Abstract
PCSK9 degrades LDL receptor (LDLR) in liver and thereby influences the circulating level of LDL cholesterol. Hence, mechanisms inhibiting PCSK9 expression have potential for cholesterol-lowering intervention. Previously, we demonstrated that oncostatin M (OM) activates LDLR gene transcription, resulting in an increased LDL uptake in HepG2 cells and a reduction of plasma LDL in hypercholesterolemic hamsters. Here we identify the suppression of PCSK9 expression by OM as one new mechanism that increases LDLR protein in HepG2 cells. Treating HepG2 cells with OM decreases PCSK9 mRNA and protein levels. Inhibition studies and small interfering RNA -targeted depletion revealed a critical role for JAK1 and JAK2 in mediating this OM inhibitory effect. Furthermore, we showed that OM induces transient phosphorylation of STAT1, STAT3, and STAT5 and sustained activation of ERK signaling molecules. While depletion of STAT members in HepG2 cells did not affect OM inhibitory activity on PCSK9 expression, blocking activation of the MEK1/ERK signaling pathway resulted in attenuation of the OM inhibitory effect. Finally, by using an anti-hamster PCSK9 antibody, we demonstrated the in vivo suppression of liver PCSK9 mRNA and protein expression by OM in hypercholesterolemic hamsters. Our study uncovered a cytokine-triggered regulatory network for PCSK9 expression that is linked to JAKs and the ERK signaling pathway.
Keywords: hamsters, LDL receptor, JAK, ERK, proprotein convertase subtilisin/kexin type 9
Recent studies have identified proprotein convertase subtilisin/kexin type 9 (PCSK9) as a critical new player in the control of plasma LDL cholesterol (LDL-C) levels through its role in mediating the posttranslational degradation of LDL receptor (LDLR) (1–4). PCSK9 is a secreted protein expressed predominantly in the liver, small intestine, and kidney (1). It is synthesized as a ∼72 kDa zymogen that undergoes intramolecular cleavage in the endoplasmic reticulum to form a heterodimer of a prosegment (122 amino acids) and a 60 kDa active form, which is secreted as a proteolytic inactive form (1, 5). In plasma, PCSK9 binds directly to the EGF-A extracellular domain of hepatic LDLR (6, 7). This binding and the subsequent internalization of the PCSK9-LDLR complex leads to the intracellular degradation of LDLR in lysosomes, thus increasing the level of circulating LDL-C. In humans, gain-of-function mutations in PCSK9 result in autosomal hypercholesterolemia and premature atherosclerosis (8). Conversely, loss-of-function mutations within PCSK9 are associated with a reduction in plasma LDL-C and protection against coronary heart disease (9, 10).
Investigations conducted in cell culture and animal models have demonstrated that the expression of PCSK9 is controlled at the transcriptional level. So far, two transcription factors have been identified as critical players in PCSK9 gene transcription. PCSK9 is a target gene of sterol regulatory element binding protein (SREBP) (4, 11). Analogous to LDLR and other cholesterol-regulated genes, the proximal region of the PCSK9 promoter contains an SRE-1 motif (12), which interacts with the nuclear forms of SREBPs under the condition of depleted intracellular sterols. The second transactivator identified for PCSK9 gene transcription is hepatic nuclear factor 1α (HNF1α), which is abundantly expressed in liver. HNF1α regulates PCSK9 transcription through a highly conserved HNF1 binding site located 28 bp upstream of the SRE-1 site in human PCSK9 promoter (13).
PCSK9 transcription has been shown to be induced by various small molecules that activate SREBP signaling pathway, including statins (12), insulin (14), and agonists of liver X receptor (LXR) (3). Statins reduce intracellular levels of sterols and activate the SREBP pathway by inhibiting HMG-CoA reductase, the rate-limiting enzyme in cholesterol biosynthesis. Insulin induces PCSK9 transcription in rodent primary hepatocytes (14) by activating SREBP1c. Treating mice with LXR agonist TO901317 elevated the level of PCSK9 mRNA in the liver, presumably through an SREBP1c-mediated activation mechanism (3). Induction of PCSK9 expression by statins has been confirmed in human studies (15, 16) and is considered a limitation of stain efficacy in treating hypercholesterolemic patients (17–19). Therefore, identification of new regulatory mechanisms that inhibit PCSK9 expression will be of clinical significance. Such mechanisms may be applied to improve statin efficacy by prolonging the upregulation of LDLR.
Our laboratory has previously shown that oncostatin M (OM), a member of the interleukin-6 (IL-6) family of cytokines, is a strong inducer of LDLR expression in HepG2 cells through an SREBP-independent mechanism (20–22). OM activates LDLR gene transcription by inducing the binding of transcription factors Egr1 and c/EBPβ to the sterol-independent regulatory element (SIRE) located downstream of the SRE-1 site of the LDLR promoter (23–25). In HepG2 cells, the level of LDLR mRNA was elevated rapidly after 1 h of OM treatment and reached a maximal level by 2 h and declined thereafter (26). Interestingly, we have observed that the LDLR uptake activity was highest after 24 h of OM treatment, when the LDLR mRNA level had already declined (27). The differences in temporal inductions between LDLR mRNA and protein suggest that the activation of LDLR transcription might not be the sole mechanism responsible for the strong and extended induction of LDLR protein expression in HepG2 cells nor for the decrease observed in plasma LDL-C of hypercholesterolemic hamsters after OM treatment (28, 29).
In this study, we examined the effects of OM on PCSK9 expression in HepG2 cells and in liver tissue of hamsters. Our results demonstrated that while LDLR mRNA expression was transiently induced by OM, the LDLR protein level was continuously increased over a 24-h treatment period in HepG2 cells. This was accompanied by a steady decline of PCSK9 mRNA and protein. We further showed that the PCSK9 mRNA stability was not changed by OM treatment, suggesting that the inhibitory effect of OM on PCSK9 expression might occur at the transcriptional level. Importantly, we demonstrated that Janus kinase 1 (JAK1) and JAK2 played a critical role in the regulation of PCSK9 expression by OM. Inhibition of JAK activity with specific inhibitors prevented the downregulation of PCSK9 by OM. This finding was corroborated by small interfering RNA (siRNA)-mediated silencing of JAK expression, which restored PCSK9 expression in OM-treated cells. We further showed that the MEK1/ERK signaling pathway downstream of JAK is involved in transmitting the OM signal that leads to the suppression of PCSK9 expression. Through cDNA cloning and generation of a highly specific anti-hamster PCSK9 antibody, we confirmed the in vivo suppression of hepatic PCSK9 mRNA and protein by OM in hypercholesterolemic hamsters. Altogether, our study uncovers a new regulatory network for PCSK9 expression that involves members of the JAK family of tyrosine kinases (Tyk) and the ERK signaling pathway. This new regulatory mechanism could be explored in a therapeutic strategy that combines a statin with an OM mimetic inhibitor of PCSK9 expression.
MATERIALS AND METHODS
Cells and reagents
The human hepatoma cell line HepG2 was obtained from American Type Culture Collection and cultured in MEM supplemented with 10% FBS (Summit Biotechnology, Fort Collins, CO), 1 mM streptomycin, and 1 mM penicillin. JAK2 inhibitor AG490 and JAK3 inhibitor Janex-1 were obtained from Cayman Chemical (Ann Arbor, MI). JAK inhibitor I (JAK I), and MEK1 inhibitor U0126, PI-3 kinase inhibitor LY294002, p38 kinase inhibitor SB203580, c-Jun N-terminal kinase (JNK) inhibitor SP600125, and protein kinase C inhibitor calphostin C were from CalBiochem (Gibbstown, NJ). Rabbit antibody against human LDLR was obtained from BioVision (Mountain View, CA). The rabbit antibodies against human recombinant PCSK9 protein expressed in Escherichia coli BL21 were prepared according to standard procedures (11). Rabbit antibodies targeting phosphorylation of ERK (phospho-ERK), phospho-STAT1 (p-STAT1), p-STAT3, p-STAT5, STAT3, ERK2, JAK1, and JAK2 were obtained from Cell Signaling. Rabbit antibodies against STAT1 and STAT5 were from Santa Cruz Biotechnology. Monoclonal anti-β-actin, actinomylin D, berberine chloride (BBR), and simvastatin (SMV) were obtained from Sigma. The full-length human recombinant oncostatin M protein expressed in Chinese hamster ovary cells was generously provided by Bristol-Myers Squibb (Princeton, NJ) and was used in studies of HepG2 cells and hamsters.
RNA isolation and real-time RT-PCR
Total RNA was extracted from HepG2 cells or liver tissues of hamsters, using UltraspecTM total RNA isolation reagent (Biotecx Laboratories). Two micrograms of total RNA was reverse-transcribed with a high-capacity cDNA reverse transcription kit (Applied BioSystems, Foster City, CA) using random primers. Real-time PCR was performed with the cDNA, using an ABI Prism 7900-HT sequence detection system with SYBR PCR Master Mix under the following conditions: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 60 s. Results are reported as means ± SEM of triplicate assays for each cDNA sample after normalization to GAPDH. The primer sequences are listed in supplementary Table I. The assessments of hamster LDLR and GAPDH mRNA levels were accomplished by using predeveloped TaqMan assay reagents (Applied BioSystems).
siRNA transfection
Silencer®-validated siRNAs targeted to human JAK2 mRNA (catalog no. AM51331; ID, 608); Silencer®-selected siRNA targeted to human JAK1, STAT1, STAT3, and MEK1 mRNA (catalog no. 4390824; IDs, s7648, s278, s745, and s11167); and Silencer-selected negative control siRNA (catalog no. 4390844) were obtained from Applied BioSystems. STAT5a and STAT5b siRNA were obtained from Dharmacon (STAT5a, catalog no. L-005169-00-0005; STAT5b, catalog no. L-010539-00-0005). HepG2 cells in suspension were mixed with siRNA in the Silencer siRNA transfection reagent for 10 min and plated in 12 well plates at a density of 9 × 104 cells/well. The final concentration of siRNA was 30 nM. Twenty-four h later, culture medium was replaced with medium containing 0.5% FBS and incubated overnight. Cells were treated with OM for 24 h prior to cell lysis for RNA or protein isolation.
Western blot analysis
Lysates of HepG2 cells and liver tissue protein were prepared as previously described (13, 28). Protein concentration was measured using a bicinchoninic acid assay kit (Pierce). Equal amounts of sample protein in Laemmli buffer were separated on 4%–20% Tris-HCl polyacrylamide SDS gels (Bio-Rad) and transferred to Whatman nitrocellulose membranes (Whatman GmbH, Dassel, Germany). Blots were placed in blocking buffer (5% non-fat milk in TBS with 0.05% Tween-20) for 1 h and then incubated with various primary antibodies in blocking solution for 1 h at room temperature or overnight at 4°C. Blots were washed three times with TBS with 0.05% Tween-20 and incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. Specific immunoreactive bands were visualized using an ECL Plus kit (GE Healthcare Life Sciences, Piscataway, NJ) and quantified with Kodak molecular imaging software (Kodak, New Haven, CT).
PCSK9 promoter–reporter plasmids and analysis of PCSK9 promoter activity
The plasmid pGL3-PCSK9-D1 contains the 5′ flanking region of the PCSK9 gene, from nucleotide (nt) −1711 to −94 (relative to the ATG start codon) in front of the luciferase coding sequence. The plasmid pGL3-PCSK9-D4 contains the 5′ flanking region of the PCSK9 gene from nt −440 to −94. CL26 cells are HepG2-derived cells that stably express pGL3-PCSK9-D1 (13). To determine whether OM regulated PCSK9 promoter activity, CL26 cells were seeded in 96 well plates at a density of 1.5 × 104 cells per well. On the next day, cells were cultured in serum-reduced medium (0.5% FBS in MEM) and incubated overnight and then treated with OM, BBR, or SMV for 24 h, before they were lysed for measuring luciferase activity. Additionally, HepG2 cells were transiently transfected with the pGL3-PCSK9-D1 and pGL3-PCSK9-D4 plasmids along with a Renilla luciferase expression vector, pRL-SV40. One day posttransfection, cells were incubated in 0.5% FBS-MEM overnight and then treated with OM for 24 h. Dual luciferase activities were measured, and the firefly luciferase activity was normalized to the Renilla luciferase activity.
Cloning of hamster PCSK9 complete coding sequence
To clone the complete hamster PCSK9 coding sequence, we first compared mRNA sequences from human, mouse, and rat PCSK9 genes and then selected highly conserved regions across these species with which to design primer sets. Using primers identified by this approach, we amplified the hamster PCSK9 coding region from a hamster liver cDNA library, cloned into pCR2.1-Topo vectors (Invitrogen, Carlsbad, CA) and sequenced. The primers are listed in Supplementary Table I. The hamster PCSK9 coding region was subcloned into pcDNA4.0-HisMax-TOPO vector, resulting in the plasmid pHis-HamPCSK9 for hamster PCSK9 expression in mammalian cells.
Generation of a rabbit anti-hamster PCSK9 antibody
Rabbit polyclonal antibodies against hamster PCSK9 were raised commercially by GenScript Corporation (Piscataway, NJ). Briefly, a peptide corresponding to the C-terminal end of hamster PCSK9 (CRNRPSAKASWVHQ) was synthesized, purified, and conjugated to keyhole limpet hemocyanin. Rabbit antiserum against PCSK9 peptide antigen was obtained using a standard immunization protocol and subjected to affinity purification. The anti-PCSK9 antibody was used at 1:2,500 dilution (1 µg/ml) for Western blotting experiments.
Generation of short hairpin RNA against hamster PCSK9
A U6 promoter-based vector (pSH-hamPCSK9) that expresses short hairpin RNA (shRNA) targeting the hamster PCSK9 coding region (5′-GCAAACTGTAGCATCCATACC-3′) was generated using a Block-iT™ (Invitrogen) U6 RNA interference (RNAi) entry vector kit following the manufacturer's instructions. Briefly, shRNA target sequences on the hamster PCSK9 gene were selected with the assistance of the Block-iT™ RNAi Designer (an online tool developed by Invitrogen [http://rnaidesigner.invitrogen.com]). Two complementary single-stranded DNA oligonucleotides, one of which encoded the shRNA against the selected target region, were synthesized and annealed to generate a double-stranded oligonucleotide. The resulting double-stranded oligonucleotide was subsequently cloned into the linearized pENTR™/U6 vector. Finally, the resulting pSH-HamPCSK9 vector was sequenced to confirm the orientation and the sequence of the oligonucleotide insert. To test the efficiency of the shRNA knockdown and the specificity of the newly generated PCSK9 antibody, we cotransfected HEK293 cells with the hamster PCSK9 expression vector (pHis-hamPCSK9) and different amounts of pSH-HamPCSK9 plasmid or a negative control vector using Lipofectamine 2000 reagent (Invitrogen). The total amount of cotransfected DNA was kept constant by adjusting with carrier DNA. After 48 h of transfection, cell lysates were prepared and subjected to immunoblotting using the anti-PCSK9 antibody.
Detection of hamster plasma PCSK9 by immunoprecipitation and Western blotting
PCSK9 from hamster serum samples was detected by immunoprecipitation, followed by Western blotting, using the rabbit anti-hamster antibody described above. For each precipitation, 10 µl of serum was added to 500 µl of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH7.4, 1% NP-40, 150 mM NaCl, 0.25% sodium deoxycholate,1 mM EDTA) containing a mixture of protease inhibitors (1× Complete medium [Roche Diagnostics GmbH, Mannheim, Germany]). Two micrograms of anti-PCSK9 rabbit polyclonal antibody was added to the diluted serum sample, and samples were incubated for 1 h at 4°C. Afterward, 50 µl of preequilibrated anti-rabbit IgG beads (rabbit IgG TrueBlot set; eBioscience, San Diego, CA) was added, and the reaction was carried out at 4°C overnight with constant mixing. After the samples underwent brief centrifugation, beads were washed three times with RIPA buffer, and 100 µl of 1× SDS loading buffer was added to each tube. Samples were vortexed and boiled for 10 min. Twenty microliters of each sample was separated by 4%–20% SDS-PAGE. Proteins were transferred to Whatman nitrocellulose membranes. Nitrocellulose blots were blocked for 1 h at room temperature in TBS containing 0.1% Tween-20 and 5% dry milk. After blocking, blots were probed with rabbit anti-PCSK9 antibody (1 µg/ml, 1:2,500 dilution) in blocking buffer overnight at 4°C. Membranes were washed three times in TBS with 0.1% Tween-20 and probed with TrueBlot anti-rabbit IgG-conjugated HRP (1:1,000; eBioscience).
Statistical analysis
All results are representative of two to four separate experiments. Real-time PCR data and densitometry results are expressed as means ± SEM. Statistical analysis was performed using a two-tailed Student's t test or one-way ANOVA with a Bonferroni posttest of multiple comparisons. A P value of <0.05 was considered statistically significant.
RESULTS
Time-dependent inhibition of PCSK9 mRNA and protein expression by OM in HepG2 cells
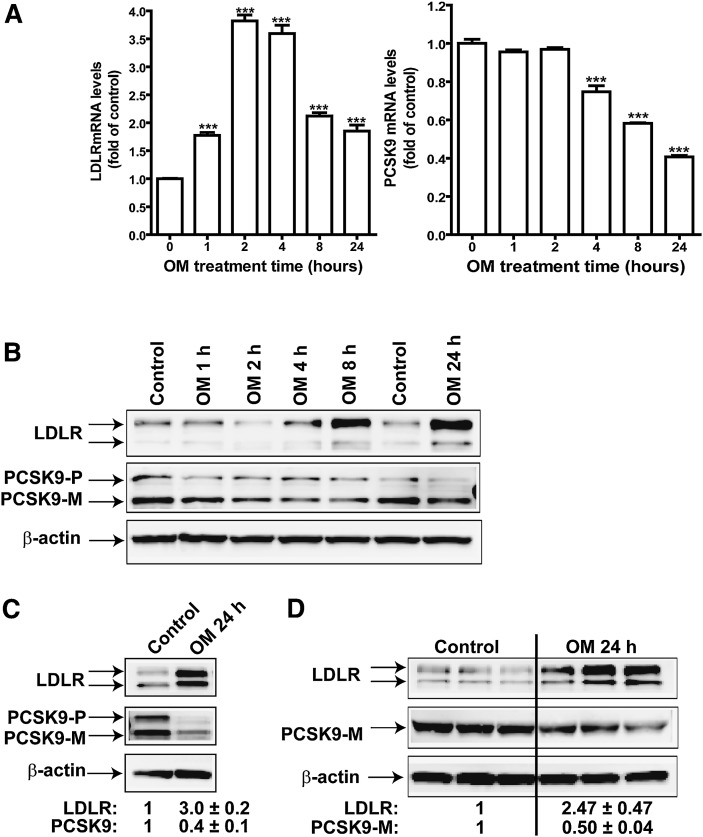
To explore the possibility that PCSK9 has a role in OM-mediated upregulation of LDLR, we first conducted a kinetics study to examine the time-dependent effect of OM on the expression of PCSK9 and LDLR mRNA by real-time RT-PCR (Fig. 1A). Consistent with our previous findings (26), OM, at a concentration of 100 ng/ml, induced a rapid increase in LDLR mRNA level. A 1.7-fold increase was detected at 1 h after OM was added to the cells; the increase reached a 3.8-fold maximal level compared to control cells by 2 h and declined to a 1.8-fold level compared to control cells at 24 h. In contrast to this rapid induction of LDLR mRNA, the level of PCSK9 mRNA steadily decreased in OM-treated cells. At 8 h, a 33% reduction was detected (P < 0.001), and the abundance of PCSK9 mRNA further declined in OM-treated cells to 40% of that of control cells at 24 h. We further investigated this observation using Western blotting to detect changes in amounts of PCSK9 and LDLR protein upon OM treatment. Figure 1B shows that an increase in LDLR protein level was noticeable at 4 h and that it continued to rise. At 24 h, the level of LDLR protein in OM-treated cells was 3-fold (3.0 ± 0.2; n = 6) higher than that in control cells (Fig. 1C). Conversely, the amount of PCSK9 protein gradually declined during the treatment to 40% of that of control cells at 24 h. We further examined the effect of OM on the amount of PCSK9 secreted in culture medium, using triplicate culture dishes with and without OM treatment. Consistently, the abundance of secreted PCSK9 protein was reduced to 50% of that of control cells (P < 0.001) by OM treatment at 24 h (Fig. 1D). These results provided evidence that the reduction in PCSK9 by OM leads to the sustained high level of LDLR protein by inhibiting LDLR degradation.
Fig. 1.
Time-dependent effects of OM on PCSK9 mRNA and protein expression in HepG2 cells. HepG2 cells were treated with 100 ng/ml OM for different times. A: Total RNA was isolated, and mRNA levels of LDLR and PCSK9 were quantified by quantitative real-time PCR. B: Total cell lysates were isolated from HepG2 cells treated with OM from 0–24 h, and the protein abundance of LDLR and PCSK9 was examined by Western blotting. PCSK9-P, PCSK9 proprotein; PCSK9-M, mature protein. C: Cells were treated with OM for 24 h and harvested for detection of LDLR and PCSK9. The LDLR and PCSK9 bands were quantified using an Kodak Image Station 4000R. Values were normalized to β-actin and graphed relative to those of untreated cells. Data are presented as means ± SEM from six separate experiments. D: HepG2 cells were untreated or treated with OM in triplicate. The total cell lysate and medium were harvested to detect LDLR protein and PCSK9.
OM may suppress PCSK9 gene transcription
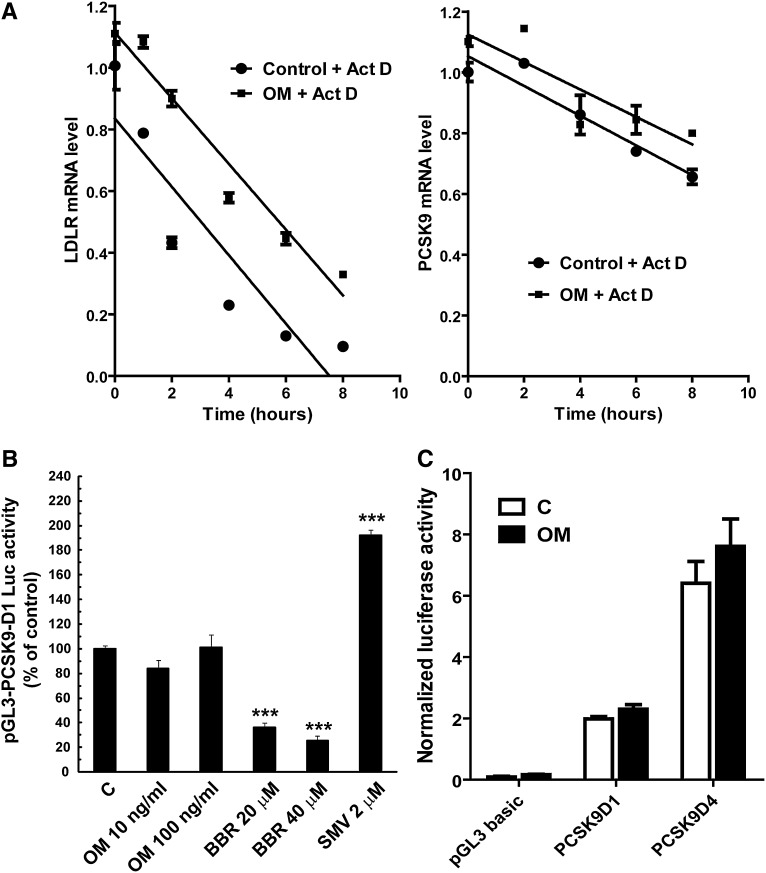
Having observed the inhibitory effect of OM on PCSK9 mRNA and protein expression, we sought to understand at what levels OM exerts its effect. We first examined PCSK9 mRNA stability in control and OM-treated cells. HepG2 cells were treated with OM for 15 h before actinomycin D was added. Total RNA was isolated after 1, 2, 4, 6, and 8 h. PCSK9 mRNA levels, along with levels of LDLR mRNA abundance, were quantified by real-time PCR. As shown in Fig. 2A (right panel), PCSK9 mRNA amounts slowly decreased in control cells over the 8 h treatment course with actinomycin D. The presence of OM in culture medium did not accelerate its decay, indicating that OM had no destabilizing effect on PCSK9 mRNA. In addition, OM did not affect LDLR mRNA stability (Fig. 2A, left panel), which agreed with its action mechanism of transcriptional activation of LDLR (21).
Fig. 2.
Examination of the OM effects on PCSK9 mRNA stability and promoter activity. A: HepG2 cells were untreated or treated with OM for 15 h. Actinomycin D (Act D) was added to cells at different intervals. Total RNA was isolated and analyzed for the amount of PCSK9 mRNA (right panel) and LDLR mRNA (left panel) by real-time PCR. Normalized PCSK9 or LDLR mRNA levels were plotted as the percentage of the mRNA remaining. Decay versus time curves were plotted. B: HepG2-derived CL26 cells were incubated with OM (100 ng/ml), BBR (20 and 40 μM), or with 2 μM of SMV for 24 h. Luciferase activities are expressed relative to that of untreated control cells. Significant differences between control and treatment groups were assessed by one-way ANOVA with a Bonferroni posttest of multiple comparisons. ***, P < 0.001 compared with untreated control cells. Data shown are representative of three separate experiments with similar results. C: HepG2 cells were transiently transfected with pGL3-PCSK9-D4 and pRL-SV40 for 1 day, and cells were treated with OM for 24 h before being lysed for dual luciferase assays.
We next examined the effect of OM on PCSK9 promoter activity in HepG2-derived CL26 cells, which stably express the luciferase reporter construct pGL3-PCSK9-D1 (13). This plasmid contains the 5′ flanking region of the PCSK9 gene, from nt −1711 to −94 (relative to the ATG start codon), in front of the luciferase coding sequence. CL26 cells were treated for 24 h with OM at concentrations of 10 and 100 ng/ml, along with BBR, an inhibitor, and SMV, an inducer of PCSK9 promoter activity (13). Luciferase activity was measured in cell lysates (Fig. 2B). Surprisingly, this PCSK9 promoter reporter was unresponsive to OM treatment. In contrast, the luciferase activity of CL26 cells was dose-dependently repressed by BBR down to 25% of that of control cells and 2-fold increased by 2 µM of SMV, consistent with our previous studies (13). To verify this finding, we transiently transfected the PCSK9 promoter constructs PCSK9-D1 and PCSK9-D4 into HepG2 cells along with a Renilla expression vector for normalization of transfection efficiency. Plasmid PCSK9-D4 contains the functional proximal PCSK9 promoter region (nt −440 to −94), in which the Sp1, HNF1, and SRE-1 sites reside, upstream of the luciferase gene (11). Again, the normalized promoter activities of PCSK9-D1 and PCSK9-D4 were not decreased by OM treatment (Fig. 2C). Collectively, these data suggest that the regulatory element responsible for OM-mediated suppression of PCSK9 transcription might lie outside the currently characterized promoter region.
Activation of JAK by OM inhibits PCSK9 expression
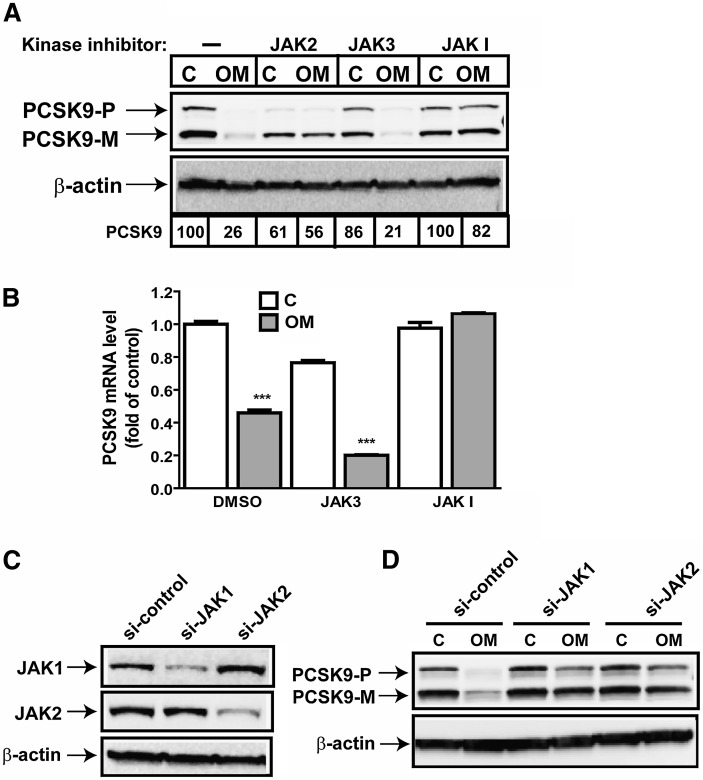
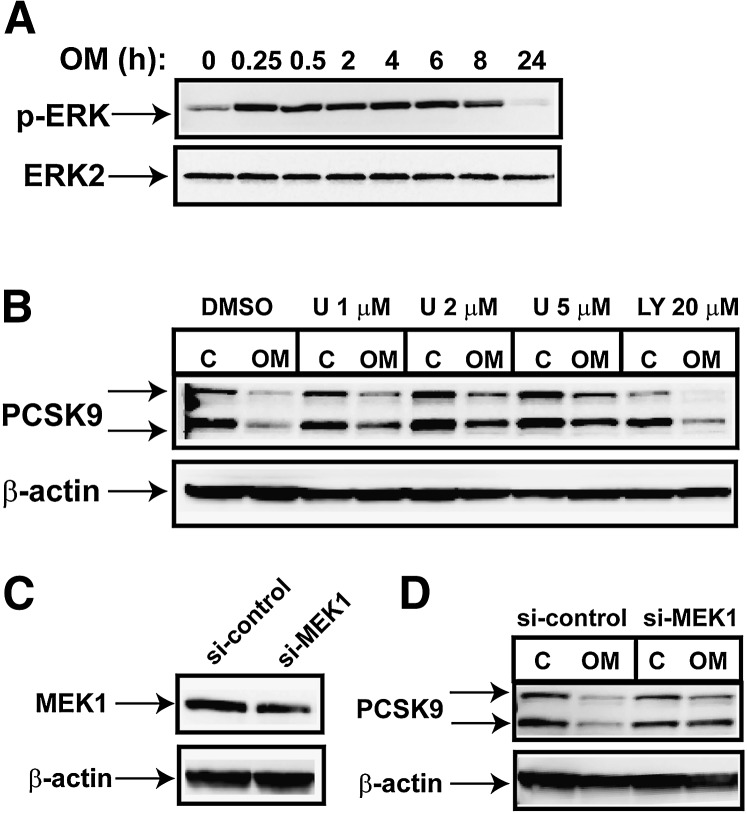
Immediately after OM binds to its cell surface receptors, the JAK family of tyrosine kinases (JAK1, JAK2, and Tyk) is activated, which transmits extracellular signals to the nucleus to alter gene expression (30). To determine which specific JAK isoform(s) mediates the OM effect on PCSK9 expression, we first used pharmacological inhibitors. HepG2 cells were pretreated for 2 h with efficacious concentrations of JAK2 inhibitor AG490, JAK3 inhibitor Janex-1, and JAK inhibitor I before OM was added. Cell lysates were isolated after 24 h, and the PCSK9 protein level was assessed by Western blotting (Fig. 3A). The JAK3 inhibitor had no effect on blocking OM activity on PCSK9 expression, thereby excluding its role from this OM-mediated biological event. Remarkably, pretreatment with JAK I, an inhibitor known to block activities of all four JAK kinases, JAK1, JAK2, JAK3, and Tyk2, resulted in complete resistance to the OM inhibitory effect on PCSK9. The JAK2 inhibitor also effectively attenuated OM activity. The combination of JAK2 inhibitor and JAK I produced attenuating effects that were comparable to those of each inhibitor alone (supplementary Fig. I) on OM-mediated suppression of PCSK9 protein expression. We further validated the effects of JAK inhibitors on PCSK9 mRNA expression in control and OM-treated cells. Again, the decrease in PCSK9 mRNA level from OM treatment was observed in DMSO-treated control cells and in JAK3 inhibitor-treated HepG2 cells but was not detected in cells cotreated with JAK inhibitor I (Fig. 3B).
Fig. 3.
OM-mediated suppression of PCSK9 expression requires JAK activation. A: HepG2 cells were preincubated for 2 h with JAK2 inhibitor (50 µM), JAK3 inhibitor (100 µM), or JAK inhibitor I (10 µM) prior to OM treatment for 24 h. Total cell lysates were harvested for Western blotting to detect PCSK9. PCSK9-P, PCSK9 proprotein; PCSK9-M, mature protein; C, control. B: HepG2 cells were preincubated for 2 h with JAK3 inhibitor (100 µM) or JAK inhibitor I (10 µM) prior to OM treatment for 24 h. Total RNA was prepared for real-time PCR analysis to quantify PCSK9 mRNA and GAPDH mRNA. C, D: HepG2 cells were transfected with si-JAK1, si-JAK2, or a control siRNA with a scrambled sequence for 36 h, followed by OM treatment for 24 h.
Because no unique inhibitor of JAK1 is available, we used specific siRNAs targeted to JAK1 and JAK2 to further investigate their individual roles in OM-induced suppression of PCSK9. Control nonspecific siRNAs and JAK siRNAs were separately transfected into HepG2 cells for 36 h before OM treatment. As shown in Fig. 3D, the decrease in amounts of PCSK9 protein in OM-treated cells was prominent in control siRNA-transfected cells but was not observed in JAK1- or JAK2-depleted cells. Figure 3C shows the immunoblots for JAK1 and JAK2, which confirm that each siRNA was effective and specific to its own target without affecting the expression of the other isoform. Altogether, these data demonstrate that activation of the JAK isoforms, particularly JAK1 and JAK2, by OM results in repression of PCSK9 expression in HepG2 cells.
STAT activation is not linked to the OM-mediated inhibition of PCSK9 expression
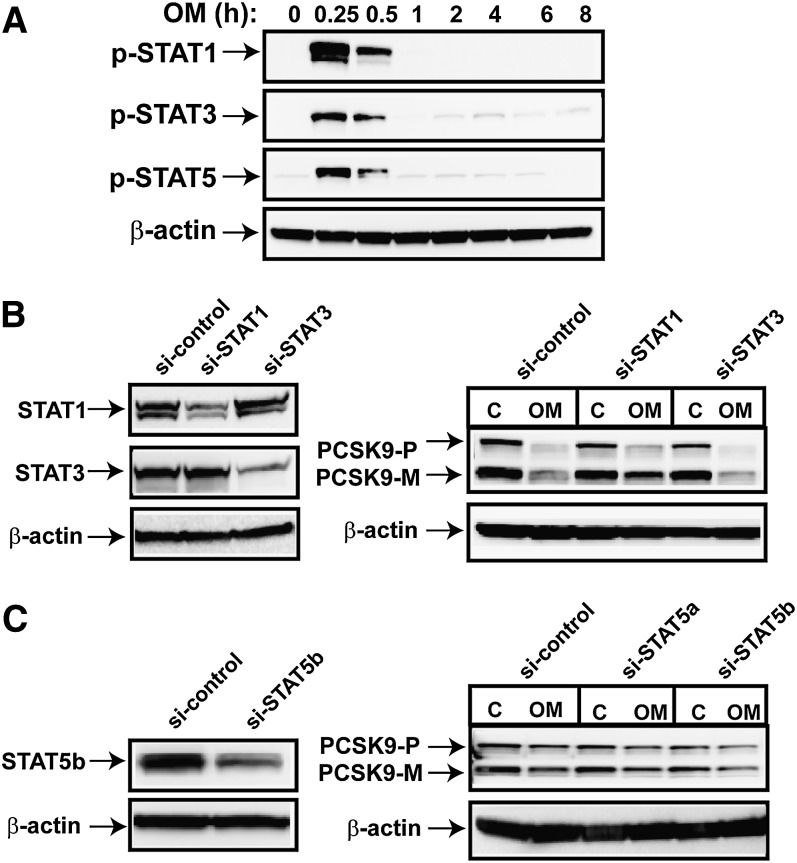
It is well known that after OM binds to its cognate receptor, STAT1, STAT3, and STAT5 are phosphorylated and activated by members of the JAK family (30). Once activated, the STATs dimerize and translocate to the nucleus and modulate the expression of target genes. We probed p-STAT1, p-STAT3, and p-STAT5 in OM-treated cell lysates as a means of assaying STAT activation by OM. Figure 4A shows that levels of p-STAT1, p-STAT3, and p-STAT5 were markedly increased after 15 min of OM treatment, maintained the activated state for 30 min, and then rapidly declined to near-basal levels after 1 h of OM treatment. To determine whether the JAK/STAT pathway is involved in the regulation of PCSK9 expression, we transfected siRNAs targeted to STAT1, STAT3 (Fig. 4B), and STAT5a and -5b (Fig. 4C) into HepG2 cells to deplete these signal transducers. After 36 h of siRNA transfection, OM was added to the cells, and total cell lysates were isolated after 24 h. Decreased cellular protein levels of STAT1, STAT3, and STAT5b in siRNA-transfected cells were clearly observed (Fig. 4B, C, left panels). The silencing of STAT5a was confirmed at the mRNA level by real-time PCR (data not shown). However, the inhibitory effect of OM on PCSK9 expression was persistent regardless of the expression status of the STATs (Fig. 4B, C, right panels). These results are consistent with the hypothesis that the transient activation of STATs is not required for OM to suppress PCSK9 expression.
Fig. 4.
Examination of the involvement of STAT activation in OM-mediated suppression of PCSK9 protein expression. A: HepG2 cells were treated with OM at different intervals. Total cell lysates were prepared and probed for p-STAT1, p-STAT3, and p-STAT5. B, C: HepG2 cells were transfected with si-STAT1, si-STAT3, si-STAT5a, or si-STAT5b or a control (C) siRNA for 36 h, and then treated with OM for 24 h. PCSK9-P, PCSK9 proprotein; PCSK9-M, mature protein. Depletion of STAT1, STAT3, and STAT5b were detected by Western blotting with specific antibodies. The data shown are representatives of three separate transfection experiments with similar results.
ERK signaling pathway is involved in OM-mediated inhibition of PCSK9 expression
It has been shown that in addition to the activation of STATs, JAKs mediate the recruitment of other signaling molecules such as the MAP kinases and PI-3 kinase (31). Our previous studies have demonstrated that the OM-mediated activation of LDLR gene transcription is transmitted by the MEK1/ERK signaling pathway (26). To determine whether this signaling cascade is also involved in the downregulation of PCSK9 expression observed in OM-treated HepG2 cells, we first investigated the kinetics of ERK activation by OM. Figure 5A shows that the level of phosphorylated ERK rapidly increased and that the elevation was maintained for 8 h of OM treatment. This finding was in contrast to the transient activation of the STATs by OM. Next, we examined the effect of OM on PCSK9 expression in HepG2 cells that were pretreated for 2 h with different doses of U0126, a specific inhibitor of the ERK upstream kinase MEK1 (Fig. 5B). The OM-mediated decrease in the amount of PCSK9 protein was partially reversed by U0126 in a concentration-dependent manner. In contrast to inhibition by U0126, preincubation of cells with the PI-3 kinase inhibitor LY294002 at an effective concentration did not affect the OM activity on PCSK9 expression. We further confirmed the role of MEK1 in OM-inhibited PCSK9 expression by silencing its expression with a specific siRNA. Figure 5C shows that MEK1 protein level decreased 60% in MEK1 siRNA-transfected cells compared with that in cells transfected with a nonspecific control siRNA. Despite the incomplete depletion, the inhibitory effect of OM on PCSK9 protein expression was clearly reduced by MEK1 siRNA transfection. This was in line with the results generated by the MEK1 inhibitor. Collectively, these data demonstrate that the ability of OM to repress PCSK9 transcription is mediated in part by the MEK1/ERK signaling pathway.
Fig. 5.
MEK1/ERK signaling pathway is involved in mediating the OM effect on PCSK9 expression. A: HepG2 cells were treated with OM for different times, and total cell lysates were used to probe p-ERK1 and p-ERK2. Anti-ERK2 antibody was used to probe the total ERK in samples for protein loading control (C). B: HepG2 cells were pretreated for 2 h with inhibitor U0126 at the indicated concentrations or with PI-3 kinase inhibitor LY294002 at 20 µM for 2 h before OM was added. PCSK9-P, PCSK9 proprotein; PCSK9-M, mature protein. Cell lysates were harvested 24 h later for Western blotting of PCSK9. C, D: Cells were transfected with si-MEK1 or a control siRNA for 36 h before treatment with OM for 24 h. The protein amounts of MEK1 (C) and PCSK9 (D) were detected by Western blotting.
Cloning of the hamster PCSK9 coding region
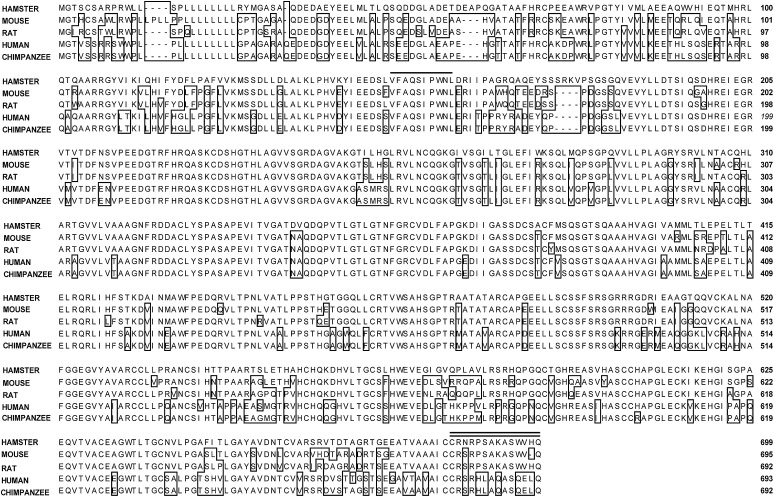
OM has been shown to reduce serum LDL-C in hypercholesterolemic hamsters (28, 29). To determine whether reduction of PCSK9 expression by OM contributed to its cholesterol-lowering activity in hamsters, we first cloned the coding region of hamster PCSK9 cDNA from a hamster liver cDNA library and then compared its protein sequence with those of PCSK9 homologs of mouse, rat, human, and chimpanzee (Fig. 6). The hamster PCSK9 cDNA encodes the hamster precursor protein of 699 amino acids with a calculated molecular mass of 75 kilodaltons. The hamster PCSK9 amino acid sequence has an overall identity of 83.7% to mouse, 82.8% to rat, and 75% to human and chimpanzee PCSK9. Although the divergence in amino acids spreads throughout most regions of the protein, the site of intracellular molecular cleavage (Val-Phe-Ala-Gln↓Ser-Ile-Pro-Trp-Asn-Leu) is identical in all species.
Fig. 6.
Comparison of hamster, mouse, rat, human, and chimpanzee PCSK9 protein sequences. The amino acid residues of mouse, rat, human, and chimpanzee PCSK9 that differed from those of the hamster sequence are boxed. Sequences with a single underline are the consensus cleavage sites. Antigen peptide residues are indicated by a double underline.
Generation and characterization of anti-hamster PCSK9 antibody
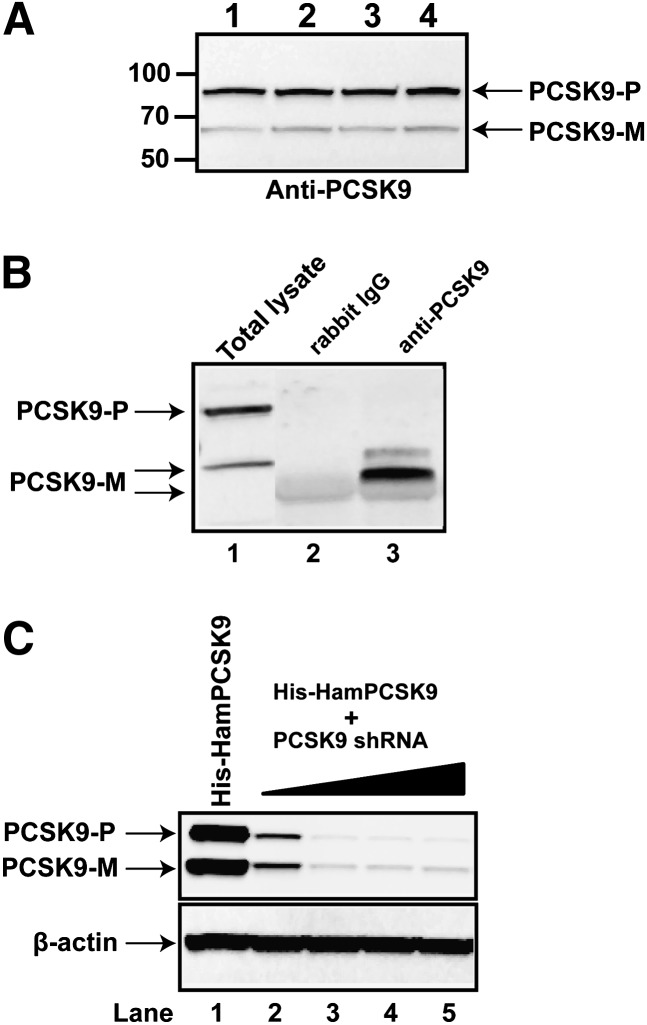
Based upon the amino acid sequence, we generated a rabbit polyclonal antibody against hamster PCSK9 residues 686–699 at the C terminus, a peptide sequence that is unique to hamster PCSK9. To test whether the antibody can specifically recognize the PCSK9 protein, we performed Western blotting of protein extracts prepared from livers of normal-diet-fed hamsters (Fig. 7A). Two bands with approximate molecular masses of 75 and 66 kDa were observed in hamster liver samples, representing the proprotein form of PCSK9 (proPCSK9) and the processed mature form of PCSK9, respectively. Intensities of the 75 kDa bands were 4-fold higher than that of 66 kDa bands, indicating that the antibody predominantly recognized the proprotein in liver extracts. To detect the secreted PCSK9 in serum, we performed immunoprecipitation and Western blotting using the rabbit anti-PCSK9 antibody or normal rabbit IgG as the negative control (Fig. 7B), using freshly isolated serum samples from hamsters fed a normal diet. In the Western blotting results, a sample of total cell lysate of hamster primary hepatocytes was included to mark the positions of proPCSK9 and mature PCSK9 (Fig. 7B, lane 1). Two immunoreactive bands at approximate molecular masses of 66 kDa and 53 kDa were detected in the immunocomplex formed with the anti-PCSK9 antibody (Fig. 7B, lane 3), but these two bands were not detected in the complex formed with normal rabbit IgG (Fig. 7B, lane 2). Interestingly, in contrast to human plasma samples, in hamster serum, the mature protein formed the minor band (∼66 kDa) with an intensity of less than 10% of the lower band (∼53 kDa), suggesting that the majority of secreted PCSK9 is in the short degradation form (Fig. 7B) (32, 33).
Fig. 7.
Detection of hamster PCSK9 in liver and plasma. A: Tissue protein extracts prepared from four individual hamster livers were probed with a rabbit polyclonal anti-PCSK9 antibody. Protein markers shown on the left indicate the protein molecular mass. PCSK9-P, PCSK9 proprotein; PCSK9-M, mature protein. B: A hamster serum sample was analyzed via PCSK9 immunoprecipitation and Western blotting. Total cell lysates prepared from hamster primary hepatocytes were analyzed in lane 1, the immunocomplex of rabbit normal IgG was run in lane 2, and the immunocomplex of anti-PCSK9 antibody was run in lane 3. C: HEK 293 cells were transiently transfected with pHis-HamPCSK9 alone (lane 1) or cotransfected with pSH-HamPCSK9 at the plasmid ratio of 1:1 (lane 2), 1:5 (lane 3), 1:10 (lane 4), or 1:20 (lane 5). Plasmid DNA of the empty cloning vector was used to normalize the total amount of transfected DNAs for each condition. Cell lysates were prepared 48 h after transfection, and the hamster PCSK9 protein was detected by Western blotting using the anti-hamster PCSK9 antibody.
To further test the specificity of the anti-PCSK9 antibody, we transfected HEK293 cells with a hamster PCSK9 expression vector (pHis-hamPCSK9) and different amounts of plasmid pSH-PCSK9 that expressed a shRNA targeting hamster PCSK9 (Fig. 7C). After 48 h of transfection, cell lysates were prepared and subjected to immunoblotting using the anti-PCSK9 antibody. Results showed that the antibody recognized 75 kDa and 66 kDa bands with similar intensities in 293 cells and that this strong signal was markedly diminished in cells cotransfected with increasing amounts of plasmid pSH-PCSK9. Altogether, these experiments confirmed that the anti-PCSK9 antibody specifically recognized the hamster PCSK9 protein in liver extract and plasma.
Downregulation of PCSK9 expression by OM in vivo
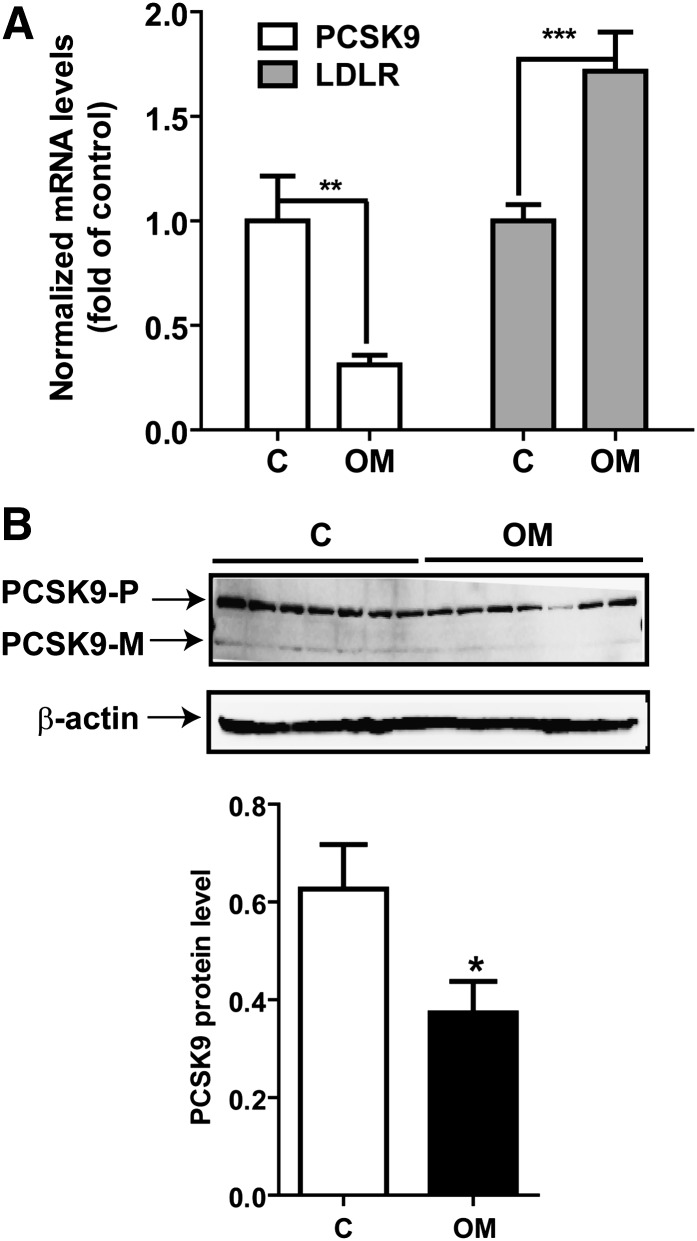
We previously conducted a study to examine the effects of OM on plasma lipid parameters. Hamsters were fed a cholesterol-enriched diet for 2 weeks and then treated with PBS or OM at a dose of 0.2 mg/kg of body weight for 8 days (29). OM treatment lowered serum total cholesterol levels to 53.7% ± 10.4% of that of untreated control cells (P < 0.001) and lowered LDL-C to 71.2% ± 8% of that of untreated control cells (P < 0.001) (supplementary Fig.II). Now, with both the coding sequence and the specific antibody at hand, we assessed the expression of PCSK9 mRNA and protein in liver samples of vehicle (PBS)-treated hamsters and those of OM-treated hamsters. Results of real-time PCR revealed that the level of LDLR mRNA was 1.7-fold increased (P < 0.001) and that the level of PCSK9 mRNA was reduced to 31% of that of control cells (P < 0.01) by the OM treatment (Fig. 8A). Examination of amounts of PCSK9 protein in hamster liver samples by Western blotting corroborated the results of mRNA and showed a 41% decrease in PCSK9 protein abundance (P < 0.05) in OM-treated livers compared with that of the control. Because all serum samples were used for lipid measurements in the previous study (29), we were unable to confirm the reduction of secreted PCSK9 by OM treatment. We further performed Western blotting to detect changes in LDLR protein levels in hamster liver samples. In contrast to HepG2 cells, where both the 160 kDa glycosylated mature form and the 120 kDa unglycosylated precursor form were detected, in hamster liver samples, the anti-LDLR antibody detected only a single band of molecular mass of ∼130 kDa. OM treatment did not appear to increase the abundance of this form of LDLR (supplementary Fig. III). Nevertheless, collectively, these data, including plasma LDL-C reduction, increased LDLR mRNA abundance, and reductions of PCSK9 mRNA and protein provided substantial evidence to support our in vitro findings in HepG2 cells and suggested that the regulatory mechanisms used by OM in the cell culture system also operate in the liver tissue under in vivo conditions.
Fig. 8.
Inhibition of PCSK9 mRNA and protein expression in liver cells of hypercholesterolemic hamsters. A: Total RNA was isolated from 50 mg of liver tissue from hamsters that were untreated or treated with 0.2 mg/kg OM for 8 days (29). Individual levels of LDLR and PCSK9 mRNA were assessed by quantitative real-time PCR using hamster-specific primers. Results are means ± SEM from 5–7 animals per group. **, P < 0.01; and ***, P < 0.001, compared with the vehicle control (C) group. B: Seven individual liver protein extracts from the control group and the OM-treated group were analyzed for PCSK9 protein expression by Western blotting. The PCSK9 protein signals were normalized to the signal intensities of β-actin individually. Values are means ± SEM. *, P < 0.05 compared with the control group. PCSK9-P, PCSK9 proprotein; PCSK9-M, mature protein.
DISCUSSION
We set out to understand the differential kinetics of OM actions on LDLR mRNA and protein expression. This investigation led us to uncover a cytokine-triggered regulatory network that downregulated the expression of PCSK9, a natural inducer of LDLR protein degradation and a promising new therapeutic target in LDL-C metabolism.
We have shown that OM induces a time-dependent reduction of PCSK9 mRNA levels in HepG2 cells. This decrease could result from an accelerated mRNA decay or a reduced rate of transcription. As our data showed that OM did not affect the mRNA half-life of PCSK9, we were able to exclude the possibility of increasing PCSK9 mRNA degradation. Thus, it is conceivable that OM exerts an inhibitory effect on PCSK9 gene transcription; however, the molecular mechanism by which OM inhibits PCSK9 gene transcription is not yet clear.
Previous studies have shown that PCSK9 transcription is controlled through cis regulatory elements located in the proximal promoter region of the PCSK9 gene where the Sp1 sites and HNF1 and SRE-1 are located (11, 13). The induction of PCSK9 transcription by statins (12) and insulin (14) are mediated through SRE-1 and SRE-1-binding proteins. The SRE-1 motif is also involved in the downregulation of PCSK9 transcription by the fibrate class of compounds through a PPAR-mediated mechanism (34). Our previous studies have identified the HNF1 motif as the primary regulatory site for the repression of PCSK9 transcription by BBR, a natural cholesterol-lowering compound (13). To understand how OM exerts its effect on PCSK9 transcription, we analyzed PCSK9 promoter activities in HepG2 cells that either transiently or stably expressed the PCSK9 promoter-driven luciferase reporters PCSK9-D1 and PCSK9-D4. Unexpectedly, we found that both PCSK9-D1, the longer promoter reporter (nt −1711 to −94), and PCSK9-D4, the short functional promoter reporter (nt −455 to −94), were unresponsive to OM treatment, while both promoter activities were induced by SMV and were suppressed by BBR in the same experimental settings.
To identify potential mechanisms underlying the disparity between OM regulation of PCSK9 mRNA and the promoter, we analyzed genomic sequences encompassing the 5′ flanking sequence, exon 1, and the first intron. We noticed that the sequences surrounding exon 1 and intron 1 contain a CpG island (nt +35 to +562, relative to the ATG start codon). OM could potentially induce DNA methylation in this region to inactivate the transcription, thereby lowering PCSK9 mRNA levels. DNA methylation has been implied in the OM-mediated suppression of the synuclein gamma gene expression in breast cancer cells (35). Therefore, we conducted genomic sequencing of bisulfite-modified DNA isolated from untreated and OM-treated HepG2 cells (data not shown). However, we did not find methylated CpG sites within the proximal promoter region, exon1, or intron 1 of the PCSK9 gene, regardless of OM treatment. Thus, we excluded the possibility that PCSK9 transcription was inactivated through epigenetic modification of the PCSK9 gene by OM. In addition, we constructed a luciferase reporter that extended from the 3′ end of the PCSK9 promoter flanking sequence, from nt −94 to + 455 (relative to the ATG start site), to include intron 1 to examine potential regulatory sequences in this region. Again, the promoter reporter was unresponsive to OM treatment. These negative findings open up the possibility that the OM response sequences are possibly located upstream of the currently characterized promoter region (nt −1171 to + 455).
While the precise mechanisms underlying the OM regulation of PCSK9 transcription at promoter levels await further investigation, our results using JAK inhibitors in this study clearly demonstrated this regulation is a JAK-mediated event. We showed that a JAK3-specific inhibitor did not affect the OM activity, but a JAK2-specific inhibitor and a general inhibitor of all JAKs prevented the reduction of PCSK9 expression in OM-treated cells. The specific roles of JAK1 and JAK2 were further confirmed by silencing their expressions. Transfection of specific siRNAs targeted to JAK1 and JAK2 restored the OM-reduced PCSK9 protein levels to that of untreated cells. We noticed that the JAK2 inhibitor lowered PCSK9 mRNA and protein levels in the absence of OM treatment. This effect might not be related to JAK2 activity as depletion of JAK2 by siRNA transfection did not affect PCSK9 protein levels in untreated control cells. The effect of the JAK2 inhibitor on PCSK9 expression in control cells might be caused by unknown off-target effects of the compound.
The JAK/STAT pathway is the principal signaling mechanism by which the IL-6 cytokine family transduces a multitude of signals for regulating various cellular functions (30). Our previous studies have identified STAT3 as an essential transactivator for mediating the OM-induced growth inhibition of breast cancer cells (36). In this study, we showed that depletion of STAT3 as well as STAT1 and STAT5 had no effects on OM-induced suppression of PCSK9 protein (Fig. 4) and mRNA expression (data not shown). Interestingly, the OM effect on PCSK9 can be attenuated by preincubation of cells with U0126, the inhibitor of the ERK upstream kinase MEK1 (37). To a similar extent, the MEK1 siRNA transfection that reduced MEK1 expression also blocked the OM inhibitory activity on PCSK9 expression. The fact that both the inhibitor and the siRNA that target the MEK1/ERK signaling pathway only partially blocked the OM activity implies that additional signaling cascades are used by OM in this biological process. We have used the PI-3 kinase inhibitor LY294002, the p38 MAP kinase inhibitor SB203580, the JNK inhibitor SP600125, and the protein kinase C inhibitor calphostin C in this study. These inhibitors failed to block the effects of OM activity on PCSK9 expression, thereby lessening the likelihood of the involvement of these signaling pathways. Further studies are needed to identify additional signaling pathways that may cross-talk with ERK to fully execute the OM action on PCSK9 transcription.
It is well known that OM has pleiotropic effects that include inflammation, neurogenesis, hematopoiesis, regulation of cell proliferation, and fibrosis. In addition, OM plays a crucial, nonredundant role in liver regeneration after partial hepatectomy or CCl4 treatment (38). OM gene therapy has attenuated liver damage induced by dimethyl nitrosamine (39). Some of these functions of OM overlap with those of other IL-6-type cytokines. However, we have observed the effect of OM in the regulation of lipid metabolism was stronger than that of leukemia inhibitory factor (LIF) (26), a cytokine closely related to OM. Furthermore, the requirement for ERK activation in the regulation of PCSK9 is distinct from the effects of OM on the induction inflammatory genes in which STAT3 activation is required (36). The OM-specific type II receptor is abundantly expressed in liver cells. The interaction of OM with its specific receptor, versus that of LIF-OM with its shared type I receptor, may transduce the OM signaling in liver cells that leads to alterations in lipid metabolism. Currently, it is unknown whether and how the regulation of the LDLR-PCSK9 pathway is related to the beneficial effects of OM in liver development and regeneration. This area of research could be explored in future investigations to gain a better understanding of the physiological roles of OM and the activation of the JAK pathway in liver tissue.
Knockout and transgenic animal models have been applied to study the function of PCSK9 in the control of plasma LDL-C levels (40–44). One advantage of extending those studies to hamsters is the close similarity between lipid metabolism in humans and in hamsters. It is well recognized that the cholesterol lipoprotein distribution in hamsters is close to that in humans (45, 46). Our laboratory has been using hypercholesterolemic hamsters to study the sterol-independent regulation of LDLR by OM under in vivo conditions (28, 29). We have now extended our previous studies based on new findings for PCSK9 regulation in this hamster model. We showed that while LDLR mRNA was 1.7-fold increased compared to that of control cells, the PCSK9 mRNA level in liver was indeed decreased by OM treatment. Through cloning of hamster PCSK9 and generation of a robust anti-hamster PCSK9 antibody, for the first time, we were able to detect PCSK9 protein in liver tissue as well as in sera of hamsters. Because the antibody was generated using the C-terminal peptide of hamster PCSK9, it recognized both the proPCSK9 and the mature PCSK9 in liver extracts with high specificity. Interestingly, with serum samples, the anti-PCSK9 antibody detected the mature PCSK9 as the low-intensity band and the short degradation form as the predominant form of PCSK9 in plasma samples of hamsters fed a normal diet. This pattern was different from those of human and mouse plasma samples in which the mature 60 kDa protein was the major form detected (32, 33, 44). Western blot analysis of PCSK9 protein in liver samples of control and OM-treated hamsters demonstrated a reduction of PCSK9 protein abundance by OM treatment. Unfortunately, the serum samples were not available for detecting plasma PCSK9 in this study. The detection of plasma PCSK9 will be incorporated into our future studies of the effect of OM alone or in combination with statins to examine the antagonism of OM on statin-induced increases in PCSK9 production in vitro and in vivo. In this study of hyperlipidemic hamster liver samples, we did not observe a noticeable increase in LDLR protein abundance by OM treatment. Considering the fact that LDLR mRNA was significantly elevated and that the plasma levels of total cholesterol (TC) and LDL-C were markedly decreased by OM treatment, it is conceivable that the level of functional LDLR protein was increased in OM-treated livers despite the lack of changes in the signal intensity of the approximate 130 kDa band detected by anti-human LDLR antibody. Currently, it is not clear whether the 130 kDa band represents the mature functional form of LDLR. This question will be addressed in our future investigation by developing specific antibodies recognizing hamster LDLR protein.
Supplementary Material
Acknowledgments
The authors thank Dr. Bin Dong for technical help with real-time PCR data analysis and Dr. Michael R. Briggs for critical review of the manuscript.
Footnotes
Abbreviations:
- BBR
- berberine
- HNF1α
- hepatic nuclear factor 1α
- JAK
- Janus kinase
- LDL-C
- LDL cholesterol
- LDLR
- LDL receptor
- LXR
- liver X receptor
- PCSK9
- Abbreviations: proprotein convertase subtilisin/kexin type 9
- RIPA
- radioimmunoprecipitation assay
- RNAi
- RNA interference
- shRNA
- short hairpin RNA
- SIRE
- sterol-independent regulatory element
- siRNA
- short interfering RNA
- SREBP
- sterol regulatory element binding protein
- Tyk
- tyrosine kinase
This study was supported by the Department of Veterans Affairs Office of Research and Development, Medical Research Service, and by National Center for Complementary and Alternative Medicine Grants 1RO1 AT002543-01A1 and 1R21AT003195-01A2.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures and one table.
REFERENCES
- 1.Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Bélanger J., Stifani S., Basak A., Prat A., Chrétien M. 2003. The secretory proprotein convertase neutal apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. U S A. 100: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34: 154–156. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell K. N., Soccio R. E., Duncan E. M., Sehayek E., Breslow J. L. 2003. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J. Lipid Res. 44: 2109–2119. [DOI] [PubMed] [Google Scholar]
- 4.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U S A. 100: 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., et al. 2004. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 279: 48865–48875. [DOI] [PubMed] [Google Scholar]
- 6.Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. 2007. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282: 18602–18612. [DOI] [PubMed] [Google Scholar]
- 7.McNutt M. C., Kwon H. J., Chen C., Chen J. R., Horton J. D., Lagace T. A. 2009. Antagonism of secreted PCSK9 increases low density lipoprotein receptor expression in HepG2 cells. J. Biol. Chem. 284: 10561–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timms K. M., Wagner S., Samuels M. E., Forbey K., Goldfine H., Jammulapati S., Skolnick M. H., Hopkins P. N., Hunt S. C., Shattuck D. M. 2004. A mutation in PCSK9 causing autosomal-dominant hypercholesterolemia in a Utah pedigree. Nat. Genet. 114: 349–353. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J. C., Boerwinkle E., Mosley T. H., Jr., Hobbs H. H. 2006. Sequence variations in PCSK9, LDL, and protection against coronary heart disease. N. Engl. J. Med. 354: 1264–1272. [DOI] [PubMed] [Google Scholar]
- 10.Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., Hobbs H. H. 2005. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37: 161–165. [DOI] [PubMed] [Google Scholar]
- 11.Jeong H. J., Lee H-S., Kim K-S., Kim Y-K., Yoon D., Park S. W. 2008. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J. Lipid Res. 49: 399–409. [DOI] [PubMed] [Google Scholar]
- 12.Dubuc G., Chamberland A., Wassef H., Davignon J., Seidah N. G., Bernier L., Prat A. 2004. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 24: 1454–1459. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Dong B., Park S. W., Lee H-S., Chen W., Liu J. 2009. Hepatocyte nuclear factor 1alpha plays a critical role in PCSK9 gene transcription and regulation by the natural hypocholesterolemic compound berberine. J. Biol. Chem. 284: 28885–28895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costet P., Cariou B., Lambert G., Lalanne F., Lardeux B., Jarnoux A-L., Grefhorst A., Staels B., Krempf M. 2006. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. J. Biol. Chem. 281: 6211–6218. [DOI] [PubMed] [Google Scholar]
- 15.Careskey H. E., Davis R. A., Alborn W. E., Troutt J. S., Cao G., Konrad R. J. 2008. Atovastatin increases human serum levels of proprotein convertase subtilisin/kexin type 9. J. Lipid Res. 49: 394–398. [DOI] [PubMed] [Google Scholar]
- 16.Mayne J., Dewpura T., Raymond A., Cousines M., Chaplin A., Lahey K., Lahaye S. A., Mbikay M., Ooi T. C., Chrétien M. 2008. Plasma PCSK9 levels are significantly modified by statins and fibrates in humans. Lipids Health Dis. 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidah N. G. 2009. PCSK9 as a therapeutic target of dyslipidemia. Expert Opin. Ther. Targets. 13: 19–28. [DOI] [PubMed] [Google Scholar]
- 18.Cao G., Qian Y-W., Kowala M. C., Konrad R. J. 2008. Further LDL cholesterol lowering through targeting PCSK9 for coronary artery disease. Endocr. Metab. Immune Disord. Drug Targets. 8: 238–243. [DOI] [PubMed] [Google Scholar]
- 19.Hedrick J. A. 2009. Targeting PCSK9 transcription for the treatment of hypercholesterolemia. Curr.Opin.in Inves. Drugs. 10: 938–946. [PubMed] [Google Scholar]
- 20.Liu J., Grove R. I., Vestal R. E. 1994. Oncostatin M activates the LDL receptor transcription in sterol-repressed liver cells. Cell Growth Differ. 5: 1333–1338. [PubMed] [Google Scholar]
- 21.Liu J., Streiff R., Zhang Y. L., Vestal R. E., Spence M. J., Briggs M. R. 1997. Novel mechanism of transcriptional activation of hepatic LDL receptor by oncostatin M. J. Lipid Res. 38: 2035–2048. [PubMed] [Google Scholar]
- 22.Liu J., Briggs M. R., Kraemer F. B. 2006. Elucidation of an SRE-1/SREBP-independent cellular pathway for LDL-receptor regulation: from the cell surface to the nucleus. Future Cardiol. 2: 605–612. [DOI] [PubMed] [Google Scholar]
- 23.Liu J., Ahlborn T. E., Briggs M. R., Kraemer F. B. 2000. Identification of a novel sterol-independent regulatory element in the human low density lipoprotein receptor promoter. J. Biol. Chem. 275: 5214–5221. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F., Ahlborn T. E., Li C., Kraemer F. B., Liu J. 2002. Identification of Egr1 as the oncostatin M-induced transcription activator that binds to the sterol-independent regulatory element of the human LDL receptor promoter. J. Lipid Res. 43: 1477–1485. [DOI] [PubMed] [Google Scholar]
- 25.Zhang F., Lin M., Abidi P., Thiel G., Liu J. 2003. Specific interaction of Egr1 and c/EBP-beta leads to the transcriptional activation of the human low density lipoprotein receptor gene. J. Biol. Chem. 278: 44246–44254. [DOI] [PubMed] [Google Scholar]
- 26.Li C., Kraemer F. B., Ahlborn T. E., Liu J. 1999. Induction of low density lipoprotein receptor (LDLR) transcription by oncostatin M is mediated by the extracellular signal-regulated kinase signaling pathway and the repeat 3 element of the LDLR promoter. J. Biol. Chem. 274: 6747–6753. [DOI] [PubMed] [Google Scholar]
- 27.Liu J., Zhang F., Li C., Lin M., Briggs M. R. 2003. Synergistic activation of human LDL receptor expression by SCAP ligand and cytokine oncostatin M. Arterioscler. Thromb. Vasc. Biol. 23: 90–96. [DOI] [PubMed] [Google Scholar]
- 28.Kong W., Abidi P., Jiang J. D., Liu J. 2005. In vivo activities of cytokine oncostatin M in the regulation of plasma lipid levels. J. Lipid Res. 46: 1163–1171. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y., Abidi P., Kim A., Chen W., Huang T-T., Kraemer F. B., Liu J. 2007. Transcriptional activation of hepatic ACSL3 and ACSL5 by oncostatin M reduces hypertriglyceridemia through enhanced beta-oxidation. Arterioscler. Thromb. Vasc. Biol. 27: 2198–2205. [DOI] [PubMed] [Google Scholar]
- 30.Heinrich P. C., Behrmann I., Muller-Newen G., Schaper F., Graeve L. 1998. Interleukin-6-type cytokine signaling through the gp130/Jak/STAT pathway. Biochem. J. 334: 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth D. C., Kerr C., Li Y., Tang D., Richard C. D. 2008. Oncostatin M induction of eotaxin-1 expression requires the convergence of PI3′K and ERK1/2 MAPK signal transduction pathways. Cell. Signal. 20: 1142–1150. [DOI] [PubMed] [Google Scholar]
- 32.Benjannet S., Rhainds D., Hamelin J., Nassoury N., Seidah N. G. 2006. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications. J. Biol. Chem. 281: 30561–30572. [DOI] [PubMed] [Google Scholar]
- 33.Dubuc G., Tremblay M., Paré G., Jacques H., Hamelin J., Benjannet S., Boulet L., Genest J., Bernier L., Seidah N. G., et al. 2010. A new method for measurement of total plasma PCSK9: clinical applications. J. Lipid Res. 51: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kourimate S., May C. L., Langhi C., Jarnoux A. L., Ouguerram K., Zaïr Y., Nguyen P., Krempf M., Cariou B., Costet P. 2008. Dual mechanisms for the fibrate-mediated repression of proprotein convertase subtilisin/kexin type 9. J. Biol. Chem. 283: 9666–9673. [DOI] [PubMed] [Google Scholar]
- 35.Gupta A., Godwin A. K., Vanderveer L., Lu A., Liu J. 2003. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 63: 664–673. [PubMed] [Google Scholar]
- 36.Zhang F., Li C., Halfter H., Liu J. 2003. Delineating an oncostatin M-activated STAT3 signaling pathway that coordinates the expression of genes involved in cell cycle regulation and extracellular matrix deposition of MCF-7 cells. Oncogene. 22: 894–905. [DOI] [PubMed] [Google Scholar]
- 37.Favata M. F., Horiuchi K. Y., Manos E. J., Daulerio A. J., Stradley D. A., Feeser W. S., Van Dyk D. E., Pitts W. J., Earl R. A., Hobbs F., et al. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273: 18623–18632. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura K., Nonaka H., Saito H., Tanaka M., Miyajima A. 2004. Hepatocyte proliferation and tissue remodeling is impaired after liver injury in oncostatin M receptor knockout mice. Hepatology. 39: 635–644. [DOI] [PubMed] [Google Scholar]
- 39.Hamada T., Sato A., Hirano T., Yamamoto T., Son G., Onodera M., Torii I., Nishigami T., Tanaka M., Miyajima A., et al. 2007. Oncostatin M gene therapy attenuates liver damage induced by dimethylnitrosamine in rats. Am. J. Pathol. 171: 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rashid S., Curtis D. E., Garuti R., Anderson N. N., Bashmakov Y., Ho Y. K., Hammer R. E., Moon Y-A., Horton J. D. 2005. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. U S A. 102: 5374–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le May C., Kourimate S., Langhi C., Chétiveaux M., Jarry A., Comera C., Collet X., Kuipers F., Krempf M., Cariou B., et al. 2009. Proprotein convertase subtilisin/kexin type 9 null mice are protected from postprandial triglyceridemia. Arterioscler. Thromb. Vasc. Biol. 29: 684–690. [DOI] [PubMed] [Google Scholar]
- 42.Luo Y., Warren L., Xia D., Jensen H., Sand T., Petras S., Qin W., Miller K. S., Hawkins J. 2009. Function and distribution of circulating human PCSK9 expressed extrahepatically in transgenic mice. J. Lipid Res. 50: 1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herbert B., Patel D., Waddington S. N., Eden E. R., McAleenan A., Sun X. M., Soutar A. K. 2010. Increased secretion of lipoproteins in transgenic mice expressing human D374Y PCSK9 under physiological genetic control. Arterioscler. Thromb. Vasc. Biol. 30: 1333–1339. [DOI] [PubMed] [Google Scholar]
- 44.Zaid A., Roubtsova A., Essalmani R., Marcinkiewicz J., Chamberland A., Hamelin J., Tremblay M., Jacques H., Jin W., Davignon J., et al. 2008. Proprotein convertase subtilisin/kexin type 9 (PCSK9): hepatocyte-specific low-density lipoprotein receptor degradation and critical role in mouse liver regeneration. Hepatology. 48: 646–654. [DOI] [PubMed] [Google Scholar]
- 45.Ohtani H., Hayashi K., Hirata Y., Dojo S., Nakashima K., Nishio E., Kurushima H., Saeki M., Kajiyama G. 1990. Effects of dietary cholesterol and fatty acids on plasma cholesterol level and hepatic lipoprotein metabolism. J. Lipid Res. 31: 1413–1422. [PubMed] [Google Scholar]
- 46.Sullivan M. P., Cerda J. J., Robbins F. L., Burgin C. W., Beatty R. J. 1993. The gerbil, hamster, and guinea pig as rodent models for hyperlipidemia. Lab. Anim. Sci. 43: 575–578. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.