Abstract
Cranial neural crest (CNC) is a multipotent migratory cell population that gives rise to most of the craniofacial bones. An intricate network mediates CNC formation, epithelial-mesenchymal transition, migration along distinct paths, and differentiation. Errors in these processes lead to craniofacial abnormalities, including cleft lip and palate. Clefts are the most common congenital craniofacial defects. Patients have complications with feeding, speech, hearing, and dental and psychological development. Affected by both genetic predisposition and environmental factors, the complex etiology of clefts remains largely unknown. Here we show that Fas-associated factor-1 (FAF1) is disrupted and that its expression is decreased in a Pierre Robin family with an inherited translocation. Furthermore, the locus is strongly associated with cleft palate and shows an increased relative risk. Expression studies show that faf1 is highly expressed in zebrafish cartilages during embryogenesis. Knockdown of zebrafish faf1 leads to pharyngeal cartilage defects and jaw abnormality as a result of a failure of CNC to differentiate into and express cartilage-specific markers, such as sox9a and col2a1. Administration of faf1 mRNA rescues this phenotype. Our findings therefore identify FAF1 as a regulator of CNC differentiation and show that it predisposes humans to cleft palate and is necessary for lower jaw development in zebrafish.
Introduction
The formation of the jaw, a pharyngeal arch derivative, depends on the migration of cranial neural crest cells from one part of the embryo to another.1 Defects in this process cause cleft clip and/or palate (OFC1 [MIM 119530]), a common craniofacial birth defect (1/700 live births).2 Clefts are notable for their lifelong morbidity. Patients often present complex problems, and they have to undergo several surgical and nonsurgical treatments that can considerably reduce the quality of life and have a major impact on the patients and their families. According to epidemiological and genetic studies, clefts are divided into cleft lip with or without palate (CL/P) and cleft palate only (CPO).2 They are mainly nonsyndromic (70% of CL/P and 50% of CPO) and have a complex, largely unknown etiology.3 Genetic factors involved in isolated clefts have been identified on the basis of association studies, animal models, and role in known human cleft syndromes. The Interferon regulatory factor-6 gene (IRF6 [MIM 607199]) has been shown to be associated with isolated CL/P across populations,4,5 and loci such as 8q24,6 10q25.37 and 17q227 are predisposing factors. Nevertheless, no gene has been shown to be associated with isolated CPO. The Pierre Robin Sequence (PRS [MIM 261800]) is a subgroup of cleft palate. It is characterized by CPO, micrognathia, and glossoptosis8 and was first described by Robin (1923) as a condition in which the tongue tends to obstruct the airway and lead to feeding and respiratory difficulties during the early postnatal period. Some insights into its etiology have been obtained by studies showing association with conserved noncoding elements flanking the SRY-box 9 gene (SOX9 [MIM 608160]).9
In this study, we characterize the role of FAF1 [MIM 604460] in orofacial development. We show that FAF1 haploinsufficiency causes CPO in one PRS family. Sporadic CPO patients have significantly decreased FAF1 expression. Furthermore, association studies conducted on 7597 individuals show that the locus is strongly associated with CPO; on average, relative risk is increased 1.47-fold in individuals with the associated allele. Because the role of FAF1 during development was not known, we characterized its function in zebrafish embryos. Results show that Faf1 is needed for the development of pharyngeal arches and for CNC differentiation into cartilage. We conclude that FAF1 plays a role in the etiopathogenesis of cleft palate and is a key molecule in development.
Material and Methods
Families
Data from families in the Belgian cohort were collected in collaboration with the CL/P Center, Cliniques Universitaires Saint-Luc, Brussels, Belgium and the CHU de Lille, Hôpital Jeanne de Flandre, Lille, France. We obtained informed consent from participants prior to conducting the study, as approved by the institutional review board and local ethics committees. Patients were evaluated by contributing physicians and asked about familial cleft history. Venous blood samples were drawn for each participant. DNA was extracted from whole blood and/or buccal cell swabs.10 For the family with a translocation, no DNA was available from the aunt or the grandparents. RNA was extracted from lymphocytes with the TriPure Isolation Reagent (Roche), and cDNA synthesis was performed with the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer's instructions.
Cytogenetics
BAC and fosmid clones were identified via the Ensembl and UCSC genome browser databases and obtained from the BACPAC Resource Center (CHORI). Karyotyping and metaphase spreading for fluorescence in situ hybridization (FISH) analysis were made from a fixed cell suspension of 3-day-cultured, phytohemagglutinin-stimulated peripheral blood lymphocytes. DNA probes were differentially labeled with the BioPrime DNA labeling System (Invitrogen). Probes were denatured, pre-annealed with human Cot-1 DNA (Invitrogen), and hybridized onto the denatured slides overnight at 37°C in a moist chamber. Signals were detected with FITC-avidin and rhodamine- antidigoxigenin monoclonal antibody. Slides were air-dried and mounted with Vectashield and DAPI. At least five metaphases were analyzed for each hybridization with a Zeiss Axioplan 2 epifluorescence microscope (Carl Zeiss). Metaphases were captured with Isis software (Metasystems GmbH).
Microdissection, Reverse Array, and Chromosome Painting
Chromosome microdissection, reverse array, and chromosome painting were done as previously described.11 In brief, fresh cells were dropped on a coverslip before trypsin-Giemsa banding was performed. Under an inverted microscope, rearranged chromosomes (1 and 2) were identified and dissected with a glass needle. Four copies were separately transferred into a collection tube and incubated with Proteinase K at 37°C for 2 hr. DNA was amplified by DOP-PCR, tested for amplification efficiency, and labeled by a second-round DOP-PCR with the SpectrumOrange-dUTP fluorescent dye. Chromosomal microdissection was tested by reverse hybridization to normal human metaphases as a control, and by array-based comparative genomic hybridization using arrays described elsewhere.11
Multiplex Polymerase Chain Reaction/Liquid Chromatography Assay
A 316 bp and a 377 bp cDNA fragment from FAF1 (MIM 604460) and a Ras p21 protein activator 1 (RASA1 [MIM 139150]) control gene, respectively, were amplified via multiplex polymerase chain reaction (PCR). Aliquots of 5 μl of PCR products were injected on a semi-automated 3500 Wave HS system (Transgenomic). DNA was eluted, mixed with the Wave HS Staining Solution I, and detected via fluorescence. Data were analyzed with Navigator 1.6.4 software.
Association Studies
Association studies were conducted on a cohort of 7597 available individuals of different geographic origins (2530 trios, each including a patient with nonsyndromic cleft and that patient's two parents) (Table 1). Cleft patients from Belgium, the Eurocran and Italcleft study (see Web Resources), Germany, Texas, Iowa, and the Philippines were genotyped for a total of ten SNPs capturing as much information as possible: tagging SNPs (r2 ≥ 0.8), a high coverage of FAF1, and/or a minor allele frequency cutoff of 0.2 (Figure S1). We analyzed the different cohorts separately to replicate FAF1 association results. We then merged populations to increase the statistical power of the calculations. We performed a transmission disequilibrium test (TDT) by subdividing the data on the basis of proband cleft type. Genotypes were considered from complete parent-child trios (all incomplete trios or trios with incomplete genotypes were excluded). Association between markers and clefts was tested with FBAT and APL (See Table 1 for SNPs genotyped).12,13 Relative risks (RR) associated with the genotype of the child were calculated via a log-linear approach under the assumption of a recessive model (risk of TT genotypes compared to TC + CC).14
Table 1.
Association Results
| Population Origin (in Collaboration with) | Genotyped SNPs (Major/Minor allele) | Cleft Type | Number of Individuals (T = 7597) | Number of Affected Individuals | Associated SNP (Associated Allele) | Familial History | TDT p Value rs1149795 | TDT p Value rs3827730 | Relative risk of CPO Conferred by TT Genotype of rs3827730; p value |
|---|---|---|---|---|---|---|---|---|---|
| Belgium | rs6588392 (C/G), rs11205768 (T/C), rs17387761(G/A), rs3827730 (T/C) | mixed | 1072 | 421 | rs3827730 (T) | S | N/A | 0.004 | |
| CPO | 325 | 126 | rs3827730 (T) | S | N/A | 0.001 | 2.72 (1.49-4.98); p = 0.001 | ||
| CL | 227 | 94 | none | S | N/A | 0.23 | |||
| CLP | 520 | 202 | none | S | N/A | 0.45 | |||
| CL/P | 747 | 296 | none | S | N/A | 0.15 | |||
| European descent- Iowa, USA (J. Murray) | rs11205760 (C/T), rs1149795 (T/C), rs3827730 (C/T), rs2784143 (C/T), rs2455636 (G/A) | mixed | 981 | 350 | rs1149795 (C) | F + S | 0.006 | 0.16 | |
| CPO | 246 | 90 | rs1149795 (C) | F + S | 0.02 | 0.34 | 1.23 (0.6–2.53); p = 0.569 | ||
| CL | 275 | 91 | none | F + S | 0.67 | 0.78 | |||
| CLP | 412 | 146 | none | F + S | 0.61 | 0.34 | |||
| CL/P | 42 | 22 | none | F + S | 0.1 | 0.65 | |||
| European descent- Texas, USA (J. Hecht) | rs3827730 (T/C) | CL/P | 499 | N/A | rs3827730 (T) | F + S | N/A | 0.38 | |
| Mexican Hispanics- Texas, USA (J. Hecht) | rs3827730 (T/C) | CL/P | 177 | N/A | rs3827730 (T) | F + S | N/A | 0.14 | |
| Philippines (J. Murray) | rs11205760 (C/T), rs1149795 (T/C), rs3827730 (T/C), rs2784143 (C/T), rs2455636 (G/A) | mixed | 2511 | 570 | none | F | 0.73 | 0.62 | |
| CPO | 22 | 4 | none | F | NI | 0.52 | |||
| CL | 429 | 98 | none | F | 1 | 0.93 | |||
| CLP | 1203 | 246 | none | F | 0.86 | 0.78 | |||
| CL/P | 823 | 212 | none | F | 0.51 | 0.29 | |||
| Germany (E. Mangold) | rs11205760 (C/T), rs17387761 (G/A), rs11587909 (C/T), rs17382596 (T/G), rs3827730 (T/C) | mixed | 1514 | 679 | none | N/A | N/A | 0.16 | |
| CPO | 242 | 99 | none | N/A | N/A | 0.78 | 1.17 (0.63-2.17); p = 0.624 | ||
| CL | 225 | 99 | none | N/A | N/A | 1 | |||
| CLP | 1047 | 481 | none | N/A | N/A | 0.12 | |||
| CL/P | 1272 | 580 | none | N/A | N/A | 0.16 | |||
| Eurocran and Italcleft: IT, NL, UK, HU, SL, ET, SK, and BU (M. Rubini) | rs1149795 (T/C), rs3827730 (C/T), rs17382596 (T/G) | CPO | 843 | 281 | rs3827730 (T) | S | 0.67 | 0.007 | 1.38 (1.02-1.87); p = 0.036 |
| NL | CPO | 165 | 55 | none | S | N/A | 0.42 | 1.02 (0.51-2.05); p = 0.955 | |
| IT | CPO | 183 | 61 | rs3827730 (T) | S | N/A | 0.02 | 2.04 (1.06-3.96); p = 0.034 | |
| UK | CPO | 201 | 67 | none | S | N/A | 0.43 | 1.44 (0.77-2.70); p = 0.252 | |
| Eurocran Western Europe (NL + IT + UK) | CPO | 549 | 183 | rs3827730 (T) | S | N/A | 0.03 | 1.43 (0.98- 2.08); p = 0.064 | |
| Eurocran Eastern Europe (HU, SL, ET, SK, BU) | CPO | 294 | 98 | none | S | N/A | 0.09 | 1.38 (0.82-2.30); p = 0.224 | |
| Belgium + Germany + Eurocran and Italcleft | rs3827730 (C/T) | CPO | 1410 | 506 | rs3827730 (T) | S+N/A | N/A | 0.0007 | 1.51 (1.18-1.93); p = 0.001 |
| Belgium + Germany + Eurocran and Italcleft + Iowa | rs3827730 (C/T) | CPO | 1656 | 596 | rs3827730 (T) | F+S | N/A | 0.0003 | 1.47 (1.17-1.86); p = 0.001 |
TDT was done with the FBAT program for all populations except the European-descent cohort from Texas (APL). Notations are as follows: mixed, all cleft types together (CPO + CL + CLP); CPO, cleft palate only; CL, cleft lip; CLP, cleft lip and palate; CL/P, ceft lip with or without cleft palate; F, familial; S, sporadic; N/A, not available; NI, not informative; T, total; IT, Italy; NL, The Nerherlands; UK, The United Kingdom; HU Hungary; SL Slovenia; ET, Estonia; SK, Slovakia; and BU, Bulgaria. Note that in the CPO cohort from Belgium, 38 patients had isolated PRS (CPO + retrognatia), all others had CPO.
Mutation Analysis
A pilot cohort of available patients was screened for mutations in FAF1 (NM_007051.2). In total, 228 individuals with different cleft types were analyzed. Because mutations in IRF6 account for 63% of patients with van der Woude syndrome (VWS [MIM 119300]),15 we included VWS patients for whom IRF6 was screened and excluded (n = 14).16 The cohort also included patients with CPO (n = 65), PRS (n = 34), and CL/P (n = 115). Exons 1–21 and corresponding exon-intron boundaries of FAF1 were PCR-amplified from genomic DNA and subjected to mutation analysis via denaturing high-performance liquid chromatography (DHPLC) on a semi-automated 3500 Wave HS system (Transgenomic). Each sample with an abnormal profile was purified (QIAGEN) and sequenced on a CEQ2000 fluorescent capillary sequencer (Beckman Coulter). We used DHPLC to assess cosegregation of identified changes with the phenotype.
Real-Time Quantitative PCR
Two controls and nine patients (5 CLP, 4 CPO) from whom RNA from lymphocytes was available were chosen for FAF1 expression studies. Real-time qPCR was performed in an iCycler thermocycler and MyiQ detection System (Bio-Rad) with the SYBR Green PCR Master Mix (Applied Biosystems). Quantification of FAF1 was based on standard curves constructed from 5-fold serially diluted normal genomic DNA. The relative copy number of FAF1 was calculated against ACTB.
Zebrafish Whole-Mount In Situ Hybridization and Alcian Blue Staining
Dechorionated embryos were fixed overnight in 4% paraformaldehyde at 4°C. Whole-mount in situ hybridization was performed as described17 with antisense probes for faf1, cmlc2,18 islet1,19 myoD,20 snail1b,21 crestin,22 sox10,23 foxd3,24 dlx2,25 sox9a,26 and col2a1.27 Cartilages were visualized with Alcian blue according to a modified protocol.28 In brief, 5 dpf larvae were fixed and washed several times in phosphate-buffered saline with 0.1% Tween-20 (PBST). Subsequently, embryos were transferred into a filtered Alcian blue solution (1% concentrated hydrochloric acid, 70% ethanol, and 0.1% Alcian blue) and stained overnight at room temperature. Specimens were cleared in acidic ethanol (5% concentrated hydrochloric acid and 70% ethanol) for 4 hr, rehydrated in an ethanol series, and stored in 70% glycerol/PBS. Stained embryos were imaged with a Zeiss Lumar V12 stereomicroscope.
Zebrafish faf1 Knockdown Analysis
Tg(fli1:EGFP)y1 zebrafish29 were maintained under standard laboratory conditions. The following morpholino oligonucleotides were purchased from Gene Tools: 5′- TGTCCATGTTGCTGTCGGAGGCCAT-3′ (faf1-MO-ATG), 5′- ATGTGAGAAAACCCACCTGAAAGTC-3′ (faf1-MO-Splice), both used for knockdown of faf1 expression, and 5′- TGTgCATcTTGCTcTCGGAcGCgAT-3′, mismatched at five base pairs, as a control. Different doses of morpholinos were injected into single- to four-cell stage zebrafish embryos via procedures previously described.17 All data shown were obtained after injection of 2–6 ng faf1-MO-ATG or 8–16 ng faf1-MO-Splice per embryo. At least 30–40 injected embryos were analyzed per experiment,and each experiment was repeated at least three times. We scored the penetrance of the phenotype by counting the affected embryos. To block apoptosis, we coinjected a p53 targeting morpholino (5′- GACCTCCTCTCCACTAAACTACGAT-3′) at 1× to 1.5× the concentration of the ATG or splice-blocking morpholino.30 To test efficacy of faf1-MO-ATG, we cloned the 25 nucleotides of the 5′ UTR, the ATG, and 40 nucleotides of the coding sequence of faf1 (containing the ATG) in an expression vector, upstream of and in frame with the luciferase cDNA (lacking the initiator ATG codon). Expression of this chimeric reporter protein in the erythrocyte lysate assay was monitored via luminometry. We tested the efficacy of faf1-MO-Splice by RT-PCR on extracts of morphant embryos by using primers allowing size-discrimination between correctly and erroneously spliced mRNA.
Phenotypic Rescue with Zebrafish faf1 mRNA
For rescue experiments, zebrafish faf1 full-length coding sequence was subcloned into the pCS2+ vector for in vitro transcription. The mRNA was synthesized with Sp6 RNA polymerase and the message machine kit (Ambion). Two hundred pg of mRNA was injected into 1-cell-stage embryos together with faf1-MO-Splice.
Results
We identified a translocation present in a multigenerational family affected by CPO and PRS. We characterized the breakpoints and demonstrated that they occur within a gene. We investigated the role of this gene in cleft cohorts by conducting expression and association studies, as well as relative risk analysis. We performed a series of morpholino knockdown and rescuing assays, whole-embryo staining, and in situ hybridization to unravel the role of this gene during zebrafish embryological development.
FAF1 Haploinsufficiency in Syndromic CPO
The proband is a 4-year-old boy who is of European descent and has Pierre Robin sequence. The boy presented a cleft of the secondary palate and associated micro/retrognathia, microstomia, malar hypoplasia, and oligodontia (Figures 1A, 1B, and 1C). His father and aunt were similarly affected (Figure 1A). The karyotypes of the father and the child were 46,XY,t(1;2)(p34;q33), and Affymetrix 50K SNP Chip analysis revealed no copy-number variation (data not shown). FISH analysis, chromosome microdissection followed by reverse array, chromosome painting, and PCR on derivative chromosome 1 revealed that the 1p breakpoint interrupted FAF1 within its first intron (Figures 1D, 1E, and 1G). A mulitplex PCR/liquid chromatography (MP/LC) assay demonstrated lowered FAF1 expression in lymphocytes of the child compared to controls (Figure 1F and data not shown). Analysis of the 2q breakpoint showed that the region is not well conserved between humans and mice, and it has no sites for microRNAs or regulatory elements. It contains no genes, UTRs, LINEs, SINEs, or other repetitive interspersed or tandem-repeat elements. Thus, it is unlikely to play a role in the observed inherited phenotype.
Figure 1.
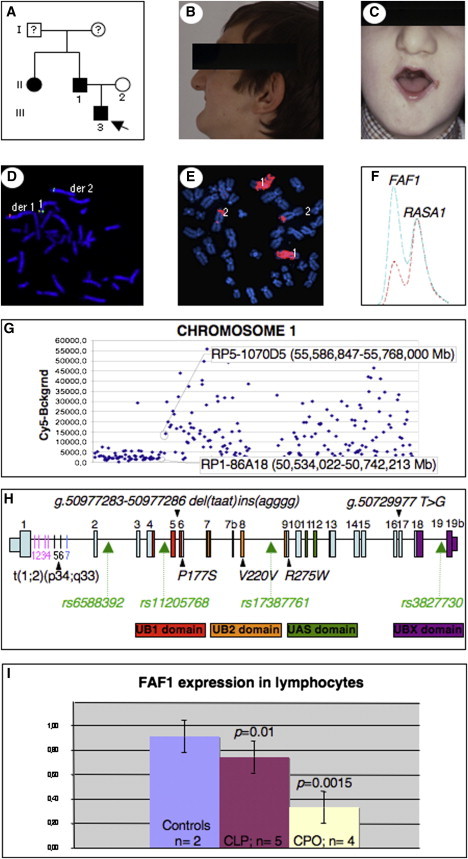
Identification of t(1;2) Breakpoint of the 46,XY,t(1;2)(p34;q33) Translocation
(A) Pedigree of family. An arrow indicates the proband; filled symbols indicate affected individuals.
(B and C) Father (during adolescence, after surgery), with micro/retrognathia, microstomia, and translocation.
(D) FISH with fosmid G248P84517F11 (green) spanning 1p breakpoint and fosmid G248P81885B3 (red) centromeric to 1p breakpoint.
(E) Reverse chromosome painting of microdissected derivative chromosome 1 red-labeled and hybridized to normal metaphases.
(F) MP/LC multiplex chromatogram for FAF1/RASA1. Chromatograms represent fluorescence intensity and are normalized to the RASA1 control amplicon. Blue indicates a control individual; red indicates a patient with a translocation. RASA1 is the Ras p21 protein activator 1 gene.
(G) Mapping of amplified microdissected aberrant chromosome 1 by array CGH. Dark squares indicate log-2-transformed intensity ratios. Clones are ordered from 1p tel to 1q tel. The first clones with decreased and increased intensities are indicated.
(H) FAF1 structure. Exons are positioned to scale. For colored exons, the FAF1 domains are denoted below. Probes 1–4 (pink bars in intron 1) were amplified and sequenced from aberrant chromosome 1; probes 5 and 6 (black) were not amplifiable from either derived chromosome, and probe 7 (blue) was amplified and sequenced from aberrant chromosome 2. Translocation breaks FAF1 in intron 1. Arrowheads indicate the position of identified changes and polymorphism; green arrows indicate SNPs genotyped in the Belgian cohort.
(I) qPCR of FAF1 on lymphocytes from controls, CL/P, and CPO patients.
FAF1 Association with Clefts
In the overall dataset of isolated CPO trios, the T allele of rs3827730 was significantly overtransmitted from heterozygous parents to affected children (p = 0.0003), although there were wide differences between countries (Table 1). Over-transmission of the T allele was significant in Belgium (p = 0.001). Positive association was replicated in the Italian cohort (p = 0.02) and the Western European cohort (p = 0.03). A trend of overtransmission was observed in all countries except for Germany. The TT genotype of rs3827730 in the child was associated with an overall 1.47-fold increased risk of CPO (when all 474 trios were considered) (Table 1). The association was significant in Belgium and Italy (2.72- and 2.04-fold increased risks, respectively), but it was absent in Germany and the Netherlands. Eastern Europe and the United Kingdom had an approximately 1.4-fold increased risk. The Iowan CPO subgroup showed association with the C allele of rs1149795 (p = 0.02). When all European trios were considered, parental imprinting was not significant (data not shown).
Decreased FAF1 Expression in Patients with Clefts but without a Mendelian Mutation
Screening for FAF1 mutations in the cohort of 228 individuals with cleft revealed four changes, including two in the coding region (Figure 1H and Table S1). A p.Pro177Ser (c.529C>T) substitution was found in one PRS family, and a p.Arg275Trp (c.823C>T) substitution was found in two unrelated individuals with CL/P and CL. Screening age-matched controls revealed 3/600 individuals with p.Pro177Ser (c.529C>T) and 3/400 with p.Arg275Trp (c.823C>T). A synonymous change, p.Val220Val (c.660A>G), was present in 28.3% of 400 unaffected controls and 26.3% of patients. Neither change was statistically significantly associated with clefts. Affymetrix SNP Chips did not unravel any copy-number variation at 1p34 in 140 cleft patients (data not shown).
Because changes in regulatory regions play an important role in complex diseases, such as clefts, FAF1 expression was studied by quantitative RT-PCR on peripheral blood lymphocytes from control individuals (n = 2) versus cleft patients (n = 9). A significant decrease in expression was observed in CPO (p = 0.0015) and CL/P (p = 0.01) (Figure 1I). Genotyping the patients for rs3827730 revealed an enrichment of the T allele (75% of alleles in CPO and 60% of alleles in CL/P patients).
Severe Jaw Defects in faf1-Knockdown Zebrafish Embryos
In zebrafish embryos, at 24 and 30 hr post-fertilization (hpf), faf1 transcripts were detected by whole-mount in situ hybridization in the pharyngeal arch primordia that give rise to cranial cartilages (Figures 2A and 2B). At 56 hpf, when the pharyngeal cartilages are forming, faf1 was strongly expressed in all arches; the most prominent expression was in the first (mandibular) and second (hyoid) arch (Figures 2C and 2D).
Figure 2.
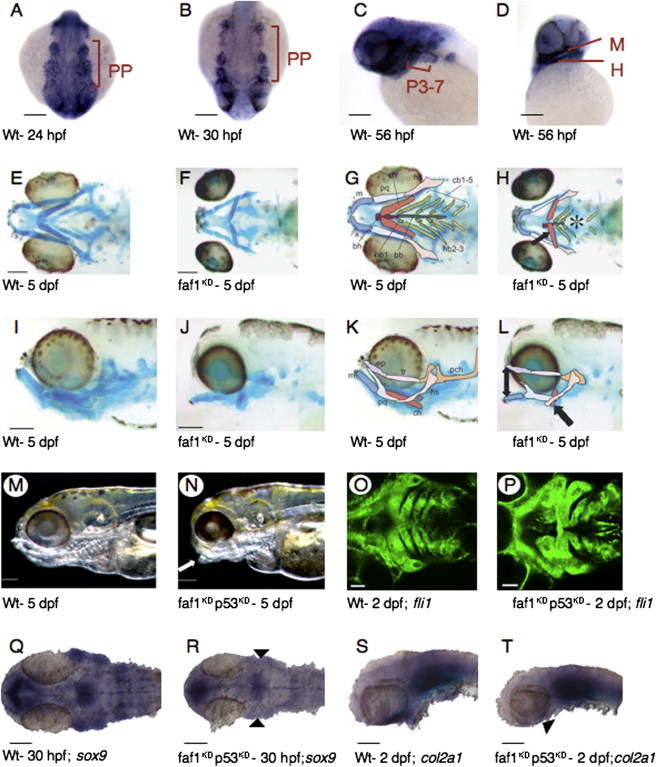
faf1 Expression Studies and Pharyngeal Arch Development in Zebrafish Larvae
(A–D) Expression pattern of faf1 in control zebrafish of different stages is shown by whole-mount in situ hybridization. At 24 hpf and 30 hpf, strong faf1 expression in pharyngeal arch primordia (PP) (A and B). Dorsal views of the pharyngeal region are shown; the rostral side is down. At 56 hpf, there is strong faf1 expression in all arches, and the most prominent expression is in the first (mandibular, M) and second (hyoid, H) arches (C and D). Scale bars represent 100 μm.
(E–L) Ventral and side views of Alcian-blue-stained 5 dpf wild-type (E, G, I, and K) and faf1KD (F, H, J, and L) larvae; the focus is on pharyngeal cartilage. At 6 ng/embryo, all cartilages are generally smaller, ceratohyal cartilage is oriented more medially (black arrows), branchial cartilages are partially missing (black asterisk), and Meckel's cartilage is oriented ventrally (double black arrow), resulting in an open-mouth-like appearance. Abbreviations for cartilege are as follows: bb, basibranchial; bh, basihyal; cb, ceratobranchial; ch, ceratohyal; ep, ethmoid plate; hb, hypobranchial; hs, hyosymplectic; m, Meckel's; pch, parachordal; pq, palatoquadrate; and tr, trabecula. Scale bars represent 200 μm.
(M and N) Macroscopic side views of the head of 5 dpf control (M) and faf1KD (N) zebrafish embryos. Faf1KD embryos have an open-mouth-like appearance (white arrow). Scale bars represent 200 μm.
(O and P) Green fluorescent protein under fli1 promoter in 2 dpf Tg(fli1:eGFP)y1 wild-type (O) and faf1KDp53KD (P) zebrafish larvae. The CNC of pharyngeal arches defective in faf1KDp53KD zebrafish larvae is shown. Scale bars represent 50 μm.
(Q–T) sox9a (Q and R) and col2a1 (S and T) expression in wild-type (Q and S) and faf1KDp53KD (R and T) larvae. Sox9a expression is reduced in pharyngeal arches of 30 hpf faf1KDp53KD larvae (black arrowheads); a dorsal view is shown. Col2a1 expression is absent in the mandible of 2 dpf faf1KDp53KD larvae (black arrowhead); a dorsal view is shown. Scale bars represent 100 μm.
To block faf1 expression, we injected antisense morpholino oligonucleotides (designed to inhibit translation or splicing of pre-mRNA) into 1- to 4-cell-stage zebrafish embryos. Effective knockdown of faf1 was demonstrated with a chimeric faf1/Luciferase fusion reporter assay: translation of the reporter in vitro was suppressed by the translation blocking morpholino faf1-MO-ATG in a dose-dependent manner (Figure S2A). In contrast, a control mismatched at five base pairs was ineffective, demonstrating specificity of the faf1-MO-ATG morpholino (Figure S2A). For the splice-blocking morpholino, faf1-MO-Splice, RT-PCR analysis showed that it inhibited splicing of the faf1 pre-mRNA in morphant embryos in a dose dependent manner (Figure S2B).
Macroscopic inspection revealed that faf1-knockdown (faf1KD) embryos had smaller heads and an underdeveloped jaw region from 2 dpf onward, so that at 5 dpf they had an “open mouth” phenotype (Figures 2M and 2N). The majority of morphant embryos appeared otherwise normal, although embryos with smaller heads and bent, shortened bodies were observed, but only at the highest morpholino dose. In situ hybridization of whole-mount 30 hpf embryos with probes specific for the developing somites (myoD), (motor) neurons (islet1), and heart (cmlc2) demonstrated normal development of these tissues (Figure S3). The penetrance and severity of the jaw phenotype increased with the doses of morpholino used (Figure S4A and Table S2). These defects were not induced by the mismatch control.
Because the open-mouth phenotype suggested defective cartilage development, we used whole-mount Alcian blue staining to visualize the cartilage, especially in the facial region. Upon injection of faf1-MO-ATG at a dose of 4 ng per embryo, Alcian blue staining revealed a mild cartilage defect in most embryos: there was altered orientation of the ceratohyal cartilage, which was pointing more inward than in controls (Figure S5B and Table S2). After a more complete knockdown (6 ng per embryo), all pharyngeal cartilages were underdeveloped. The ceratohyal cartilage was even more inwardly oriented, and the Meckel's cartilage was positioned more ventrally, resulting in an open-mouth phenotype (Figures 2E–2L and Figure S5C). The neurocranial cartilages were also underdeveloped (Figures S5D, S5E, and S5F). Knockdown with the splice-blocking morpholino also resulted in facial cartilage defects. These defects were qualitatively indistinguishable from those induced by faf1-MO-ATG (Figure S4B and Table S2).
Because morpholinos can, in certain cases, induce off-target effects of apoptosis through nonspecific p53 activation, we wished to exclude the possibility that the faf1KD phenotype was merely a consequence of increased CNC apoptosis, and we therefore assessed whether the phenotypic changes induced by the faf1-targeting morpholinos could be blocked by coknockdown of p53, an established procedure in the field.30,31 At a dose of 4 ng faf1-MO-ATG, TUNEL staining showed more apoptosis in the neural plate and tube and in the spinal cord in faf1KD than control embryos from about 12 hpf (early somitogenesis) to 5 dpf (Figure S6). Because the neural-plate border, neural plate, and early neural tube of vertebrate embryos give rise to CNC cells, which subsequently populate the pharyngeal arches and give rise to all lower jaw and many other cranial cartilages,32,33 we coinjected a p53-targeting morpholino that blocks morpholino-induced apoptosis in zebrafish embryos.30 Apoptosis in the CNS was reduced in faf1KDp53KD morphant embryos (Figures S6C and S6F and Table S3), but—importantly—Alcian blue staining of 3–5 dpf coinjected embryos still showed the cartilage defects (Figures S8A and S8B and Table S3). Thus, the faf1KD phenotype was not dependent on off-target effects of the morpholinos used. There was also no difference in cell proliferation in faf1KDp53KD morphant embryos (data not shown). Nevertheless, to eliminate an off-target increase in apoptosis as an influencing factor, we used the double-knockdown strategy in all subsequent experiments (and obtained similar findings in faf1KD embryos).
Altered Expression of Chondrogenic Markers in CNCs from faf1KD Zebrafish Embryos
We then analyzed how faf1 silencing induced the cartilage defects. In situ hybridization using snail1b, foxd3, dlx2, and sox10 (markers of neural-crest induction and differentiation), as well as crestin (a pan-CNC marker), showed no difference in expression between control and faf1KDp53KD embryos (Figures S7A–S7G and data not shown) and demonstrated that earlier CNCs were initially correctly patterned. In contrast, sox9a (a marker of CNCs that differentiate to cartilage) was detectable in mesenchymal condensations of pharyngeal arches in control but not in faf1KD or faf1KDp53KD 30 hpf embryos (Figures 2Q and 2R). The observed reduction in sox9a expression in faf1KDp53KD embryos was attributable to selective silencing of faf1 because it was rescued by coinjection of zebrafish faf1 mRNA with the faf1-MO-Splice and p53 morpholino (Table S4). Of 86 faf1KDp53KD embryos, only 11 (13%) had normal sox9a expression in the first two CNC migratory streams at 22 hpf. Upon coinjection with faf1 mRNA in a total of 126 embryos, 77 (61%) had normal levels of sox9a expression (p < 10−7) (Figures S8C, S8D, and S8E). Similarly, collagen, type II, alpha-1 (col2a1 [MIM 120140]) expression, which marks differentiated cartilage, was reduced in the mandible of morphant embryos at 48 hpf (Figures 2S and 2T). Furthermore, analysis of the expression pattern of EGFP in the transgenic line Tg(fli1:eGFP)y1, which marks (late) neural crest cells, showed that the arches were condensed and underdeveloped in faf1KDp53KD embryos (Figures 2O and 2P). Lack of cartilage formation was evident at 3 dpf (Figures S8A and S8B). This led us to investigate the role of these molecules in the proband with the 46,XY,t(1;2)(p34;q33) translocation. Despite large variability, a tendency toward a decrease in SOX9 expression was observed in the patient (Figure S8F). Because COL2A1 expression is absent in lymphoblasts, it could not be assessed in this cohort.
Discussion
In this study, we demonstrate that FAF1 is disrupted in one family affected by autosomal-dominant PRS. Although we did not identify coding-region mutations in 368 cleft patients, we generated compelling evidence that FAF1 is strongly associated with CPO. We were able to replicate those results across several populations. Overall, the TT genotype of rs3827730 confers an approximately 1.47-fold increase in relative risk of CPO. Although rs3827730 is not in linkage disequilibrium (LD) with the tested SNPs, the decreased expression of FAF1 in CPO patients suggests that changes in noncoding regions, probably in LD with associated SNPs, play an important role, in agreement with current theories on the basis of complex diseases.9
During facial formation, as CNC cells migrate from the hindbrain and posterior midbrain to form the pharyngeal skeleton, they express a series of molecules specific to delamination, migration, and fate determination.1 Sox9a is expressed in mesenchymal condensations prior to chondrogenesis, and Col2a1, the major collagen of cartilage, marks the formation of the latter.34 The friend leukemia integration 1 gene (fli1 [MIM 193067]) is expressed in CNC derivatives, such as the developing cartilages of the jaw and the mesenchyme of aortic arches.35–37 In our faf1KDp53KD zebrafish model, the expression of these three major CNC markers was affected in the lower jaw and the mandible. This suggests that the lack of Faf1 may disrupt chondrogenesis by altering differentiation of CNC cells derived from the first pharyngeal arch.
Cleft palate might be caused by the failure of the lower jaw to develop normally. Rapid growth of the lower jaw is needed for the tongue to descend from between the two palatal shelves. If not, the tongue prevents the palate from closing, resulting in a cleft palate.38 Because FAF1 is expressed in the Meckel's cartilage of zebrafish embryos, is disrupted in a Pierre Robin sequence family with decreased SOX9 expression, and associates with CPO, it might primarily play a role in the development of the lower jaw.
FAF1 is a Fas-binding protein whose role in development is unknown. Several lines of evidence link FAF1 to NF-kB signaling. Overexpression of FAF1 prevents NF-kB translocation to the nucleus and inhibits its activity induced by TNF-alpha and IL-1.39 Moreover, IL-1 and TNF-alpha downregulate Sox9 in chondrocytes, leading to downregulation of Col2a1 expression, an effect mediated by the NF-kB pathway.40 Our results thus raise the question of whether the lack of FAF1 leads to a decrease of Sox9 and Col2a1 as a result of insufficient inhibition of the NF-kB inhibitory pathway (Figure 3).
Figure 3.
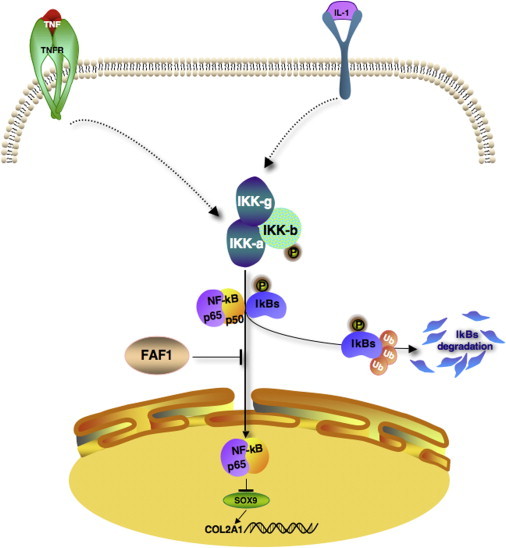
Proposed FAF1 Role in Chondrogenesis
NF-κB binds to IκB inhibitor in cytoplasm in an inactive form. In the presence of FAF1, NF-κB translocation to the nucleus is inhibited, and SOX9 and COL2A1 are normally expressed. In the absence of FAF1, upon NF-κB activation by IL-1 and TNF, subunit kinases IKK-α, -β, and -γ are activated. Phosphorylated IκB is ubiquitinated and degraded. Free NF-κB translocates from the cytoplasm to the nucleus and inhibits expression of SOX9 and COL2A1, leading to defective chondrogenesis (e.g., CPO). Abbreviations are as follows: TNF, tumor necrosis factor; TNFR, tumor necrosis factor receptor; IL-1, interleukin-1; IκB, kappa light polypeptide [arrowheads]).
NF-kB translocation to the nucleus is impaired in the stratum spinosum in Ikka−/− mice,41,42 the abnormalities of which are similar to embryos null for Irf6, a gene mutated in a syndrome involving cleft lip and palate.43 Moreover, mice deficient for IKK1 (IkB kinase 1) have a cleft of the secondary palate.44 Interestingly, IKK1 inhibits the NF-kB pathway, underscoring its role in skeletal development.44 The disruption of SOX9 or COL2A1 leads to a cleft palate in humans and mice.9,45–47 Moreover, autosomal-dominant and recessive osteochondrodysplasias are associated with mutations in COL11A1 and COL11A2, known to encode proteins involved in COL2A1 fibril size regulation.48,49 These facts corroborate our data for an early role of FAF1 in facial chondrogenic development.
In conclusion, we show that FAF1 is a key player in jaw development and that disruptions in this gene are associated with orofacial abnormalities. This expands the repertoire of FAF1 functions from apoptosis to development and suggests an interconnection between these two processes.
Acknowledgments
We are grateful to all the families for their participation in the study. The studies were partially supported by the Interuniversity Attraction Poles initiated by the Belgian Federal Science Policy, networks 6/05 and 7/11; Concerted Research Actions (A.R.C.), Conventions 02/07-276 and 07/12-005 of the Belgian French Community Ministry; and the F.R.S.-FNRS (Fonds de la Recherche Scientifique) (all to M.V.). P.C. is supported by long-term structural funding “Methusalem funding by the Flemish Government”; E.M. was supported by the Deutsche Forschungsgemeinschaft (FOR 423 and individual grants MA 2546/3-1, KR 1912/7-1, NO 246/6-1, and WI 1555/5-1); M.G. was a postdoctoral researcher of F.R.S.-FNRS; M.G. and L.D. were supported by a fellowship from F.R.I.A. (Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture); M.G. was also supported by a “Patrimoine de la Faculté de Médecine” fellowship from the Université catholique de Louvain (UCL); and K. H. was supported by a predoctoral fellowship of IWT (Institute for the Promotion of Innovation by Science and Technology in Flanders). The authors thank Khadija Bahloula, Bieke Tembuyser, and Anne Van Egeren for their expert technical assistance and Liliana Niculescu for secretarial help.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Ensembl, http://www.ensembl.org/
Eurocran, www.eurocran.org
Family-Based Association Test T (FBAT), http://www.biostat.harvard.edu/∼fbat/default.html
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
University of California, Santa Cruz (UCSC), http://genome.ucsc.edu/
References
- 1.Sauka-Spengler T., Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 2.Ghassibe M., Bayet B., Revencu N., Desmyter L., Verellen-Dumoulin C., Gillerot Y., Deggouj N., Vanwijck R., Vikkula M., CL/P Study Group Orofacial clefting: Update on the role of genetics. B-ENT. 2006;2(Suppl 4):20–24. [PubMed] [Google Scholar]
- 3.Murray J.C. Gene/environment causes of cleft lip and/or palate. Clin. Genet. 2002;61:248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- 4.Zucchero T.M., Cooper M.E., Maher B.S., Daack-Hirsch S., Nepomuceno B., Ribeiro L., Caprau D., Christensen K., Suzuki Y., Machida J. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N. Engl. J. Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- 5.Ghassibé M., Bayet B., Revencu N., Verellen-Dumoulin C., Gillerot Y., Vanwijck R., Vikkula M. Interferon regulatory factor-6: a gene predisposing to isolated cleft lip with or without cleft palate in the Belgian population. Eur. J. Hum. Genet. 2005;13:1239–1242. doi: 10.1038/sj.ejhg.5201486. [DOI] [PubMed] [Google Scholar]
- 6.Birnbaum S., Ludwig K.U., Reutter H., Herms S., Steffens M., Rubini M., Baluardo C., Ferrian M., Almeida de Assis N., Alblas M.A. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 7.Mangold E., Ludwig K.U., Birnbaum S., Baluardo C., Ferrian M., Herms S., Reutter H., de Assis N.A., Chawa T.A., Mattheisen M. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 8.Jakobsen L.P., Knudsen M.A., Lespinasse J., García Ayuso C., Ramos C., Fryns J.P., Bugge M., Tommerup N. The genetic basis of the Pierre Robin sequence. Cleft Palate Craniofac. J. 2006;43:155–159. doi: 10.1597/05-008.1. [DOI] [PubMed] [Google Scholar]
- 9.Benko S., Fantes J.A., Amiel J., Kleinjan D.J., Thomas S., Ramsay J., Jamshidi N., Essafi A., Heaney S., Gordon C.T. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 10.Ghassibé M., Revencu N., Bayet B., Gillerot Y., Vanwijck R., Verellen-Dumoulin C., Vikkula M. Six families with van der Woude and/or popliteal pterygium syndrome: All with a mutation in the IRF6 gene. J. Med. Genet. 2004;41:e15. doi: 10.1136/jmg.2003.009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backx L., Van Esch H., Melotte C., Kosyakova N., Starke H., Frijns J.P., Liehr T., Vermeesch J.R. Array painting using microdissected chromosomes to map chromosomal breakpoints. Cytogenet. Genome Res. 2007;116:158–166. doi: 10.1159/000098181. [DOI] [PubMed] [Google Scholar]
- 12.Laird N.M., Horvath S., Xu X. Implementing a unified approach to family-based tests of association. Genet. Epidemiol. 2000;19(Suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 13.Horvath S., Xu X., Laird N.M. The family based association test method: Strategies for studying general genotype—phenotype associations. Eur. J. Hum. Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 14.Weinberg C.R., Wilcox A.J., Lie R.T. A log-linear approach to case-parent-triad data: Assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am. J. Hum. Genet. 1998;62:969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desmyter L., Ghassibe M., Revencu N., Boute O., Lees M., François G., Verellen-Dumoulin C., Sznajer Y., Moncla A., Benateau H. IRF6 screening of syndromic and a priori non-syndromic cleft lip and palate patients: Identification of a new type of minor VWS sign. Mol. Syndromol. 2010;1:67–74. doi: 10.1159/000313786. Published online June 9, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lima R.L., Hoper S.A., Ghassibe M., Cooper M.E., Rorick N.K., Kondo S., Katz L., Marazita M.L., Compton J., Bale S. Prevalence and nonrandom distribution of exonic mutations in interferon regulatory factor 6 in 307 families with Van der Woude syndrome and 37 families with popliteal pterygium syndrome. Genet. Med. 2009;11:241–247. doi: 10.1097/GIM.0b013e318197a49a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stalmans I., Lambrechts D., De Smet F., Jansen S., Wang J., Maity S., Kneer P., von der Ohe M., Swillen A., Maes C. VEGF: A modifier of the del22q11 (DiGeorge) syndrome? Nat. Med. 2003;9:173–182. doi: 10.1038/nm819. [DOI] [PubMed] [Google Scholar]
- 18.Yelon D., Horne S.A., Stainier D.Y. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- 19.Appel B., Korzh V., Glasgow E., Thor S., Edlund T., Dawid I.B., Eisen J.S. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development. 1995;121:4117–4125. doi: 10.1242/dev.121.12.4117. [DOI] [PubMed] [Google Scholar]
- 20.Weinberg E.S., Allende M.L., Kelly C.S., Abdelhamid A., Murakami T., Andermann P., Doerre O.G., Grunwald D.J., Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- 21.Thisse C., Thisse B., Postlethwait J.H. Expression of snail2, a second member of the zebrafish snail family, in cephalic mesendoderm and presumptive neural crest of wild-type and spadetail mutant embryos. Dev. Biol. 1995;172:86–99. doi: 10.1006/dbio.1995.0007. [DOI] [PubMed] [Google Scholar]
- 22.Luo R., An M., Arduini B.L., Henion P.D. Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Dev. Dyn. 2001;220:169–174. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 23.Dutton K.A., Pauliny A., Lopes S.S., Elworthy S., Carney T.J., Rauch J., Geisler R., Haffter P., Kelsh R.N. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- 24.Kelsh R.N., Dutton K., Medlin J., Eisen J.S. Expression of zebrafish fkd6 in neural crest-derived glia. Mech. Dev. 2000;93:161–164. doi: 10.1016/s0925-4773(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 25.Akimenko M.A., Ekker M., Wegner J., Lin W., Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: Part of a homeobox gene code for the head. J. Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang E.F., Pai C.I., Wyatt M., Yan Y.L., Postlethwait J., Chung B. Two sox9 genes on duplicated zebrafish chromosomes: Expression of similar transcription activators in distinct sites. Dev. Biol. 2001;231:149–163. doi: 10.1006/dbio.2000.0129. [DOI] [PubMed] [Google Scholar]
- 27.Yan Y.L., Hatta K., Riggleman B., Postlethwait J.H. Expression of a type II collagen gene in the zebrafish embryonic axis. Dev. Dyn. 1995;203:363–376. doi: 10.1002/aja.1002030308. [DOI] [PubMed] [Google Scholar]
- 28.Neuhauss S.C., Solnica-Krezel L., Schier A.F., Zwartkruis F., Stemple D.L., Malicki J., Abdelilah S., Stainier D.Y., Driever W. Mutations affecting craniofacial development in zebrafish. Development. 1996;123:357–367. doi: 10.1242/dev.123.1.357. [DOI] [PubMed] [Google Scholar]
- 29.Lawson N.D., Weinstein B.M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 30.Robu M.E., Larson J.D., Nasevicius A., Beiraghi S., Brenner C., Farber S.A., Ekker S.C. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisen J.S., Smith J.C. Controlling morpholino experiments: Don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 32.Kulesa P., Ellies D.L., Trainor P.A. Comparative analysis of neural crest cell death, migration, and function during vertebrate embryogenesis. Dev. Dyn. 2004;229:14–29. doi: 10.1002/dvdy.10485. [DOI] [PubMed] [Google Scholar]
- 33.Schilling T.F., Kimmel C.B. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 1994;120:483–494. doi: 10.1242/dev.120.3.483. [DOI] [PubMed] [Google Scholar]
- 34.Hargus G., Kist R., Kramer J., Gerstel D., Neitz A., Scherer G., Rohwedel J. Loss of Sox9 function results in defective chondrocyte differentiation of mouse embryonic stem cells in vitro. Int. J. Dev. Biol. 2008;52:323–332. doi: 10.1387/ijdb.072490gh. [DOI] [PubMed] [Google Scholar]
- 35.Mager A.M., Grapin-Botton A., Ladjali K., Meyer D., Wolff C.M., Stiegler P., Bonnin M.A., Remy P. The avian fli gene is specifically expressed during embryogenesis in a subset of neural crest cells giving rise to mesenchyme. Int. J. Dev. Biol. 1998;42:561–572. [PubMed] [Google Scholar]
- 36.Meyer D., Wolff C.M., Stiegler P., Sénan F., Befort N., Befort J.J., Remy P. Xl-fli, the Xenopus homologue of the fli-1 gene, is expressed during embryogenesis in a restricted pattern evocative of neural crest cell distribution. Mech. Dev. 1993;44:109–121. doi: 10.1016/0925-4773(93)90061-2. [DOI] [PubMed] [Google Scholar]
- 37.Thompson M.A., Ransom D.G., Pratt S.J., MacLennan H., Kieran M.W., Detrich H.W., 3rd, Vail B., Huber T.L., Paw B., Brownlie A.J. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev. Biol. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- 38.Sadewitz V.L. Robin sequence: Changes in thinking leading to changes in patient care. Cleft Palate Craniofac. J. 1992;29:246–253. doi: 10.1597/1545-1569_1992_029_0246_rscitl_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- 39.Park M.Y., Jang H.D., Lee S.Y., Lee K.J., Kim E. Fas-associated factor-1 inhibits nuclear factor-kappaB (NF-kappaB) activity by interfering with nuclear translocation of the RelA (p65) subunit of NF-kappaB. J. Biol. Chem. 2004;279:2544–2549. doi: 10.1074/jbc.M304565200. [DOI] [PubMed] [Google Scholar]
- 40.Murakami S., Lefebvre V., de Crombrugghe B. Potent inhibition of the master chondrogenic factor Sox9 gene by interleukin-1 and tumor necrosis factor-alpha. J. Biol. Chem. 2000;275:3687–3692. doi: 10.1074/jbc.275.5.3687. [DOI] [PubMed] [Google Scholar]
- 41.Hu Y., Baud V., Delhase M., Zhang P., Deerinck T., Ellisman M., Johnson R., Karin M. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 42.Takeda K., Takeuchi O., Tsujimura T., Itami S., Adachi O., Kawai T., Sanjo H., Yoshikawa K., Terada N., Akira S. Limb and skin abnormalities in mice lacking IKKalpha. Science. 1999;284:313–316. doi: 10.1126/science.284.5412.313. [DOI] [PubMed] [Google Scholar]
- 43.Ingraham C.R., Kinoshita A., Kondo S., Yang B., Sajan S., Trout K.J., Malik M.I., Dunnwald M., Goudy S.L., Lovett M. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat. Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q., Lu Q., Hwang J.Y., Büscher D., Lee K.F., Izpisua-Belmonte J.C., Verma I.M. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 1999;13:1322–1328. doi: 10.1101/gad.13.10.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bi W., Huang W., Whitworth D.J., Deng J.M., Zhang Z., Behringer R.R., de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc. Natl. Acad. Sci. USA. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faber J., Winterpacht A., Zabel B., Gnoinski W., Schinzel A., Steinmann B., Superti-Furga A. Clinical variability of Stickler syndrome with a COL2A1 haploinsufficiency mutation: Implications for genetic counselling. J. Med. Genet. 2000;37:318–320. doi: 10.1136/jmg.37.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbieri O., Astigiano S., Morini M., Tavella S., Schito A., Corsi A., Di Martino D., Bianco P., Cancedda R., Garofalo S. Depletion of cartilage collagen fibrils in mice carrying a dominant negative Col2a1 transgene affects chondrocyte differentiation. Am. J. Physiol. Cell Physiol. 2003;285:C1504–C1512. doi: 10.1152/ajpcell.00579.2002. [DOI] [PubMed] [Google Scholar]
- 48.Vikkula M., Mariman E.C., Lui V.C., Zhidkova N.I., Tiller G.E., Goldring M.B., van Beersum S.E., de Waal Malefijt M.C., van den Hoogen F.H., Ropers H.H. Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell. 1995;80:431–437. doi: 10.1016/0092-8674(95)90493-x. [DOI] [PubMed] [Google Scholar]
- 49.Myllyharju J., Kivirikko K.I. Collagens and collagen-related diseases. Ann. Med. 2001;33:7–21. doi: 10.3109/07853890109002055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


