Abstract
Turner syndrome (TS) is a highly prevalent genetic condition caused by partial or complete absence of one X-chromosome in a female and is associated with a lack of endogenous estrogen during development secondary to gonadal dysgenesis. Prominent cognitive weaknesses in executive and visuospatial function in the context of normal overall IQ also occur in affected individuals. Previous neuroimaging studies of TS point to a profile of neuroanatomical variation relative to age and sex matched controls. However, there are no neuroimaging studies focusing on young girls with TS before they receive exogenous estrogen treatment to induce puberty. Information obtained from young girls with TS may help to establish an early neural correlate of the cognitive phenotype associated with the disorder. Further, univariate analysis has predominantly been the method of choice in prior neuroimaging studies of TS. Univariate approaches examine between-group differences on the basis of individual image elements (i.e., a single voxel’s intensity or the volume of an a priori defined brain region). This is in contrast to multivariate methods that can elucidate complex neuroanatomical profiles in a clinical population by determining the pattern of between-group differences from many image elements evaluated simultaneously. In this case, individual image elements might not be significantly different between groups but can still contribute to a significantly different overall spatial pattern. In this study, voxel-based morphometry (VBM) of high-resolution magnetic resonance images was used to investigate differences in brain morphology between 13 pediatric, pre-estrogen girls with monosomic TS and 13 age-matched typically developing controls (3.0T imaging: mean age 9.1 ± 2.1). A similar analysis was performed with an older cohort of 13 girls with monosomic TS and 13 age-matched typically developing controls (1.5T imaging: mean age 15.8 ± 4.5). A multivariate, linear support vector machine analysis using leave-one-out cross-validation was then employed to discriminate girls with TS from typically developing controls based on differences in neuroanatomical spatial patterns and to assess how accurately such patterns translate across heterogeneous cohorts. VBM indicated that both TS cohorts had significantly reduced gray matter volume in the precentral, postcentral, and supramarginal gyri and enlargement of the left middle and superior temporal gyri. Support vector machine (SVM) classifiers achieved high accuracy for discriminating brain morphology patterns in TS from typically developing controls and also displayed spatial patterns consistent with VBM results. Furthermore, the SVM classifiers identified additional neuroanatomical variations in individuals with TS, localized in the hippocampus, orbitofrontal cortex, insula, caudate, and cuneus. Our results demonstrate robust spatial patterns of altered brain morphology in developmentally dynamic populations with TS, providing further insight into the neuroanatomical correlates of cognitive-behavioral features in this condition.
Keywords: Turner Syndrome, Voxel-Based Morphometry (VBM), Support Vector Machine (SVM), Pattern Classification, Neuroimaging
1. Introduction
Genetic and developmental disorders are commonly characterized by a unique cognitive-behavioral phenotype. Turner syndrome (TS), a common X-chromosome aneuploidy syndrome affecting approximately 1/2000 live female births (Gravholt, 2005), is caused by the partial or complete deletion of an X-chromosome. Individuals with TS are at risk for developing impairments in social awareness and fear recognition (Lawrence et al., 2003; Mazzocco et al., 1998), as well as cognitive deficits related to attention, memory, and visuospatial and executive processing (Kesler et al., 2006; Tamm et al., 2003). Verbal ability is often preserved or relatively enhanced (Temple and Carney, 1996). However, the underlying neural correlates for these traits are only recently being investigated through the use of neuroimaging techniques.
Several magnetic resonance imaging (MRI) studies have identified multiple brain regions exhibiting aberrant morphology in adolescent and adult TS populations compared to typically developing (TD) controls. Using univariate structural analyses of brain MRI (e.g., volumetric and voxel-based morphometry [VBM]), the most consistent findings in these TS populations include reduction in parieto-occipital gray matter volume (GMV) (Brown et al., 2004; Cutter et al., 2006; Molko et al., 2004; Reiss et al., 1995) and enlargement of the amygdala (Good et al., 2003; Kesler et al., 2004). Structural and functional abnormalities have also been reported in the hippocampus, caudate, orbitofrontal cortex, and superior temporal gyrus (Cutter et al., 2006; Haberecht et al., 2001; Kesler et al., 2003; Tamm et al., 2003). Furthermore, a recent whole brain surface-based analysis showed significant group differences related to cortical thickness and surface area in parietal, occipital, temporal, and pre-frontal regions (Raznahan et al., 2010). However, there is little information regarding such neuroanatomical characteristics in young pre-estrogen populations with TS, which may help establish an early morphological correlate to the cognitive-behavioral phenotype associated with the disorder.
Univariate analyses have predominantly been the methods of choice in neuroimaging studies, including those focused on TS, which typically evaluate voxel-wise or regional measures. These methods, however, do not provide information into the complex spatial organization of neuroanatomical characteristics. As a result, multivariate methods (e.g. multivoxel pattern analysis [MVPA]) are gaining popularity due to their sensitivity to spatial patterns encoded across brain regions (i.e., patterns in between-group differences as discerned by the simultaneous evaluation of a large number of image elements). This is of particular interest in the study of brain disorders, which commonly involve multiple anatomical structures of the brain, yet present complex morphological spatial characteristics that may not be easily observed and/or understood using univariate approaches.
Support vector machine (SVM) is a widely used MVPA method and has previously been employed in the context of structural MRI to discriminate between diagnostic states in both pediatric and adult populations (Ecker et al., 2010; Hoeft et al., 2008). In this study, we investigated the voxel-wise whole brain morphology of young girls with TS who had not yet received estrogen therapy, relative to sex and age-matched TD controls, and subsequently used a linear SVM to determine if morphological spatial patterns could accurately differentiate TS from TD in a completely independent and older population.
Based on previous morphological studies, we hypothesized that univariate VBM analyses would most strongly display group differences in parieto-occipital GMV in both TS cohorts as well as in the amygdala and superior temporal gyrus. However, we were unsure of the overall extent to which these studies would translate to our young pre-estrogen population. We also hypothesized that differences in morphometric patterns between TS and TD subjects for each cohort could accurately be differentiated by a binary linear SVM classifier from the alternate cohort. Equivalently, this would indicate the presence of neuroanatomical patterns in the TS brain that persist throughout several developmental stages leading into young adulthood.
2. Materials & Methods
2.1 Recruitment and Exclusion Criteria
The local Institutional Review Board at the Stanford University School of Medicine approved this study. Informed written consent was obtained from each participant. If the subject was a minor, consent was obtained from the legal guardian. Prior to enrollment in this study, all participants were screened with standard forms and interviews for MRI contraindications as well as past medical history to ensure that there were no instances of neurological injury, psychiatric illness or disease (except for social anxiety or attention deficit hyperactivity disorder), or gross physical impairments. Individuals born prematurely (i.e. < 34 weeks gestation) were also not included, nor were those exhibiting a mosaic karyotype of the X-chromosome. TS participants were recruited through the National Turner Syndrome Society and Foundation, a local network of physicians, and advertisement on the Stanford University School of Medicine website. TD controls were recruited through local print media and parent networks.
2.2 Participants
The primary cohort (3.0T imaging) was comprised of 13 females with TS (monosomic [45X]; mean age 9.1 ± 2.4; range 5.0 – 12.8) and 13 individually age-matched female controls (mean age 9.1 ± 2.7; range 3.8 – 12.5), whose participation occurred between 2005 and 2009. Of those with TS, 11 were receiving growth hormone (GH) only, one was receiving topical estrogen cream only for the treatment of labial lesions, and the status of one was unknown. Data regarding dosages and age at GH therapy initiation were unavailable. The secondary cohort (1.5T imaging) was comprised of 13 females with TS (monosomic [45X], mean age 15.8 ± 4.5; range 7.6 – 24.0) and 13 individually age-matched female controls (mean age 14.5 ± 3.7; range 6.7 – 20.4), whose participation occurred between 1999 and 2006. Of those with TS, two were receiving GH only, five were receiving exogenous estrogens only, three were receiving both GH and exogenous estrogens, and the status of three was unknown. Data regarding dosages, prior GH therapy, and age at GH/estrogen therapy initiation were unavailable. There was no union of subjects between the two cohorts; however, previous studies from our colleagues have reported findings using subjects from the secondary cohort (Brown et al., 2002; Brown et al., 2004; Kesler et al., 2003; Kesler et al., 2004). Additionally, seven subjects overlapped in age between the two cohorts (3.0T: 1 TS, 1 TD; 1.5T: 2 TS, 3 TD).
2.3 Cognitive Assessment
All participants under age 17 were administered cognitive assessments based on the Wechsler Intelligence Scale for Children [Primary cohort: fourth edition (WISC-IV), (Wechsler, 2003); secondary cohort: third edition (WISC-III), (Wechsler, 1991)]. The Wechsler Adult Intelligence scale [third edition (WAIS-III), (Wechsler, 1997)] was administered to all participants in the secondary cohort age 17 and older. Participants demonstrating a full-scale IQ (FSIQ) below 70 or above 130 were excluded from further analyses to avoid confounding effects. The mean FSIQ for TS and TD subjects in the primary cohort was 94 ± 11 and 116 ± 9 (significant at p<0.001), respectively. Similarly, the mean verbal comprehension index (VCI) was 109 ± 16 and 116 ± 15 (non-significant), and the mean perceptual reasoning index (PRI) was 93 ± 12 and 115 ± 7 (significant at p<0.001). For the secondary cohort, the mean FSIQ for TS and TD subjects was 106 ± 10 and 115 ± 7 (significant at p<0.05), respectively. Similarly, the mean verbal IQ (VIQ) was 115 ± 12 and 118 ± 8 (non-significant), and the mean performance IQ (PIQ) was 96 ± 9 and 111 ± 10 (significant at p=0.001).
2.4 MRI Preparation
All subjects were introduced to a mock MRI scanner prior to their real scan in an effort to desensitize them to the sights and sounds of an actual MRI environment. Each subject practiced lying still in the bore of the mock scanner to become familiarized with the experience of receiving a brain MRI and underwent behavioral training to help reduce motion related artifacts. Visual inspection of the data was also employed to eliminate scans/subjects with significant head motion artifact from further analysis.
2.5 MR Image Acquisition
All MR images from the younger cohort were acquired at the Stanford University Lucas Center for Medical Imaging. 3.0T imaging was performed between 2005 and 2009 on a 3.0T GE Signa HDx whole body MR system (GE Medical Systems, Milwaukee, WI) using a standard birdcage head coil. A spoiled gradient recalled (SPGR) echo pulse sequence was employed to obtain a high-resolution T1 anatomical brain image of each subject (124 coronal slices, repetition time [TR] / echo time [TE] = 6.4/2 msec, inversion time [TI] = 300 msec, flip angle = 15°, NEX = 3, FOV = 22 × 22 cm, matrix = 256 × 256, 1.5 mm thickness, acquisition time = 15 min 45 sec). 1.5T imaging was performed between 1999 and 2006 on 1.5T GE Signa (Horizon/Excite) whole body MR systems (GE Medical Systems, Milwaukee, WI) using a standard birdcage head coil. An SPGR pulse sequence was also employed to obtain a high-resolution T1 anatomical brain image of each subject (124 coronal slices, TR/TE = 35/6 msec, flip angle = 45°, NEX = 1, FOV = 24 × 24 cm, matrix = 256×256, 1.5 mm thickness, acquisition time = 14 min 24 sec). Previous neuroimaging studies have validated the compatibility of 1.5T GE Signa scanners in both longitudinal and multisite studies (Kloppel et al., 2008b; Stonnington et al., 2008) and have included data from our 1.5T Horizon subset (Patwardhan et al., 2001).
2.6 Image Preprocessing
All image data were visually inspected to eliminate scans with significant head motion artifact in an effort to prevent confounds due to systematic bias. Accordingly, 16 scans were eliminated from the 3.0T cohort (6 TS, 10 TD) and 10 scans were eliminated from the 1.5T cohort (2 TS, 8 TD) due to visible artifacts. Each native image volume was then manually aligned onto the axis of the anterior and posterior commissures (Talairach and Tournoux, 1988). Next, using Statistical Parametric Mapping software (SPM5, Wellcome Department of Imaging Neuroscience, University College London, London, UK; www.fil.ion.ucl.ac.uk/spm) in the MATLAB computing environment (The MathWorks, Natick, MA), corrections for inhomogeneity of the magnetic field were applied followed by unified segmentation of the gray matter (GM), white matter (WM), and cerebrospinal fluid (Ashburner and Friston, 2005).
Accordingly, female pediatric a priori tissue probability maps (CCHMC2_girls_5-18, Cincinnati Children’s Hospital Medical Center; irc.cchmc.org/software/pedbrain), were used in conjunction with iterative weighting of Hidden Markov Random Fields to encode spatial information based on constraints of neighboring voxels (Zhang et al., 2001). A custom GM template was then generated for each cohort based on registration of each subject’s image volume (Ashburner, 2007) using the Diffeomorphic Anatomical Registration Through Lie Algebra toolbox (DARTEL, Wellcome Department of Imaging Neuroscience, University College London, London, UK; www.fil.ion.ucl.ac.uk/spm). Warped and modulated GM and WM images were subsequently created in Montreal Neurological Institute (MNI) space. Finally, all images were spatially smoothed using a 3-dimensional 8mm full-width-at-half-maximum (FWHM) Gaussian smoothing kernel.
2.7 Whole Brain Volumetric Analysis
Differences in total GMV and white matter volume (WMV) between TS and TD were evaluated using a two-sample t-test (adjusted for age).
2.8 VBM
For each cohort, between-group morphological differences of regional GMV between TS and TD subjects were investigated by applying the general linear model (GLM) in SPM5. Accordingly, a group analysis was performed for each cohort using a voxel-wise two-sample t-test, while covarying for the effects of age and GMV. Statistical inference of significant clusters was then evaluated using the VBM5 toolbox (Christian Gaser, University of Jena; dbm.neuro.uni-jena.de/vbm/vbm5-for-spm5/) at a height of p<0.01, spatial extent p<0.05 (family-wise-error [FWE] corrected), while applying non-stationary cluster extent correction to account for non-uniform smoothness across the data (Hayasaka et al., 2004; Worsley et al., 1999). A similar statistical analysis was performed based on data that were spatially smoothed at 4mm (3-D FWHM Gaussian kernel) in an attempt to isolate distinct anatomical regions within significant clusters. However, doing so generally resulted in comparable cluster extent and spatial localization at various statistical thresholds.
2.9 SVM Feature Extraction & Dimensionality Reduction
Supervised MVPA was performed using a linear SVM algorithm (C=1) to investigate how differences in spatial patterns between TS and TD controls from one cohort can translate to the other cohort. This was achieved using an in-house MATLAB toolbox (Hoeft et al., 2008) based on SVM software from Fraunhofer FIRST (http://www.first.fraunhofer.de/en/home) and principal component analysis (PCA) software by Samar Khatiwala (Columbia University; www.ldeo.columbia.edu/~spk/Code/Matlab/PCA). All processed images (i.e., those used in our VBM analyses) were first down-sampled to 4mm isotropic voxels. Common voxels between the two cohorts (i.e., voxels that were non-zero for every subject) were subsequently mapped into the feature space as an n-dimensional vector for each subject (where n represents the total number of features common to all subjects and each vector represents the GMV spatial patterns for a single subject’s n voxels). After using linear multiple regression to calculate the residuals (with GMV as a dependent variable and each feature as an independent variable), data were normalized for both rows and columns of the feature space such that mean = 0 and standard deviation = 1. Dimensionality reduction was then performed using PCA, and the resulting principal component (PC) maps were generated by selecting the eigenvectors accounting for 80% of the variance in the data. After these steps, each dataset was treated separately.
2.10 SVM Classifier Training & Parameter Optimization
A binary linear classifier was trained for each cohort, which represents morphometric spatial patterns that discriminate TS (class label = 1) from TD controls (class label = −1). The classifier was obtained by leave-one-out (LOO) cross-validation, performing n iterations of classifier training using n-1 subjects (where n is the number of subjects) as training data and validating the classifier on the nth subject (i.e., a single TS or TD subject for each iteration). Concurrently, the least relevant features (i.e., PCs) were recursively identified (in 30% increments) and discarded during each training iteration (Guyon et al., 2002). This was done in an effort to optimize the classification parameters (i.e., weight vectors) and subsequently overall classification performance. The average classification accuracy for n classifiers within each cohort was then calculated, as well as specificity (correct identification of TS) and sensitivity (correct identification of TD), and the corresponding weight vectors were transformed back into voxel space (Mourao-Miranda et al., 2006).
2.11 Pattern Classification Across Cohorts
In order to investigate how differences in spatial patterns between TS and TD controls from one cohort translate to data from the other, the PC map from the second data set was applied to the first data-set’s n classifiers. Overall classification accuracy was calculated by identifying whether TS was correctly identified as TS and TD as TD. This procedure was repeated in both directions (i.e., classifier from 3.0T cohort applied to 1.5T cohort and classifier from1.5T cohort applied to 3.0T cohort). Additional classifiers were trained and tested after excluding the 7 subjects that overlapped in age between the two cohorts (3.0T: 1 TS, 1 TD; 1.5T: 2 TS, 3 TD) to verify that across cohort classification performance was not driven by these subjects. In doing so, overall performance showed a small increase for both classifiers.
2.12 Permutation Analyses
Permutation analyses were performed for each classifier 2000 times each to empirically determine whether classification accuracies were significantly greater than chance. A statistical threshold of p = 0.05 was used.
3. Results
3.1 Whole Brain Volumetric Analyses
Whole brain volumetric measures of GMV and WMV showed no significant differences (adjusted for age) between TS and TD populations within either cohort (Table 1).
Table 1.
Demographics, Intelligence Quotients, and Volumetric Measures
| Metric | 1.5T |
3.0T |
||||
|---|---|---|---|---|---|---|
| TS (n=13) | TD (n=13) | p-value | TS (n=13) | TD (n=13) | p-value | |
| Age (years) | 15.8 ± 4.5 | 14.5 ± 3.7 | 0.433 | 9.1 ± 2.4 | 9.1 ± 2.7 | 0.97 |
| FSIQ | 106 ± 10b | 115 ± 7 | 0.02 | 94 ± 11 | 116 ± 9 | <0.001 |
| VIQ/VCIa | 115 ±12b | 118 ± 8 | 0.433 | 109 ± 16 | 116 ± 15c | 0.272 |
| PIQ/PRIa | 96 ± 9b | 111 ± 10 | 0.001 | 93 ± 12 | 115 ± 7c | <0.001 |
| GMV (mL) | 796 ± 91 | 824 ± 57 | 0.36 | 880 ± 70 | 897 ± 67 | 0.541 |
| WMV (mL) | 405 ± 44 | 397 ± 33 | 0.61 | 377 ± 46 | 387 ± 43 | 0.565 |
VIQ/PIQ apply to 1.5T cohort; VCI/PRI apply to 3.0T cohort.
Data for 1 TS subject were missing.
Data for 2 TD subjects were missing
TS, Turner Syndrome; TD, Typically Developing; FSIQ, Full Scale Intelligence Quotient; VIQ, Verbal Intelligence Quotient; VCI, Verbal Comprehension Index; PIQ, Performance Intelligence Quotient; PRI, Perceptual Reasoning Index; GMV, Gray Matter Volume; WMV, White Matter Volume.
3.2 Whole Brain Univariate VBM Analyses
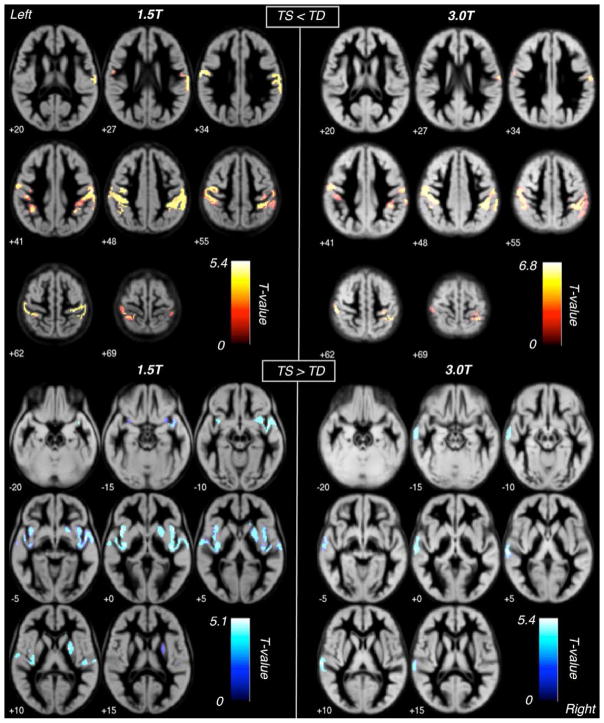
Univariate analysis of the voxel-wise whole brain morphology of TS (Figure 1; Table 2) in both the primary (3.0T) and secondary (1.5T) cohorts displayed significantly reduced GMV compared to TD controls bilaterally in the precentral gyrus (BA4 & BA6), postcentral gyrus (BA1, BA2, & BA3), and inferior parietal lobule (BA40). Reduction in GMV also extended into the right superior parietal lobule and supramarginal gyrus. Increases in GMV were seen in the in the left temporal lobe of the primary cohort and bilaterally in the secondary cohort, including both the middle and superior temporal gyri (BA21, BA22, & BA42). Additional increases in GMV were seen in the secondary cohort in the insula bilaterally as well as in the right putamen. Significant clusters exhibiting reduced GMV (TS < TD) for both cohorts were overlaid graphically to qualitatively evaluate the extent of spatial agreement (Figure 2).
Figure 1. Voxel-based morphometry results.
Axial slice representations of significant GMV clusters from whole brain VBM (p<0.05, corrected for FWE and non-stationary smoothness) overlaid on custom GM templates. Significant regions displaying TS < TD are shown at 1.5T (upper left quadrant) and 3.0T (upper right quadrant). Significant regions displaying TS > TD are also shown at 1.5T (lower left quadrant) and 3.0T (lower right quadrant).
Table 2.
Clusters displaying significant group differences in voxel-based morphometry
| Region | Brodmann Area(s) | Talairach Coordinates (peak voxel) | T-score (maximum) | Cluster Size | p-value (cluster-level) | ||
|---|---|---|---|---|---|---|---|
| x | y | z | |||||
|
3.0T |
|||||||
| TS > TD | |||||||
| Left superior temporal gyrus middle temporal gyrus | 21,22,42 | −67 | −23 | 7 | 5.42 | 1206 | 0.025 |
| TS < TD | |||||||
| Left postcentral gyrus precentral gyrus inferior parietal lobule | 1,2,3,4,6,40 | −51 | −24 | 56 | 6.78 | 1737 | 0.009 |
| Right postcentral gyrus precentral gyrus inferior parietal lobule supramarginal gyrus superior parietal lobule | 1,2,3,4,6,40 | 32 | −38 | 63 | 6.15 | 2196 | 0.004 |
|
1.5T |
|||||||
| TS > TD | |||||||
| Left inferior frontal gyrus superior temporal gyrus insula | 22 | −44 | 16 | 1 | 4.84 | 1718 | 0.009 |
| Right insula superior temporal gyrus putamen | 22 | 46 | −80 | 24 | 5.07 | 2060 | 0.005 |
| TS < TD | |||||||
| Left precentral gyrus postcentral gyrus inferior parietal lobule | 1,2,3,4,6,40 | −63 | −6 | 33 | 5.43 | 958 | 0.043 |
| Right precentral gyrus postcentral gyrus inferior parietal lobule supramarginal gyrus superior parietal lobule | 1,2,3,4,6,40 | 32 | −30 | 57 | 5.61 | 2434 | 0.003 |
Clusters displaying significant GMV differences between TS and TD from whole brain VBM (p<0.05, corrected for FWE and non-stationary smoothness) are identified by the Talairach coordinates and T-score of the peak voxel and the cluster level p-value. Additional regions contained within each cluster are also listed.
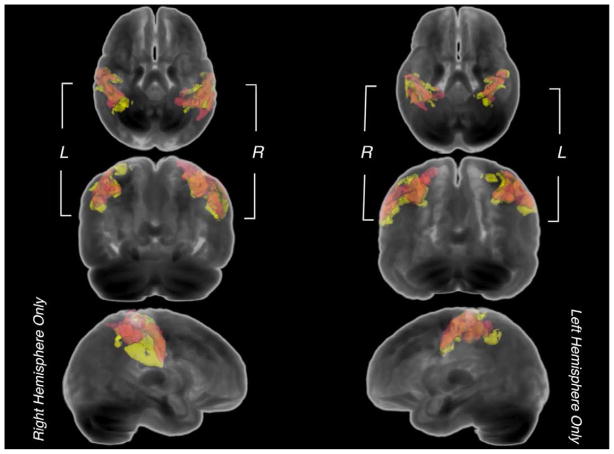
Figure 2. Voxel-based morphometry spatial overlap between cohorts.
3-dimensional renderings of significant clusters exhibiting reduced GMV from univariate VBM analyses (p<0.05, corrected for FWE and non-stationary smoothness). Spatially overlapping voxels between a young pediatric cohort (3.0T, red) and an older cohort (1.5T, yellow) are displayed as orange. Negative weight clusters are not included due to a low extent of overlap.
3.3 SVM Classification Performance
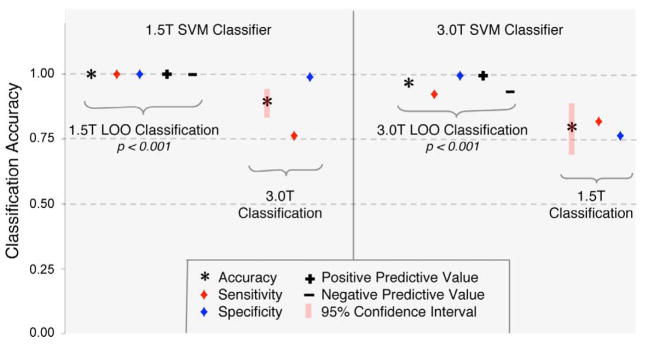
A separate binary linear classifier was trained for each cohort using LOO cross-validation and was subsequently applied to the alternate cohort (Figure 3). The classifier based on the 3.0T (younger) cohort correctly discriminated TS from TD controls with 96.2% average accuracy (p<0.001) during LOO cross-validation (sensitivity/specificity: 92.3%/100%) and subsequently identified TS and TD controls in the 1.5T cohort with 78.6% accuracy (sensitivity/specificity: 82.0%/75.2%). Similarly, the classifier based on the 1.5T (older) cohort correctly discriminated TS from TD controls with 100% accuracy (p<0.001) during LOO cross-validation (sensitivity/specificity: 100%/100%) and subsequently identified TS and TD controls in the 3.0T cohort with 87.6% average accuracy (sensitivity/specificity: 76.3%/98.8%). A subsequent analysis based on classifiers using age as a regressor yielded comparable across-cohort classification performance. Specifically, the classifier based on the 3.0T (younger) cohort achieved an accuracy/sensitivity/specificity of 81.8%/92.0%/71.6%, and the classifier based on the 1.5T (older) cohort achieved an accuracy/sensitivity/specificity of 84.8%/84.6%/84.9%.
Figure 3. Support vector machine pattern classification results.
Leave-one-out (LOO) SVM classifier training and pattern classification results based on common GM voxels (using GMV as a regressor) between the 1.5T and 3.0T cohorts to discriminate between TS and TD subjects. PCA and RFE (30% increments) were applied for dimensionality reduction and parameter optimization. Left: Linear SVM classifier training results based on the 1.5T cohort (p<0.001, 2000 permutations) and resulting classifier accuracies when applied to the 3.0T cohort. Right: Linear SVM classifier training results based on the 3.0T cohort (p<0.001, 2000 permutations) and resulting classifier accuracies when applied to the 1.5 cohort.
3.4 SVM Classifier Weight Vectors
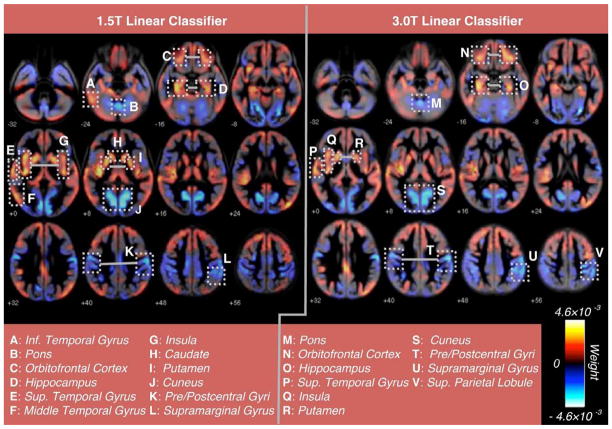
Figure 4 displays a visual representation of the weight vectors for both optimal classifiers, which represent spatial patterns among regions that are anatomically relevant in distinguishing TS from TD. Both classifiers assigned strong negative classification weights to areas that included the precentral and postcentral gyri, supramarginal gyrus, superior parietal lobule, and cuneus. Strong positive classification weights were assigned to areas that included the middle and superior temporal gyri, orbitofrontal cortex, insula, hippocampus, putamen and caudate.
Figure 4. Whole-brain representation of pattern classifiers.
Visual representation of whole brain pattern classifiers discriminating TS from TD using only GMV voxels common to both cohorts. Axial slice representations of the weight vectors (representing the degree to which each voxel contributes to the discrimination between TS and TD based on GMV measurements) are displayed from a leave-one-out linear SVM (employing PCA and RFE) using training data from the1.5T cohort (left) and using training data from the 3.0T cohort (right). Notable regions exhibiting relatively stronger classification weights (negative or positive) are identified for each classifier.
4. Discussion
4.1 Study Overview
In this study, we employed a traditional univariate VBM approach (Ashburner and Friston, 2000) to examine the voxel-wise whole brain neuroanatomical characteristics of TS and TD subjects in a young pediatric population 3 to 12 years of age and evaluated the subsequent morphological consistency with an older sample. To date, there is little information regarding such characteristics in pre-estrogen TS populations, which may help establish an early morphological correlate to the cognitive-behavioral phenotype associated with the disorder. Specifically, we observed abnormally low GMV in parietal and precentral regions for both cohorts with a high extent of spatial overlap in the postcentral gyrus, as well as increased GMV in middle and superior temporal regions. Our older cohort also exhibited bilateral enlargement of the insula. Additionally, we demonstrated that GMV spatial patterns can discriminate girls with TS from TD controls with high accuracy, and can effectively translate across heterogeneous cohorts.
4.2 Voxel-Based Morphometry
Our VBM-based finding of reduced GMV in inferior and superior parietal regions of both cohorts (Figure 1; Table 2) is consistent with the neuroimaging literature in adolescent and adult TS populations, and suggests a putative neuroanatomical correlate of impairments in visuospatial and verbal memory skills. The relatively pervasive bilateral reduction in GMV of the postcentral gyrus replicates a previous finding from our group (Brown et al., 2004), though the observation of reduced GMV extending into precentral regions is a new observation. GMV reductions in these regions provide potential morphological correlates for motor and sensory impairments in TS (Nijhuis-van der Sanden et al., 2000). Increases in middle and superior temporal GMV also replicate recent neuroimaging findings in TS (Cutter et al., 2006; Kesler et al., 2003); however, we did not observe a large amount of spatial consistency between the two cohorts, which may be related to several factors such as age, hormonal status, and hardware / scan parameter differences. Nonetheless, enlargement of these temporal regions might provide a neuroanatomical correlate of strong verbal fluency skills observed in individuals with TS (Temple, 2002).
The large extent of spatial overlap observed in reduced GMV between the two cohorts (Figure 2) is likely to be the result of genetic influences specific to X-monosomy. Supporting this hypothesis, several studies have reported excess X-linked gene expression in brain tissues (Nguyen and Disteche, 2006; Xu and Disteche, 2006) to which transcriptional deficiencies related to X monosomy might hinder neuroanatomical development. Post hoc analyses have also suggested possible modulatory effects based on parental X chromosome origin in TS (Cutter et al., 2006; Kesler et al., 2003; Murphy et al., 1994). However, our subject groups were too small to investigate such an effect.
Furthermore, our ability to investigate age-related correlations between the two cohorts was confounded by differences in exogenous hormone therapy status, scanner, and pulse sequence. Since the dynamics of such effects are critical towards understanding the progression of cognitive impairments in TS, a controlled longitudinal analysis on the same cohort is highly warranted.
Additionally, it is important to note that VBM analyses are limited by their dependence on a priori information that influence user-defined choices related to parameter optimization and statistical modeling. Accordingly, the use of additional analytical techniques, such as manual/automated volumetry or tensor-based algorithms, is warranted to corroborate these results.
4.3 Support Vector Machine
We also report that differences in GMV spatial patterns of girls with TS and TD controls can be accurately discriminated by a linear SVM. Previous applications have employed SVM approaches to successfully discriminate clinical populations from controls across different scanners of the same magnetic field strength (Kloppel et al., 2008a; Kloppel et al., 2008b)]). Our data, though differing in magnetic field strength, demonstrate that spatial patterns can similarly be applied to a heterogeneous clinical population. This demonstrates the robust nature of morphological spatial patterns in developmentally dynamic TS populations (i.e., in the presence of potential confounds such as age and the status of estrogen replacement therapy and/or growth hormone supplementation). Furthermore, we found that the classifier generated by our older population was both more sensitive and more accurate than the classifier generated by our younger population (Figure 3), which may indicate the presence of more defined neuroanatomical attributes in older individuals; however prospective, longitudinal studies at the same magnetic field strength are required to systematically identify age and exogenous hormone-related effects on morphological spatial patterns.
Our multivariate analysis also shows that GMV spatial patterns in TS appear to incorporate a more intricate network of brain regions than is observed using univariate VBM analysis (Figure 4). For example, the hippocampus, orbitofrontal cortex, insula, putamen, and caudate are assigned strong positive classification weights along with the superior and middle temporal gyri. Similarly, the cuneus is assigned a strong negative classification weight along with the pre/postcentral gyri, supramarginal gyrus, and superior parietal lobule. Interestingly, structural and functional abnormalities in the insula have never been reported in TS, though recent studies of other clinical populations have identified a functional network involving the insula and amygdala that may influence salience processing (Etkin et al., 2009; Seeley et al., 2007). Although we do not report significant findings in the amygdala in the current study, enlargement of the amygdala has previously been observed in TS (Good et al., 2003; Kesler et al., 2004) and has been hypothesized to be linked to autonomic arousal and impairment in fearful face expression (Skuse et al., 2005). Abnormalities in the orbitofrontal cortex, caudate, putamen, and cuneus also may account for known deficiencies in attention and working memory in TS (Tamm et al., 2003). However, hypotheses regarding neural networks in TS remain speculative at this juncture, as multi-modal and longitudinal imaging studies with larger sample size are required for more definitive observations.
The absence of amygdala abnormalities in the VBM and SVM analyses for both cohorts may be a result of methodological and biological factors. For instance, topological variation among subjects is a source of registration error during spatial normalization, and regions of mixed tissue composition (i.e., partial gray/white matter) are likely to yield suboptimal tissue segmentation. These factors may inordinately affect subcortical structures such as the amygdala. Furthermore, recent studies implicating structural abnormalities of the amygdala were based on a priori methods such as manual tracings (Kesler et al., 2004) and a VBM analysis employing small volume correction around this structure (Good et al., 2003). It has also been shown that amygdala volume is highly variable in typical children ages four to 18 (Giedd et al., 1996), which may further limit our ability to observe amygdala abnormalities in the current study.
It is also important to recognize that since TS can be diagnosed with 100% accuracy via genetic testing, the use of SVM purely as a diagnostic tool for this disorder is unnecessary. Rather, the current study uses SVM to provide evidence of robust neuroanatomical attributes of TS that are characteristic across a dynamic age range yet may remain obscured in a mass-univariate approach. Accordingly, in the context of genetic disorders, the high-dimensional predictive capacity of SVM may provide an alternative means to evaluate the fine-grained neuroanatomical differences associated with genetic variants that may otherwise be too subtle to detect via univariate approaches, though further investigation is required.
4.4 Limitations
Though we provide evidence of robust neuroanatomical characteristics inherent to TS, our analyses are limited by two important factors. Firstly, our sample sizes are relatively small and thus our results require confirmation using larger cohorts. The reduced statistical power of the current study, however, is a direct result of a thorough visual quality assessment, which we believe eliminated scans that would have otherwise introduced deleterious confounds due to motion and blood flow (dephasing) related artifacts. Secondly, the long duration of data acquisition and the use of multiple scanners and field strengths provides opportunities for several scanner-related confounds. For instance, a random sample of signal-to-noise (SNR) and contrast-to-noise (CNR) ratios were significantly different at p<0.05 between the two cohorts (3.0T SNR/CNR: 34.1 ± 5.0 / 10.4 ± 1.5; 1.5T SNR/CNR: 44.5 ± 11.3 / 12.0 ± 1.9). Furthermore, we would expect our images acquired at 3.0T with three averages (NEX=3) to have higher SNR/CNR than those at 1.5T with one average (NEX=1), yet we observed the opposite. This may be due to the fact that our 3.0T cohort was much younger than our 1.5T cohort and was more prone to subtle motion over the course of the long scan time. Regardless, the accuracy of tissue segmentation is influenced by SNR and CNR (especially near regions of mixed gray/white matter composition) and likely yielded regional differences between our two cohorts. Nonetheless, we believe that the consistency of the observed neuroanatomical abnormalities both within and across developmentally dynamic TS cohorts, using both univariate and multivariate methods, has important implications related to the understanding of neuroanatomical characteristics in this disorder.
5. Conclusions
In conclusion, voxel-wise whole brain analysis of brain anatomy in a young pediatric population with TS (3 to 12 years) indicates the presence of significantly aberrant morphology in parietal, temporal, and precentral cortical regions. The consistency of these observations with previous univariate studies of older populations, as well as good spatial agreement with an older cohort in the current study, provides valuable insight into the morphological correlates to the cognitive-behavioral phenotype in TS. Furthermore, we demonstrated relative insensitivity of a multivariate SVM algorithm to developmentally dynamic TS populations, thus suggesting the presence of robust underlying morphological spatial patterns inherent to this important condition. Furthermore, morphometric spatial patterns in TS have only recently begun to be investigated, and further study will continue to provide valuable insight into the neuroanatomical correlates of learning strengths and weaknesses in this disorder.
Acknowledgments
The authors would like to thank Kristen Sheau, M.S., and Yaena Park, B.A., for their contributions in data collection.
Funding: This work was supported by the National Institutes of Health [5-R01-HD049653 to A.L.R.]; National Institute of Child Health and Human Development [HD054720 to F.H.]; American Psychiatric Institute for Research and Education / Lilly Psychiatric Research Fellowship Award [to D.H.] National Institute of Mental Health [T32-MH19908 to A.L.R., D.H.]; National Alliance for Research on Schizophrenia and Depression Young Investigator Award [to F.H.]; and Stanford Spectrum Child Health [to F.H.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Brown WE, Kesler SR, Eliez S, Warsofsky IS, Haberecht M, Patwardhan A, Ross JL, Neely EK, Zeng SM, Yankowitz J, Reiss AL. Brain development in Turner syndrome: a magnetic resonance imaging study. Psychiatry Res. 2002;116:187–196. doi: 10.1016/s0925-4927(02)00086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WE, Kesler SR, Eliez S, Warsofsky IS, Haberecht M, Reiss AL. A volumetric study of parietal lobe subregions in Turner syndrome. Dev Med Child Neurol. 2004;46:607–609. doi: 10.1017/s0012162204001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter WJ, Daly EM, Robertson DM, Chitnis XA, van Amelsvoort TA, Simmons A, Ng VW, Williams BS, Shaw P, Conway GS, Skuse DH, Collier DA, Craig M, Murphy DG. Influence of X chromosome and hormones on human brain development: a magnetic resonance imaging and proton magnetic resonance spectroscopy study of Turner syndrome. Biol Psychiatry. 2006;59:273–283. doi: 10.1016/j.biopsych.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Ecker C, Rocha-Rego V, Johnston P, Mourao-Miranda J, Marquand A, Daly EM, Brammer MJ, Murphy C, Murphy DG. Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. Neuroimage. 2010;49:44–56. doi: 10.1016/j.neuroimage.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Etkin A, Prater KE, Schatzberg AF, Menon V, Greicius MD. Disrupted amygdalar subregion functional connectivity and evidence of a compensatory network in generalized anxiety disorder. Arch Gen Psychiatry. 2009;66:1361–1372. doi: 10.1001/archgenpsychiatry.2009.104. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Vaituzis AC, Hamburger SD, Lange N, Rajapakse JC, Kaysen D, Vauss YC, Rapoport JL. Quantitative MRI of the temporal lobe, amygdala, and hippocampus in normal human development: ages 4–18 years. J Comp Neurol. 1996;366:223–230. doi: 10.1002/(SICI)1096-9861(19960304)366:2<223::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Good CD, Lawrence K, Thomas NS, Price CJ, Ashburner J, Friston KJ, Frackowiak RS, Oreland L, Skuse DH. Dosage-sensitive X-linked locus influences the development of amygdala and orbitofrontal cortex, and fear recognition in humans. Brain. 2003;126:2431–2446. doi: 10.1093/brain/awg242. [DOI] [PubMed] [Google Scholar]
- Gravholt CH. Clinical practice in Turner syndrome. Nat Clin Pract Endocrinol Metab. 2005;1:41–52. doi: 10.1038/ncpendmet0024. [DOI] [PubMed] [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene Selection for Cancer Classification using Support Vector Machines. Machine Learning. 2002;46:389–422. [Google Scholar]
- Haberecht MF, Menon V, Warsofsky IS, White CD, Dyer-Friedman J, Glover GH, Neely EK, Reiss AL. Functional neuroanatomy of visuo-spatial working memory in Turner syndrome. Hum Brain Mapp. 2001;14:96–107. doi: 10.1002/hbm.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE. Nonstationary cluster-size inference with random field and permutation methods. Neuroimage. 2004;22:676–687. doi: 10.1016/j.neuroimage.2004.01.041. [DOI] [PubMed] [Google Scholar]
- Hoeft F, Lightbody AA, Hazlett HC, Patnaik S, Piven J, Reiss AL. Morphometric spatial patterns differentiating boys with fragile X syndrome, typically developing boys, and developmentally delayed boys aged 1 to 3 years. Arch Gen Psychiatry. 2008;65:1087–1097. doi: 10.1001/archpsyc.65.9.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Blasey CM, Brown WE, Yankowitz J, Zeng SM, Bender BG, Reiss AL. Effects of X-monosomy and X-linked imprinting on superior temporal gyrus morphology in Turner syndrome. Biol Psychiatry. 2003;54:636–646. doi: 10.1016/s0006-3223(03)00289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Garrett A, Bender B, Yankowitz J, Zeng SM, Reiss AL. Amygdala and hippocampal volumes in Turner syndrome: a high-resolution MRI study of X-monosomy. Neuropsychologia. 2004;42:1971–1978. doi: 10.1016/j.neuropsychologia.2004.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Menon V, Reiss AL. Neuro-functional differences associated with arithmetic processing in Turner syndrome. Cereb Cortex. 2006;16:849–856. doi: 10.1093/cercor/bhj028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppel S, Stonnington CM, Barnes J, Chen F, Chu C, Good CD, Mader I, Mitchell LA, Patel AC, Roberts CC, Fox NC, Jack CR, Jr, Ashburner J, Frackowiak RS. Accuracy of dementia diagnosis: a direct comparison between radiologists and a computerized method. Brain. 2008a;131:2969–2974. doi: 10.1093/brain/awn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Jr, Ashburner J, Frackowiak RS. Automatic classification of MR scans in Alzheimer's disease. Brain. 2008b;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence K, Kuntsi J, Coleman M, Campbell R, Skuse D. Face and emotion recognition deficits in Turner syndrome: a possible role for X-linked genes in amygdala development. Neuropsychology. 2003;17:39–49. [PubMed] [Google Scholar]
- Mazzocco MM, Baumgardner T, Freund LS, Reiss AL. Social functioning among girls with fragile X or Turner syndrome and their sisters. J Autism Dev Disord. 1998;28:509–517. doi: 10.1023/a:1026000111467. [DOI] [PubMed] [Google Scholar]
- Molko N, Cachia A, Riviere D, Mangin JF, Bruandet M, LeBihan D, Cohen L, Dehaene S. Brain anatomy in Turner syndrome: evidence for impaired social and spatial-numerical networks. Cereb Cortex. 2004;14:840–850. doi: 10.1093/cercor/bhh042. [DOI] [PubMed] [Google Scholar]
- Mourao-Miranda J, Reynaud E, McGlone F, Calvert G, Brammer M. The impact of temporal compression and space selection on SVM analysis of single-subject and multi-subject fMRI data. Neuroimage. 2006;33:1055–1065. doi: 10.1016/j.neuroimage.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Murphy DG, Allen G, Haxby JV, Largay KA, Daly E, White BJ, Powell CM, Schapiro MB. The effects of sex steroids, and the X chromosome, on female brain function: a study of the neuropsychology of adult Turner syndrome. Neuropsychologia. 1994;32:1309–1323. doi: 10.1016/0028-3932(94)00065-4. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. High expression of the mammalian X chromosome in brain. Brain Res. 2006;1126:46–49. doi: 10.1016/j.brainres.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Nijhuis-van der Sanden RW, Smits-Engelsman BC, Eling PA. Motor performance in girls with Turner syndrome. Dev Med Child Neurol. 2000;42:685–690. doi: 10.1017/s0012162200001262. [DOI] [PubMed] [Google Scholar]
- Patwardhan AJ, Eliez S, Warsofsky IS, Glover GH, White CD, Giedd JN, Peterson BS, Rojas DC, Reiss AL. Effects of image orientation on the comparability of pediatric brain volumes using three-dimensional MR data. J Comput Assist Tomogr. 2001;25:452–457. doi: 10.1097/00004728-200105000-00020. [DOI] [PubMed] [Google Scholar]
- Raznahan A, Cutter W, Lalonde F, Robertson D, Daly E, Conway GS, Skuse DH, Ross J, Lerch JP, Giedd JN, Murphy DD. Cortical anatomy in human X monosomy. Neuroimage. 2010;49:2915–2923. doi: 10.1016/j.neuroimage.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AL, Mazzocco MM, Greenlaw R, Freund LS, Ross JL. Neurodevelopmental effects of X monosomy: a volumetric imaging study. Ann Neurol. 1995;38:731–738. doi: 10.1002/ana.410380507. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH, Morris JS, Dolan RJ. Functional dissociation of amygdala-modulated arousal and cognitive appraisal, in Turner syndrome. Brain. 2005;128:2084–2096. doi: 10.1093/brain/awh562. [DOI] [PubMed] [Google Scholar]
- Stonnington CM, Tan G, Kloppel S, Chu C, Draganski B, Jack CR, Jr, Chen K, Ashburner J, Frackowiak RS. Interpreting scan data acquired from multiple scanners: a study with Alzheimer's disease. Neuroimage. 2008;39:1180–1185. doi: 10.1016/j.neuroimage.2007.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-Planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System - An Approach to Cerebral Imaging. Thieme Medical Publishers; New York: 1988. [Google Scholar]
- Tamm L, Menon V, Reiss AL. Abnormal prefrontal cortex function during response inhibition in Turner syndrome: functional magnetic resonance imaging evidence. Biol Psychiatry. 2003;53:107–111. doi: 10.1016/s0006-3223(02)01488-9. [DOI] [PubMed] [Google Scholar]
- Temple CM. Oral fluency and narrative production in children with Turner's syndrome. Neuropsychologia. 2002;40:1419–1427. doi: 10.1016/s0028-3932(01)00201-9. [DOI] [PubMed] [Google Scholar]
- Temple CM, Carney R. Reading skills in children with Turner's syndrome: An analysis of hyperlexia. Cortex. 1996;32:335–345. doi: 10.1016/s0010-9452(96)80055-4. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Wechsler Intelligence Scale for Children. 3. The Psychological Corporation; San Antonio, TX: 1991. (WISC-III) [Google Scholar]
- Wechsler D. Wechsler Adult Inteligence Scale. 3. The Psychological Corporation; San Antonio, TX: 1997. (WAIS-III) [Google Scholar]
- Wechsler D. Wechsler Intelligence Scale for Children. 4. The Psychological Corporation; San Antonio, TX: 2003. (WISC-IV) [Google Scholar]
- Worsley KJ, Andermann M, Koulis T, MacDonald D, Evans AC. Detecting changes in nonisotropic images. Hum Brain Mapp. 1999;8:98–101. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<98::AID-HBM5>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Disteche CM. Sex differences in brain expression of X- and Y-linked genes. Brain Res. 2006;1126:50–55. doi: 10.1016/j.brainres.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]