Tyr residues, in particular Y166 (34) and Y192 (59), are preferred targets for site-specific oxidative modification within atheroma. However, this concept alone provides little in the way of biological insight into the function of Tyr residues in apoA-I.
Although it is a somewhat controversial idea, Tyr residues have been suggested to act as antioxidants inside lipid bilayers and protect cells from oxidative destruction by interfering with lipid-oxidizing free radical chain reactions (63) through conversion to relatively stable and nonreactive phenoxyl radicals (64, 65). The re-reduction of oxidized, radicalized membrane phase antioxidants, such as vitamin E, is known to require aqueous reducing components, such as vitamin C (66, 67). As in the amphipathic helical domains of apoA-I in general, Tyr residues have a strong tendency to partition into the boundary between the polar and nonpolar regions of bilayers. Considering the extensive dynamics of lipid bilayers and apoA-I in HDL particles (52, 55, 62), Tyr seems ideally suited to form a partition at the polar and nonpolar interface of the lipid bilayer to scavenge lipid peroxyl radicals and in the aqueous phase to be re-reduced (65). With this in mind, the location of the completely conserved residues Y115 and Y192 in the most-solvent-shielded α11/3 wheel position, wp6 (Fig. 2), is remarkable.
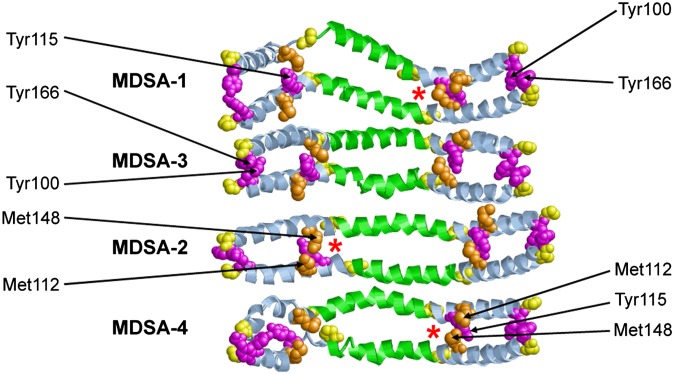
Modeling and MD simulations (68, 69) predict that the completely conserved residue Y115, in its lipid-associated form, is in a highly helical domain (Fig. 4). M112 (helix 4b, wp3), found only in primates and pigs, and M148 (helix 6a, wp6, Fig. 2), found only in humans and a few fish, interact with Tyr115 during MD simulations of the LL5/5 rotamer (Fig. 9); the driving force for this interaction is not clear. In no species, other than human, for which apoA-I sequences are available does this intriguing microenvironment occur. Oxidation of Met residues introduces hydrophilicity to the nonpolar face of the amphipathic helical structure of human apoA-I, altering its lipid affinity and local secondary structure (70). Thus, the presence of M112 and M148 in a microenvironment unique to humans, combined with the possibility that they are easily oxidized, may have a deleterious effect on the local structural environment and function (perhaps as an antioxidant?) of what should otherwise be a solvent-inaccessible residue Y115 in humans.
Fig. 9.
Ribbon diagrams of the helix4-helix 6 domain viewed from the lipid interior after four MDSAs of PL-rich apoA-I/HDL particles (52). Helix 5 is in green, proline residues are in yellow (space filling), Tyr residues are in magenta (space filling), and Met residues are in gold (space filling).
We recently reported, based upon MD simulations, that helix 8 is the least stable tandem amphipathic helix in the double-belt structure of the lipid-associating domain (residues 44–243) of apoA-I on discoidal HDL (52). In the majority of our simulations, the region of helix 8 containing the conserved E191-Y192 motif formed a solvent-exposed loop-helix-loop on the edge of the bilayer disc [Fig. 4 and (52)]. Thus, the fully conserved E191-Y192 motif appears well situated to perform an antioxidant function within HDL.
Interestingly, Y100 and Y166 (both at wp2 in helix 4a and 6a, respectively) create a unique microenvironment in most mammals, being positioned to form π-π ring stacks (Fig. 9), perhaps stabilizing interhelical interactions in this region. There are, however, no aromatic residues at either position in fish. As Y166 (34) has been suggested to be a preferred target for site-specific oxidative modification within atheroma, the uniqueness of the Y100/Y166 microenvironment to mammals is intriguing.
Myeloperoxidase oxidative pathway for generating dysfunctional HDL
While the functional role of Tyr in apoA-I is not clearly understood, there does exist extensive literature on the role of oxidative damage to HDL directed toward Tyr (as well as Met and Trp) residues of apoA-I by myeloperoxidase (MPO), a heme enzyme secreted by human artery wall macrophages (71).
Macrophages use reactive oxygen species to kill invading pathogens (72). One oxidative pathway involves MPO that is expressed in macrophages in human atherosclerotic lesions (73). The enzyme uses hydrogen peroxide, and the major end product is generally thought to be hypochlorous acid (HOCl), a potent antimicrobial agent.
A single tyrosine residue, the fully conserved Y192, has been identified as the major chlorination site when HOCl oxidizes apoA-I (74), and a strong linear association has been shown to exist between the extent of Y192 chlorination and the loss of ABCA1 transport activity (75). Chlorination of Y192 has been shown to be directed by the YXXK motif (74). The K in this motif, K195, is conserved in all but 2 of 14 mammals and in all 3 birds but is not conserved in fish or amphibians (Fig. 3C), suggesting that the YXXK motif arose at or before the branching of birds and mammals and after amphibians. This conservation suggests that the motif has a functional role, perhaps as an antioxidant.
Scanning spin label studies have suggested that an 18-residue random coil loop (amino acids 188–205) near the C terminus becomes α helical when the protein associates with lipid and energetically drives apoA-I's conformational adaptation to the presence of lipid (33). Our MD simulations suggest that the E191-Y192 motif when associated with lipid lies in the helical portion of a loop-helix-loop motif (Fig. 4). As Y192 is in the helical portion of this 18-residue region, its chlorination might alter the energy levels of protein remodeling to favor retention of the conformation of lipid-free apoA-I and thus inhibit ABCA1 transport activity (71). The complete conservation of the E191-Y192 motif may be related to the putative role of residues 188–205 in the assembly of lipidated apoA-I.
HOCl can also oxidize tryptophan residues in apoA-I (76, 77). However, oxidation failed to prevent an apoA-I mutant in which all four Trp residues were replaced with Phe from activating ABCA1 (78).
Met residues, which are not well conserved, are also extremely susceptible to oxidation by HOCl. A single methionine residue in apoA-I, M148, resides in the general vicinity of the LCAT activation domain of apoA-I (H4–H6, residues 99–164). Shao et al. (79) demonstrated that oxidation of M148 to the negatively charged methionine sulfoxide produced a loss of LCAT activity in HDL. Reversing oxidation with methionine sulfoxide reductase restored the ability of HDL to activate LCAT. Discoidal HDL prepared with apoA-I containing a mutation in which Met was replaced with Leu at position 148 (M148L) was significantly resistant to inactivation by MPO. Based on these results, it was proposed that oxidation of M148 disrupts the LCAT activation domain of lipid-associated dimeric apoA-I (79) by disrupting the central loop.
We would add that M148 also appears by MD simulations to interact strongly with the completely conserved Y115 residue. As noted later, Y115 appears to form interhelical cation-π orbital interactions with R151, thus helping to stabilize the edge of the putative presentation tunnel for LCAT activation. Stabilization of the tunnel edge would almost certainly be seriously affected by oxidation of Met to create a negatively changed residue.
Role of the prevalence of Arg residues on the right docking interface
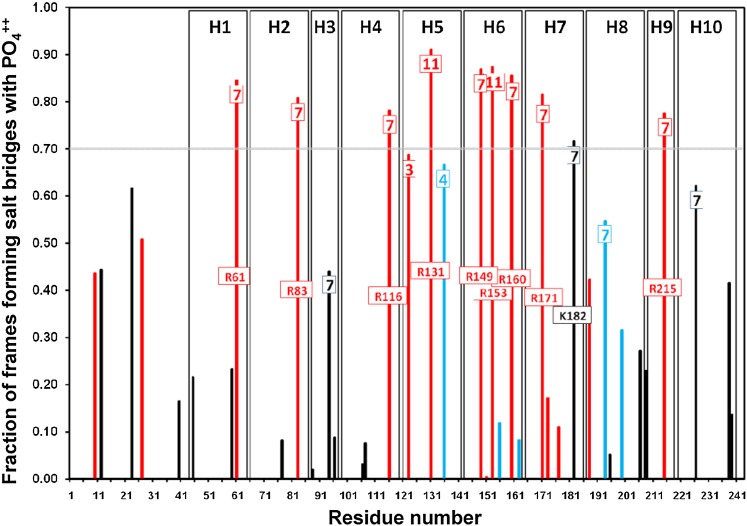
Figure 10 shows a histogram plot of the fraction of frames involved in salt bridge formation, between all basic residues and POPC during the last 10 ns of MD simulations of 16 160:24:2 particle replicas. All Arg residues located in wp7 and two of the three located in wp11 on the right docking interface (Fig. 2) are involved in salt bridge formation with the negatively charged PO4− moieties of the annular POPC molecules over 70% of the time. This is one possible explanation for why five of the six Arg residues in wp7 are well conserved (Fig. 2).
Fig. 10.
Probability of formation of salt bridges between basic residues and the PO4− moieties of POPC in 160:24:2 particles relative to wheel position and type of residue (Arg, Lys, His). The histogram plot shows the fraction of frames involved in salt bridge formation between each basic residue position and POPC during the last 10 ns of MDSAs of 16 160:24:2 particles. Arg, red; Lys, black; His, blue. Wheel positions, wp, for each basic residue in tandem helixes 1–10 located near the right docking interface (wp7, wp11, wp3, and wp4) are denoted in a box near the top of each histogram bar. The tandem helix positions are indicted by vertical rectangles. The 0.70 fraction position is denoted by a gray horizontal line. The residue identity for each basic residue whose fraction of salt bridge formation falls above 0.70 is indicated in a box near the middle of each relevant histogram bar.
One additional possible function of Arg in the right docking interface in general and in wp7 in particular is based on recent research from this laboratory showing that Arg-containing amphipathic helical peptides sequester oxidized lipids more avidly than corresponding Lys-containing peptides (80), suggesting that Arg may associate more avidly with oxidized phospholipids than Lys (see Fig. 12E for the proposed mechanism). Since the other major HDL protein, apoA-II, contains no Arg residues, perhaps the presence of Arg residues in the right docking interface of apoA-I partially explains why apoA-I-containing particles are more anti-atherogenic than apoA-II-containing particles (81).
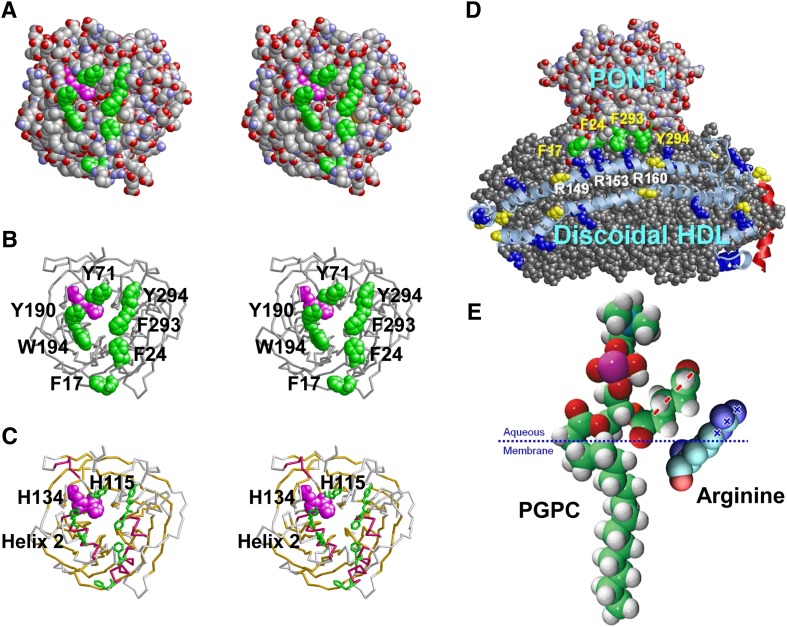
Fig. 12.
Aromatic residues may play a role in driving the binding of HDL to PON-1 (1vd4) and apoA-I to oxidized lipids through Arg-π interactions. All images were produced with RasMol software. A: Space-filling stereo view down the tunnel to the active site (H115/H143, magenta). Aromatic residues lining the opening of the tunnel are in green. B: Backbone stereo model showing the active site H115/H143 in space-filling mode and tunnel-lining aromatics in stick mode. C: Backbone stereo model showing the active site H115/H143 and tunnel-lining aromatics. β Propeller sheets are in gold and α helical domains are in red. Helix 2 is known to be part of the HDL binding site. D: Diagrammatic model of possible binding of PON-1 to discoidal HDL via Arg (blue space-filling)-π (green space-filling) interactions. Residue numbers of aromatics in PON-1 are indicated in yellow, and residue numbers of Arg in apoA-I are indicated in white. E: Model of possible binding of oxidized lipid to Arg clusters in apoA-I in which the guanidine group of Arg creates delocalized planar positive charges (+++) that interact with the SN-2 chain of oxidized PC that forms planar delocalized negative charges (- - - -). The palmitoylglutarylphosphotidylcholine (PGPC) model is a modification of the structure from the Avanti Polar Lipid catalog.
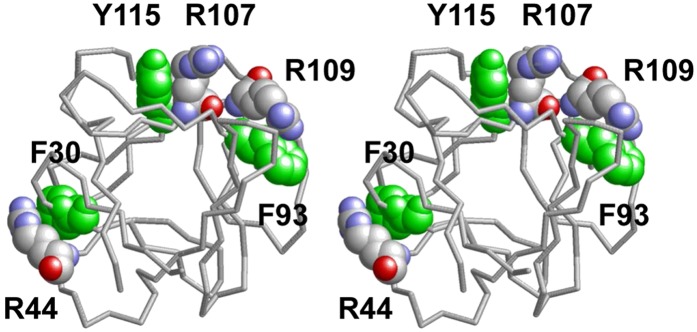
In a related possibility, it is known that the flat face of an aromatic ring has a partial negative charge due to π orbitals. It has been suggested that cationic amino acid residues, Arg or Lys, which are within 6.0 Å of the face of an aromatic ring (Phe, Tyr, or Trp), may engage in polar interactions called cation-π orbital interactions by centering themselves over the faces of aromatic rings; Arg participates in nearly twice as many cation-π interactions as Lys does; and the numbers of cation-π interactions involving Trp, Tyr, and Phe are roughly similar (82). For three examples of Arg-π interactions in a single protein, basic fibroblast growth factor, see Fig. 11. In addition, because of the conjugation between the double bond and the nitrogen lone pairs in Arg, the positive charge is delocalized, enabling Arg to form multiple H bonds. Thus, despite interactions with PO4− and/or oxidized lipids (Fig. 12E), Arg would be available for interactions with certain critical proteins, such as antioxidants.
Fig. 11.
Three examples of Arg cation-π interactions with aromatic residues in basic fibroblast growth factor 1. This stereo image of PDB coordinates taken from the Protein Database was produced with RasMol. Aromatic residues (green) and Arg residues (CPK color) forming cation-π pairs are shown in space-filling mode.
Based upon X-rays of the crystal structure of PON-1 (83), not only is the HDL binding domain of PON-1 enriched in π-electron-containing aromatic residue clusters (Fig. 12) but even more remarkably, a complete ring of aromatic residues encircles the opening of the cleft that leads to the active site of PON-1 (Fig. 12A–C). Binding of PON-1 to HDL, per se, appears to happen through the signal sequence that is still attached to PON-1 (84). It is therefore tempting to speculate that the Arg residues in wp7 on the right docking interface orient PON-1 to an oxidized lipid binding site of apoA-I by interacting with π-electron-rich clusters of the HDL binding domains of this antioxidant enzyme (Fig. 12D). One potential driving force in the creation of an oxidized lipid binding site on apoA-I is the planarity of the quanidino group of Arg that causes delocalized planar positive charges to interact with the planar delocalized negative charges of the SN-2 chain of oxidized PC (Fig. 12E).
Possible role of cation-pi orbital interactions within the double-belt structure
Another possibility involves considering whether cation-π orbital interactions could play a role in the structure of apoA-I itself. As no detailed structural information exists for lipid-free monomeric apoA-I, we will confine our discussion to the lipid-associated form.
Inspection of the four MDSAs of the 160:24:2 double-belt particle reveals that of the three completely conserved Tyr residues, Y115 and Y192 frequently form cation-π orbital interactions with basic residues within apoA-I itself.
Y115, in the majority of the eight possible pairings, forms interhelical cation-π orbital interactions with R151, suggesting that this pairing might be important in stabilizing the double belt. However, unlike Y115, R151 is only moderately well conserved (Fig. 3).
Residue Y192, on the other hand, forms intramolecular cation-π orbital interactions in the majority of the eight possible pairings with R188 (an n+4 intrahelical pairing). Perhaps this intramolecular interaction might be related to the tendency of this region of helix 8 to form a helical domain surrounded by coils (Fig. 4). Interestingly, however, R188 is conserved only in primates (and one frog species); this residue position is completely conserved as Tyr in fish. One could speculate that the uniqueness of the Y192-R188 intramolecular pairing in humans might play a role in the tendency for Y192 to act as a preferred atherogenic oxidation target in apoA-I (59).
Examination of the four Trp residues in human apoA-I, W7 (G0), W50 (H1), W72 (H2, wp7), and W108 (H4, wp10), in the four MDSAs of the 160:24:2 double-belt particle revealed that only W7 forms significant protein–protein cation-π interactions, mostly on an intramolecular level, with residue K12. Perhaps this n+4 interaction stabilizes the α helicity that begins at W7 in both lipid-associated and lipid-free apoA-I (Fig. 4).
Finally, examination of the six Phe residues in human apoA-I, F33 (G3), F57 (H1), F71 (H2, wp6), F104 (H4, wp6), and F225 and F229 (both H10), revealed that F71 forms several interhelical cation-π interactions with R188, K195, and H199, while F104 forms a few intramolecular cation-π interactions with K107. Interestingly, these two are the only Phe residues conserved in apoA-I: F71 is completely conserved in mammals and in a few fish, while F104 is almost completely conserved in land animals (Fig. 3A and B) but not in fish.
Acknowledgments
The authors thank University of Alabama Information Technology and Department of Mechanical Engineering for use of the IBM Blue Gene/L rack and the commodity cluster Cheaha that they jointly maintain.
Footnotes
Abbreviations:
- apo
- apolipoprotein
- MD
- molecular dynamics
- MDSA
- molecular dynamics simulated annealing
- MEGA 4
- molecular evolutionary genetics analysis
- PGPC
- palmitoylglutarylphosphotidylcholine
- PON-1
- serum paraoxanase-1
- POPC
- palmitoyloleoyl phosphatidylcholinepalmitoyloleoyl phosphatidylcholine
- PSI-BLAST
- position-specific iterated BLAST
- R2-2, R2-1, and R2-0
- reconstituted high high-density lipoprotein particles containing 2 molecules of apoA-I, and are approximately 106 Å, 96 Å, and 78 Å in diameter, respectively
- wp
- α11/3 helical wheel position
This work was supported by National Institutes of Health Grants P01 HL-34343 (to J. P. S.) and R01 HL-090803 (to G. M. A.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Linsel-Nitschke P., Tall A. R. 2005. HDL as a target in the treatment of atherosclerotic cardiovascular disease. Nat. Rev. Drug Discov. 4: 193–205. [DOI] [PubMed] [Google Scholar]
- 2.Zlotnick A. 2004. Viruses and the physics of soft condensed matter. Proc. Natl. Acad. Sci. U S A. 101: 15549–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiflett A. M., Bishop J. R., Pahwa A., Hajduk S. L. 2005. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J. Biol. Chem. 280: 32578–32585. [DOI] [PubMed] [Google Scholar]
- 4.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson W. S., Silva R. A., Chantepie S., Lagor W. R., Chapman M. J., Kontush A. 2009. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters. Relevance to antioxidative function. Arterioscler. Thromb. Vasc. Biol. 29: 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabana V. G., Reardon C. A., Feng N., Neath S., Lukens J., Getz G. S. 2003. Serum paraoxonase: effect of the apolipoprotein composition of HDL and the acute phase response. J. Lipid Res. 44: 780–792. [DOI] [PubMed] [Google Scholar]
- 7.Rozenberg O., Shih D. M., Aviram M. 2003. Human serum paraoxonase 1 decreases macrophage cholesterol biosynthesis: possible role for its phospholipase-A2-like activity and lysophosphatidylcholine formation. Arterioscler. Thromb. Vasc. Biol. 23: 461–467. [DOI] [PubMed] [Google Scholar]
- 8.Fuhrman B., Volkova N., Aviram M. 2002. Oxidative stress increases the expression of the CD36 scavenger receptor and the cellular uptake of oxidized low-density lipoprotein in macrophages from atherosclerotic mice: protective role of antioxidants and of paraoxonase. Atherosclerosis. 161: 307–316. [DOI] [PubMed] [Google Scholar]
- 9.Galkina E., Ley K. 2009. Immune and inflammatory mechanisms of atherosclerosis (*). Annu. Rev. Immunol. 27: 165–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segrest J. P., Jackson R. L., Morrisett J. D., Gotto A. M., Jr 1974. A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 38: 247–258. [DOI] [PubMed] [Google Scholar]
- 11.Segrest J. P., Garber D. W., Brouillette C. G., Harvey S. C., Anantharamaiah G. M. 1994. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv. Protein Chem. 45: 303–369. [DOI] [PubMed] [Google Scholar]
- 12.Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. 1992. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33: 141–166. [PubMed] [Google Scholar]
- 13.Barker W. C., Dayhoff M. O. 1977. Evolution of lipoproteins deduced from protein sequence data. Comp. Biochem. Physiol. B. 57: 309–315. [DOI] [PubMed] [Google Scholar]
- 14.Fitch W. M. 1977. Phylogenies constrained by the crossover process as illustrated by human hemoglobins and a thirteen-cycle, eleven-amino-acid repeat in human apolipoprotein A-I. Genetics. 86: 623–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLachlan A. D. 1977. Repeated helical pattern in apolipoprotein-A-I. Nature. 267: 465–466. [DOI] [PubMed] [Google Scholar]
- 16.Karathanasis S. K., Zannis V. I., Breslow J. L. 1983. Isolation and characterization of the human apolipoprotein A-I gene. Proc. Natl. Acad. Sci. U S A. 80: 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo C. C., Li W. H., Moore M. N., Chan L. 1986. Structure and evolution of the apolipoprotein multigene family. J. Mol. Biol. 187: 325–340. [DOI] [PubMed] [Google Scholar]
- 18.Li W. H., Tanimura M., Luo C. C., Datta S., Chan L. 1988. The apolipoprotein multigene family: biosynthesis, structure, structure-function relationships, and evolution. J. Lipid Res. 29: 245–271. [PubMed] [Google Scholar]
- 19.Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., Harvey S. C. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274: 31755–31758. [DOI] [PubMed] [Google Scholar]
- 20.Davidson W. S., Hilliard G. M. 2003. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: a mass spectrometry study. J. Biol. Chem. 278: 27199–27207. [DOI] [PubMed] [Google Scholar]
- 21.Bhat S., Sorci-Thomas M. G., Alexander E. T., Samuel M. P., Thomas M. J. 2005. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J. Biol. Chem. 280: 33015–33025. [DOI] [PubMed] [Google Scholar]
- 22.Li H., Lyles D. S., Thomas M. J., Pan W., Sorci-Thomas M. G. 2000. Structural determination of lipid-bound apo A-I using fluorescence resonance energy transfer. J. Biol. Chem. 275: 37048–37054. [DOI] [PubMed] [Google Scholar]
- 23.Bhat S., Sorci-Thomas M. G., Tuladhar R., Samuel M. P., Thomas M. J. 2007. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 46: 7811–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson W. S., Thompson T. B. 2007. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282: 22249–22253. [DOI] [PubMed] [Google Scholar]
- 25.Panagotopulos S. E., Horace E. M., Maiorano J. N., Davidson W. S. 2001. Apolipoprotein A-I adopts a belt-like orientation in reconstituted high density lipoproteins. J. Biol. Chem. 276: 42965–42970. [DOI] [PubMed] [Google Scholar]
- 26.Martin D. D., Budamagunta M. S., Ryan R. O., Voss J. C., Oda M. N. 2006. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J. Biol. Chem. 281: 20418–20426. [DOI] [PubMed] [Google Scholar]
- 27.Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. U S A. 94: 12291–12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koppaka V., Silvestro L., Engler J. A., Brouillette C. G., Axelsen P. H. 1999. The structure of human lipoprotein A-I. Evidence for the “belt” model. J. Biol. Chem. 274: 14541–14544. [DOI] [PubMed] [Google Scholar]
- 29.Maiorano J. N., Davidson W. S. 2000. The orientation of helix 4 in apolipoprotein A-I-containing reconstituted high density lipoproteins. J. Biol. Chem. 275: 17374–17380. [DOI] [PubMed] [Google Scholar]
- 30.Maiorano J. N., Jandacek R. J., Horace E. M., Davidson W. S. 2004. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 43: 11717–11726. [DOI] [PubMed] [Google Scholar]
- 31.Li H. H., Lyles D. S., Pan W., Alexander E., Thomas M. J., Sorci-Thomas M. G. 2002. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J. Biol. Chem. 277: 39093–39101. [DOI] [PubMed] [Google Scholar]
- 32.Silva R. A., Hilliard G. M., Li L., Segrest J. P., Davidson W. S. 2005. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 44: 8600–8607. [DOI] [PubMed] [Google Scholar]
- 33.Oda M. N., Forte T. M., Ryan R. O., Voss J. C. 2003. The C-terminal domain of apolipoprotein A-I contains a lipid-sensitive conformational trigger. Nat. Struct. Biol. 10: 455–460. [DOI] [PubMed] [Google Scholar]
- 34.Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., 3rd, Smith J. D., Gogonea V., Hazen S. L. 2007. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14: 861–868. [DOI] [PubMed] [Google Scholar]
- 35.Wu Z., Gogonea V., Lee X., Wagner M. A., Li X. M., Huang Y., Undurti A., May R. P., Haertlein M., Moulin M., Gutsche I., Zaccai G., Didonato J. A., Hazen S. L. 2009. The double super helix model of high density lipoprotein. J. Biol. Chem. 284: 36605–36619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brouillette C. G., Anantharamaiah G. M., Engler J. A., Borhani D. W. 2001. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim. Biophys. Acta. 1531: 4–46. [DOI] [PubMed] [Google Scholar]
- 37.Frank P. G., Marcel Y. L. 2000. Apolipoprotein A-I: structure-function relationships. J. Lipid Res. 41: 853–872. [PubMed] [Google Scholar]
- 38.Delcuve G. P., Sun J. M., Davie J. R. 1992. Expression of rainbow trout apolipoprotein A-I genes in liver and hepatocellular carcinoma. J. Lipid Res. 33: 251–262. [PubMed] [Google Scholar]
- 39.Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Wheeler D. L. 2006. GenBank. Nucleic Acids Res. 34: D16–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karlin S., Altschul S. F. 1990. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. U S A. 87: 2264–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff S., Henikoff J. G. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U S A. 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koonin E. V. 2005. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39: 309–338. [DOI] [PubMed] [Google Scholar]
- 44.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan J., Amend A., Borkowski J., DeMarco R., Bailey W., Liu Y., Xie G., Blevins R. 1999. MULTICLUSTAL: a systematic method for surveying Clustal W alignment parameters. Bioinformatics. 15: 862–863. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 47.Schneider K., Beck C. F. 1986. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene. 42: 37–48. [DOI] [PubMed] [Google Scholar]
- 48.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beitz E. 2006. Subfamily logos: visualization of sequence deviations at alignment positions with high information content. BMC Bioinformatics. 7: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beitz E. 2000. TEXshade: shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics. 16: 135–139. [DOI] [PubMed] [Google Scholar]
- 51.Segrest J. P., Jones M. K., Mishra V. K., Anantharamaiah G. M. 2002. Experimental and computational studies of the interactions of amphipathic peptides with lipid surfaces. Current Topics in Membranes. 52: 397–435. [Google Scholar]
- 52.Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., Segrest J. P. 2010. Structures of discoidal high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285: 4652–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Frishman D., Argos P. 1995. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Genet. 23: 566–579. [DOI] [PubMed] [Google Scholar]
- 53.Bussell R., Jr, Eliezer D. 2003. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 329: 763–778. [DOI] [PubMed] [Google Scholar]
- 54.Bussell R., Jr, Ramlall T. F., Eliezer D. 2005. Helix periodicity, topology, and dynamics of membrane-associated alpha-synuclein. Protein Sci. 14: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones M. K., Catte A., Patterson J. C., Gu F., Chen J., Li L., Segrest J. P. 2009. Thermal stability of apolipoprotein A-I in high-density lipoproteins by molecular dynamics. Biophys. J. 96: 354–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chetty P. S., Mayne L., Lund-Katz S., Stranz D., Englander S. W., Phillips M. C. 2009. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. U S A. 106: 19005–19010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka M., Vedhachalam C., Sakamoto T., Dhanasekaran P., Phillips M. C., Lund-Katz S., Saito H. 2006. Effect of carboxyl-terminal truncation on structure and lipid interaction of human apolipoprotein E4. Biochemistry. 45: 4240–4247. [DOI] [PubMed] [Google Scholar]
- 58.Fang Y., Gursky O., Atkinson D. 2003. Lipid-binding studies of human apolipoprotein A-I and its terminally truncated mutants. Biochemistry. 42: 13260–13268. [DOI] [PubMed] [Google Scholar]
- 59.Shao B., Oda M. N., Bergt C., Fu X., Green P. S., Brot N., Oram J. F., Heinecke J. W. 2006. Myeloperoxidase impairs ABCA1-dependent cholesterol efflux through methionine oxidation and site-specific tyrosine chlorination of apolipoprotein A-I. J. Biol. Chem. 281: 9001–9004. [DOI] [PubMed] [Google Scholar]
- 60.Roosbeek S., Vanloo B., Duverger N., Caster H., Breyne J., De Beun I., Patel H., Vandekerckhove J., Shoulders C., Rosseneu M., et al. 2001. Three arginine residues in apolipoprotein A-I are critical for activation of lecithin:cholesterol acyltransferase. J. Lipid Res. 42: 31–40. [PubMed] [Google Scholar]
- 61.Pauling L., Corey R. B., Branson H. R. 1951. The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. U S A. 37: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones M. K., Catte A., Li L., Segrest J. P. 2009. Dynamics of activation of lecithin:cholesterol acyltransferase by apolipoprotein a-I. Biochemistry. 48: 11196–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moosmann B., Behl C. 2000. Cytoprotective antioxidant function of tyrosine and tryptophan residues in transmembrane proteins. Eur. J. Biochem. 267: 5687–5692. [DOI] [PubMed] [Google Scholar]
- 64.Pietraforte D., Minetti M. 1997. Direct ESR detection or peroxynitrite-induced tyrosine-centred protein radicals in human blood plasma. Biochem. J. 325: 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Filipe P., Patterson L. K., Bartels D. M., Hug G. L., Freitas J. P., Maziere J. C., Santus R., Morliere P. 2007. Albumin-bound quercetin repairs vitamin E oxidized by apolipoprotein radicals in native HDL3 and LDL. Biochemistry. 46: 14305–14315. [DOI] [PubMed] [Google Scholar]
- 66.Sies H., Murphy M. E. 1991. Role of tocopherols in the protection of biological systems against oxidative damage. J. Photochem. Photobiol. B. 8: 211–218. [DOI] [PubMed] [Google Scholar]
- 67.Bisby R. H., Parker A. W. 1995. Reaction of ascorbate with the alpha-tocopheroxyl radical in micellar and bilayer membrane systems. Arch. Biochem. Biophys. 317: 170–178. [DOI] [PubMed] [Google Scholar]
- 68.Catte A., Patterson J. C., Jones M. K., Jerome W. G., Bashtovyy D., Su Z., Gu F., Chen J., Aliste M. P., Harvey S. C., et al. 2006. Novel changes in discoidal high density lipoprotein morphology: a molecular dynamics study. Biophys. J. 90: 4345–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Catte A., Patterson J. C., Bashtovyy D., Jones M. K., Gu F., Li L., Rampioni A., Sengupta D., Vuorela T., Niemela P., et al. 2008. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys. J. 94: 2306–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anantharamaiah G. M., Hughes T. A., Iqbal M., Gawish A., Neame P. J., Medley M. F., Segrest J. P. 1988. Effect of oxidation on the properties of apolipoproteins A-I and A-II. J. Lipid Res. 29: 309–318. [PubMed] [Google Scholar]
- 71.Shao B. H., Oda M. N., Oram J. F., Heinecke J. W. 2010. Myeloperoxidase: an oxidative pathway for generating dysfunctional high-density lipoprotein. Chem. Res. Toxicol. 23: 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carr A. C., McCall M. R., Frei B. 2000. Oxidation of LDL by myeloperoxidase and reactive nitrogen species: reaction pathways and antioxidant protection. Arterioscler. Thromb. Vasc. Biol. 20: 1716–1723. [DOI] [PubMed] [Google Scholar]
- 73.Daugherty A., Dunn J. L., Rateri D. L., Heinecke J. W. 1994. Myeloperoxidase, a catalyst for lipoprotein oxidation, is expressed in human atherosclerotic lesions. J. Clin. Invest. 94: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bergt C., Fu X., Huq N. P., Kao J., Heinecke J. W. 2004. Lysine residues direct the chlorination of tyrosines in YXXK motifs of apolipoprotein A-I when hypochlorous acid oxidizes high density lipoprotein. J. Biol. Chem. 279: 7856–7866. [DOI] [PubMed] [Google Scholar]
- 75.Shao B., Bergt C., Fu X., Green P., Voss J. C., Oda M. N., Oram J. F., Heinecke J. W. 2005. Tyrosine 192 in apolipoprotein A-I is the major site of nitration and chlorination by myeloperoxidase, but only chlorination markedly impairs ABCA1-dependent cholesterol transport. J. Biol. Chem. 280: 5983–5993. [DOI] [PubMed] [Google Scholar]
- 76.Panzenboeck U., Raitmayer S., Reicher H., Lindner H., Glatter O., Malle E., Sattler W. 1997. Effects of reagent and enzymatically generated hypochlorite on physicochemical and metabolic properties of high density lipoproteins. J. Biol. Chem. 272: 29711–29720. [DOI] [PubMed] [Google Scholar]
- 77.Shao B., Heinecke J. W. 2008. Using tandem mass spectrometry to quantify site-specific chlorination and nitration of proteins: model system studies with high-density lipoprotein oxidized by myeloperoxidase. Methods Enzymol. 440: 33–63. [DOI] [PubMed] [Google Scholar]
- 78.Peng D. Q., Brubaker G., Wu Z., Zheng L., Willard B., Kinter M., Hazen S. L., Smith J. D. 2008. Apolipoprotein A-I tryptophan substitution leads to resistance to myeloperoxidase-mediated loss of function. Arterioscler. Thromb. Vasc. Biol. 28: 2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shao B., Cavigiolio G., Brot N., Oda M. N., Heinecke J. W. 2008. Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A-I. Proc. Natl. Acad. Sci. U S A. 105: 12224–12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Handattu S. P., Datta G., Epand R. M., Epand R. F., Palgunachari M. N., Mishra V. K., Monroe C. E., Keenum T. D., Chaddha M., Anantharamaiah G. M., et al. 2010. Oral administration of L-mR18L, a single domain cationic amphipathic helical peptide, inhibits lesion formation in ApoE null mice. J. Lipid Res. 51: 3491–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kontush A., Chapman M. J. 2006. Functionally defective high-density lipoprotein: a new therapeutic target at the crossroads of dyslipidemia, inflammation, and atherosclerosis. Pharmacol. Rev. 58: 342–374. [DOI] [PubMed] [Google Scholar]
- 82.Gallivan J. P., Dougherty D. A. 1999. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. U S A. 96: 9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harel M., Aharoni A., Gaidukov L., Brumshtein B., Khersonsky O., Meged R., Dvir H., Ravelli R. B., McCarthy A., Toker L., et al. 2004. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 11: 412–419. [DOI] [PubMed] [Google Scholar]
- 84.Gaidukov L., Viji R. I., Yacobson S., Rosenblat M., Aviram M., Tawfik D. S. 2010. ApoE induces serum paraoxonase PON1 activity and stability similar to ApoA-1. Biochemistry. 49: 532–538. [DOI] [PubMed] [Google Scholar]