Abstract
The PGC-1 family of regulated coactivators, consisting of PGC-1α, PGC-1β and PRC, plays a central role in a regulatory network governing the transcriptional control of mitochondrial biogenesis and respiratory function. These coactivators target multiple transcription factors including NRF-1, NRF-2 and the orphan nuclear hormone receptor, ERRα, among others. In addition, they themselves are the targets of coactivator and co-repressor complexes that regulate gene expression through chromatin remodeling. The expression of PGC-1 family members is modulated by extracellular signals controlling metabolism, differentiation or cell growth and in some cases their activities are known to be regulated by post-translational modification by the energy sensors, AMPK and SIRT1. Recent gene knockout and silencing studies of many members of the PGC-1 network have revealed phenotypes of wide ranging severity suggestive of complex compensatory interactions or broadly integrative functions that are not exclusive to mitochondrial biogenesis. The results point to a central role for the PGC-1 family in integrating mitochondrial biogenesis and energy production with many diverse cellular functions.
1. Introduction
Mitochondria are membrane bound organelles that engage in many important biological functions but are best known as the major sites of oxidative energy production in eukaryotic cells [1,2]. According to the endosymbiont hypothesis, mitochondria arose from the engulfment of aerobic eubacteria by a primordial anaerobic eukaryote, an event that was possibly coincident with the origin of eukaryotic cells [3]. Thus, the organelle has its own genetic system that exhibits several prokaryotic features including a compact circular DNA genome (mtDNA), multigenic RNA transcripts and bacteria-like antibiotic sensitivity of the translational apparatus. Most of the mitochondrial genes were lost to the nucleus leaving the organelle with the capacity to encode only 13 proteins and the 22 tRNAs and 2 rRNAs required for their translation within the mitochondrial matrix [4,5].
The limited coding capacity of mtDNA necessitates that nuclear genes specify most proteins of the respiratory apparatus as well as all of the enzymes required for other oxidative and biosynthetic functions. Bi-genomic gene expression is required exclusively for the respiratory apparatus and the translational machinery. All 13 proteins encoded by mtDNA function as essential subunits of respiratory complexes I, III, IV and V. These are expressed through the bi-directional synthesis of multigenic transcripts, followed by the processing, polyadenylation and translation of individual mRNAs [4,5]. Mitochondrial transcription requires a single RNA polymerase (POLRMT), initiation (TFB2M) and stimulatory (Tfam) transcription factors and a family of termination factors (mTERFs) [6,7]. In addition, both nuclear and mitochondrial gene products depend upon a complex series of import and assembly factors to direct them to the correct mitochondrial subcompartment. These include the multisubunit outer and inner membrane receptor complexes that function in conjunction with molecular chaperones to carry out the energy-dependent import of proteins localized to both soluble and membrane compartments [8]. More specialized factors are also required for proper assembly of the respiratory complexes imbedded in the inner membrane [9].
Changes in mitochondrial mass have been documented in both normal and disease states. For example, mitochondria differentiate postnatally to acquire increased respiratory capacity as an adaptation to oxygen exposure outside of the womb [10]. Mitochondrial biogenesis increases in muscle cells upon exercise [11] or in response to contraction induced by chronic electrical stimulation [12]. Thyroid hormones have long been associated with increased mitochondrial mass and the elevated expression of key metabolic enzymes and respiratory cytochromes in responsive tissues [13,14]. Finally, developmental signals induce the proliferation of mitochondria as occurs in the brown fat of rodents and other mammals during adaptive thermogenesis [15,16]. This review will focus on selective aspects of the PGC-1 family regulatory network in mitochondrial biogenesis and the expression of the respiratory apparatus. Because of the explosion of information on the PGC-1 coactivators in metabolic regulation the review is not intended to be comprehensive.
2. DNA Binding transcription factors in mitochondrial biogenesis
The first vertebrate regulatory factors implicated in the global expression of multiple mitochondrial functions were the nuclear respiratory factors, NRF-1 and NRF-2 (Fig. 1). The properties of these proteins have been reviewed and will not be considered in detail here [7,17]. NRF-1 was identified through its binding to a palindromic sequence in the cytochrome c promoter and has subsequently been associated with the expression of many genes required for expression and function of the respiratory chain. Similarly, NRF-2 was identified as a multisubunit activator of cytochrome oxidase expression and the human homolog of mouse GABP. The two factors frequently work in conjunction in the expression of many nuclear genes specifying essential mitochondrial functions [7,17,18]. Recent experiments have implicated NRF-1 and NRF-2 in the in vivo control of all ten nucleus-encoded cytochrome oxidase subunits [19,20].
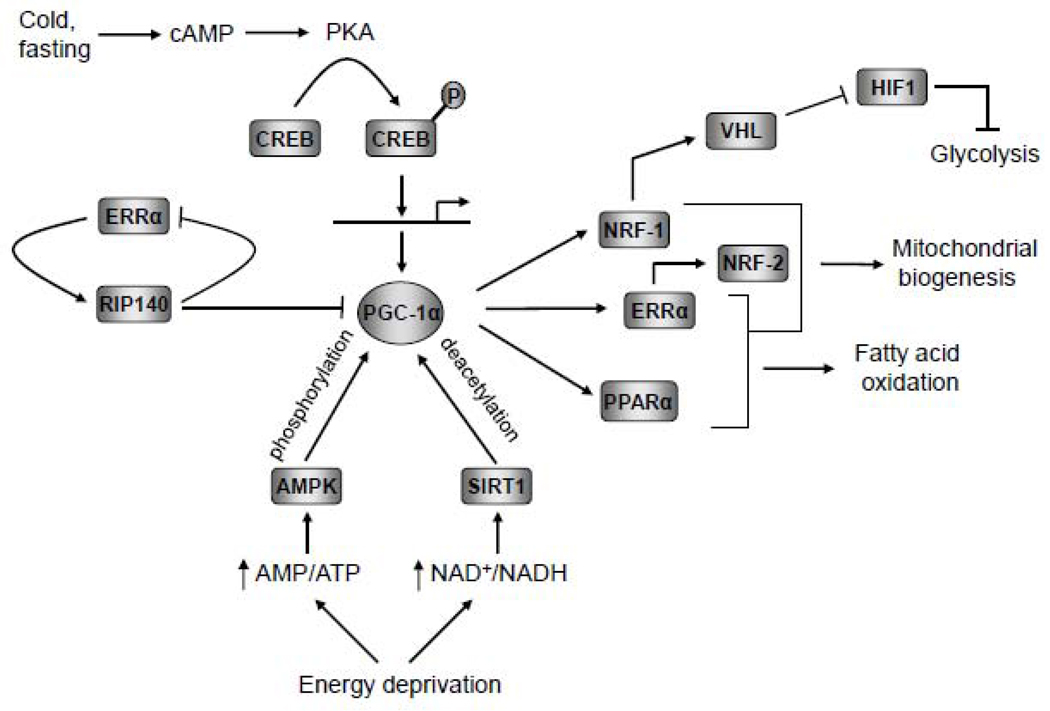
Fig. 1.
Regulatory network governing mitochondrial functions orchestrated by PGC-1α. Interactions among some key participants in the transcriptional network regulating mitochondrial biogenesis are depicted schematically. The diagram summarizes the regulation of PGC-1α by transcriptional and post transcriptional pathways and its interactions with some of its target transcription factors involved in metabolic regulation. Potential suppression of glycolysis through NRF-1 control of VHL expression and negative control by the RIP140 co-repressor are also shown. The specific action of each regulatory factor or pathway is discussed in the text.
Additional nuclear factors have been linked to the control of mitochondrial respiratory function. The cAMP response element binding protein, CREB, along with NRF-1 is involved in the growth-regulated expression of cytochrome c [21–23]. The initiator element binding factor, YY1, is engaged in both the positive and negative control of certain cytochrome oxidase subunit genes [24,25]. Muscle-specific COX genes depend upon MEF-2 and/or E-box consensus elements for their tissue-specific expression [26]. Interestingly, NRF-1 regulates the expression of MEF-2A in muscle accounting for the indirect NRF-1 control of muscle-specific COX subunits and other muscle-specific MEF-2 target genes [27]. The orphan nuclear receptor, ERRα (estrogen-related receptor), is abundantly expressed in highly oxidative tissues and promotes β-oxidation through its control of the medium chain acyl-coenzyme A dehydrogenase (MCAD) promoter [28]. As shown in Fig.1, ERRα is an important target in the control of mitochondrial biogenesis by PGC-1α [29,30] and may provide a regulatory link between fatty acid oxidation and the respiratory chain [31]. Finally, c-myc is associated with the expression of certain NRF-1 target genes [32] and myc null fibroblasts are deficient in mitochondrial content [33].
Recent studies have highlighted a potential molecular switch from c-myc-dependent mitochondrial oxidative phosphorylation to the promotion of aerobic glycolysis by hopoxia inducible factor (HIF) in cancer cells [34]. In this scheme, HIF-1, a transcriptional activator of the glycolytic pathway, functions as a negative regulator of mitochondrial biogenesis and oxygen utilization in renal carcinoma cells that lack the von Hippel-Lindau tumor suppressor (VHL). VHL is a negative regulator of HIF-1and its absence in carcinoma promotes the activation of the HIF pathway leading to enhanced glycolysis and the up regulation of pyruvate dehydrogenase kinase 1. This kinase inhibits the conversion of pyruvate to acetyl-CoA by pyruvate dehydrogenase resulting in decreased mitochondrial respiration. HIF-1 activation also promotes the HIF-dependent inhibition of c-myc transcription and degradation by the proteosome pathway [34]. Since c-myc is a positive regulator of PGC-1β, its repression by HIF-1 results in the inhibition of mitochondrial biogenesis and oxidative phosphorylation. Interestingly, the human VHL promoter region has a perfect canonical NRF-1 recognition site suggesting that its expression is under NRF-1 control [35]. Since, NRF-1 is a target for all of the PGC-1 family coactivators (see below), the PGC-1-NRF-1 pathway may promote mitochondrial oxidative metabolism in part through the up regulation of VHL expression (Fig. 1).
3. Phenotypes associated with the loss of essential mitochondrial functions
Much has been learned of the consequences associated with the loss of essential mitochondrial functions resulting from mtDNA disease mutations or from ablation of nuclear genes required for the maintenance of the respiratory apparatus. Striking proliferation of abnormal mitochondria is observed in muscle fibers from patients with certain mitochondrial myopathies and similar changes have been duplicated in mouse models of mitochondrial disease [36–38]. Moreover, homozygous knockout mice for several nuclear genes whose products are localized to mitochondria and function in the expression or maintenance of mtDNA have been generated. These include Tfam, an mtDNA transcription and maintenance factor [39], PolgA, a subunit of mtDNA polymerase [40], mTERF3, a transcription termination factor [41], and TFB1M, an RNA methyltransferase involved in ribosome assembly [42]. In each case, germ line homozygous null alleles result in embryonic lethality at approximately E8.5 accompanied by a severe respiratory chain deficiency. In addition, the Tfam and PolgA null embryos were depleted of mtDNA and the Tfam nulls exhibited increased numbers of aberrantly large mitochondria with poorly defined cristae. Cardiac-specific homozygous knockouts were also constructed and analyzed for Tfam [43], mTERF3 [41] and TFB1M [42]. In each case, the cardiac phenotype exhibited a number of shared features. These include cardiomyopathy, severe respiratory chain deficiency, defective assembly of respiratory complexes containing mtDNA-encoded subunits (complexes I, III, IV and V), and abundant, abnormal mitochondria with poorly defined cristae. Proliferation of abnormal mitochondria also occurs in mouse knockouts of the heart/muscle isoform of the adenine nucleotide translocator [36]. Thus, ablation of distinctly different nuclear genes, whose products function exclusively within the mitochondria to maintain the respiratory apparatus, results in a number of phenotypic similarities.
It is notable that homozygous null mice have also been generated for most of the nuclear transcription factors implicated in the control of mitochondrial biogenesis and none of them display the mitochondrial phenotype described above for nucleus-encoded factors that act within the mitochondria. Homozygous knockouts of several, including NRF-1 [44], NRF-2(GABP) [45] and YY1 [46], are early embryonic or preimplantation lethal, while others including ERRα [47] and PPARα [48] are viable and fertile with no global changes in gross mitochondrial number or morphology. NRF-1 null blastocysts exhibit mitochondrial DNA depletion and loss of mitochondrial membrane potential but likely have other non-mitochondrial defects that result in blastocyst arrest and failure to progress to E8.5 [44]. PPARα null mice have diminished expression of mitochondrial fatty acid oxidation enzymes [48] while ERRα knockouts have reduced lipogenesis and exhibit cold intolerance [47], but both are viable and fertile with no gross phenotypic abnormalities. More recently, the ERRα knockout mice were found to have reduced expression of mitochondrial oxidative pathways including oxidative phosphorylation in brown adipose tissue accompanied by decreased mitochondrial mass [49]. This was associated with an inability to maintain body temperature in response to cold exposure. The failure to observe global changes in basal energy metabolism in these animals may result from compensation by other regulators such as PGC-1α or ERRγ [50]. C-Myc homozygous null mice are embryonic lethal between E9.5 and E10.5 [51] and c-myc has been implicated in the control of cell cycle progression and metabolic networks [52]. These results suggest that the individual effects of these transcription factors on mitochondrial biogenesis are either part of a broad, highly integrated function, as in the case of early embryonic lethals, or serve to modulate the existing mitochondrial complement in response to physiological stress.
4. PGC-1α
PGC-1α is the founding member of a small family of structurally related transcriptional coactivators comprised of PGC-1α, PGC-1β and PRC (Fig. 2). PGC-1α and β share sequence similarities along their entire lengths and have been linked to a wide range of biological processes. Here, we will focus on their proposed role in nucleo-mitochondrial interactions. PGC-1α was identified on the basis of its induction during adaptive thermogenesis in brown fat and its interaction with PPARγ, an important regulator of adipocyte differentiation [53]. It lacks histone-modifying enzymatic activities but it interacts, through a potent amino-terminal activation domain, with an increasing number of coactivator complexes which contain intrinsic chromatin remodeling activities [54,55]. These include SRC-1, CBP/p300 and GCN5, among others. The complexity of these interactions, while poorly understood, is consistent with the hormonal and nutrient signaling pathways associated with PGC-1α (Fig. 1). In addition to PPARγ, PGC-1α binds a large complement of transcription factors and nuclear hormone receptors. Among those directly associated with mitochondrial respiratory function are NRF-1, ERRα, YY1, PPARα and MEF2C. Some of these act directly on the expression of the respiratory chain (NRF-1, ERRα, YY1, MEF2C) while others support the respiratory apparatus through their control of fatty acid oxidation (PPARα, ERRα).
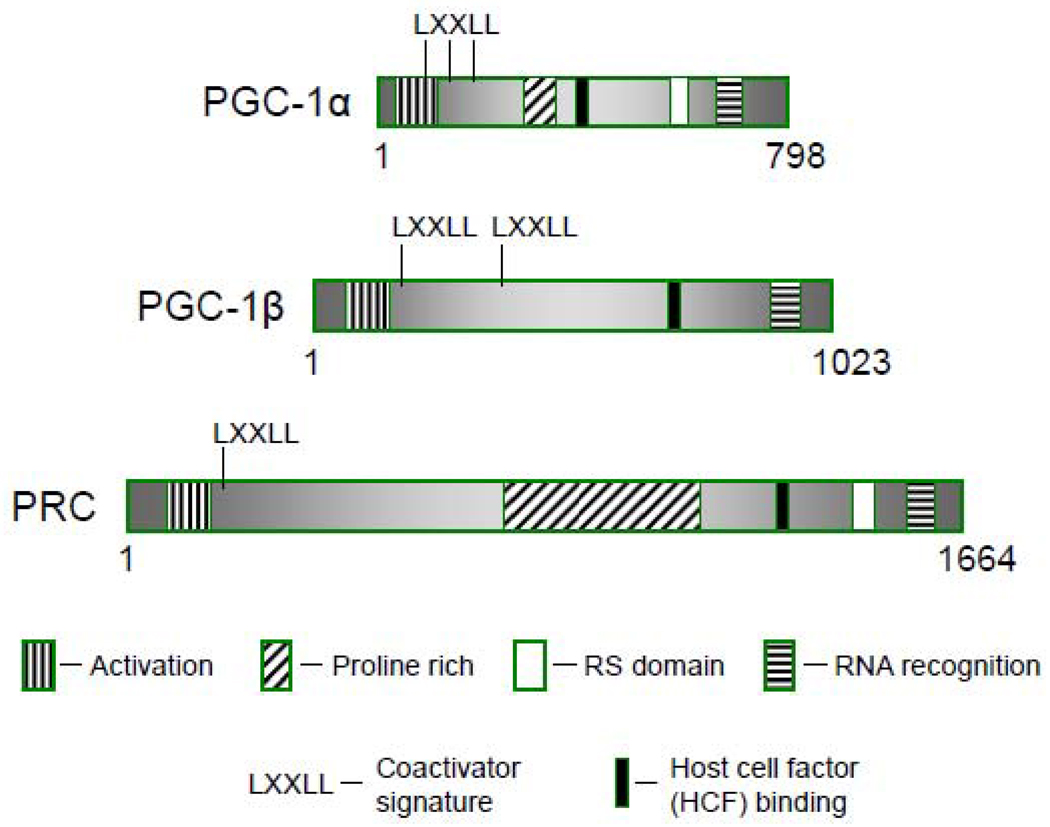
Fig. 2.
Arrangement of conserved domains among the PGC-1 family coactivators, PGC-1α, PGC-1β and PRC. Schematic comparison of PGC-1α, PGC-1β and PRC with the identities of the conserved sequence motifs shown in the key at the bottom.
NRF-1 was an initial transcription factor target identified for the induction of mitochondrial biogenesis by PGC-1α (Fig. 1). PGC-1α can trans-activate NRF-1 target genes and a dominant negative allele of NRF-1 blocks the effects of PGC-1α on mitochondrial biogenesis [56]. PGC-1α also targets estrogen-related receptor α (ERRα) in conjunction with GABPα (NRF-2α) in regulating respiratory genes including cytochrome c and β-ATP synthase [29,30]. NRF-1 and NRF-2α (GABPα) are thought to act downstream from both PGC-1α and ERRα in mediating respiratory gene expression [30,57]. ERRα may also exert direct control over GABPα transcriptional expression [30]. ERRα acts downstream from PGC-1α in orchestrating the program of adaptive thermogenesis [29] and cooperates with ERRγ and other transcription factors in driving the expression of a number of metabolic genes essential to proper cardiac function [57]. Interestingly, ERRα also stimulates the expression of the nuclear receptor corepressor RIP140 during adipogenesis [58], suggesting a role for ERRα in negative feedback control [59] (Fig. 1). RIP140 is known to suppress both oxidative metabolism and mitochondrial biogenesis through ERRα [60]. RIP140 also engages in a direct interaction with PGC-1α in suppressing the ERRα and NRF-1-dependent expression of CIDEA, a gene involved in both programmed cell death and metabolism [61]. Thus, PGC-1α utilizes a network of positive and negative transcriptional regulators in establishing and modulating metabolic function (Fig. 1).
The proposed role for PGC-1α as a regulator of mitochondrial biogenesis is supported by gain of function experiments in both cultured cells [56] and transgenic mice [62]. In cultured cells, ectopic PGC-1α expression increases COXIV and cytochrome c protein levels as well as the steady-state level of mtDNA [56]. In mice, cardiac-specific over expression in transgenic animals results in massive increases in mitochondrial content in cardiac myocytes leading to edema and dilated cardiomyopathy [62]. Over expression of PGC-1α or β may induce respiratory gene expression by overriding the ligand-dependence of target nuclear receptors. In particular, thyroid hormone receptors act as potent transcriptional repressors in the absence of ligand through recruitment of a corepressor complex. This repression is normally relieved by ligand binding resulting in the recruitment of a coactivator complex that includes CBP/p300 [63]. Strong binding of over expressed PGC-1α or β to these receptors may serve to displace the corepressor complex in the absence of ligand leading to the induction of the many metabolic genes controlled by thyroid hormones. Regardless of the mechanism, ectopic PGC-1α expression may prove useful in ameliorating the bioenergetic deficiency in mitochondrial myopathy [64].
More recently, metabolic signaling through PGC-1α was found to occur through posttranslational modifications (Fig. 1). For example, nutrient sensing through the NAD+ -dependent deacetylase, SITR1, may function, in part, by deacetylating multiple lysine residues in PGC-1α thereby promoting mitochondrial fatty acid oxidation in response to low glucose [65]. Similarly, AMP-activated protein kinase (AMPK), an enzyme sensor that is activated upon energy depletion in muscle [66], phosphorylates PGC-1α on specific serine and threonine residues. This results in increased mitochondrial gene expression supporting the idea that AMPK can mediate at least some of its effects through PGC-1α [67]. It is also apparent that the SIRT and AMPK pathways can work together in mediating calcium dependent mitochondrial proliferation in myocytes [68,69].
5. PGC-1β
A homologue of PGC-1α was designated as PGC-1β or PERC [70,71]. PGC-1α and β are closely related (Fig. 2) but PGC-1β lacks the arginine/serine (R/S) domain that is associated with RNA processing [70,72]. They are both enriched in tissues that contain abundant mitochondria such as heart, skeletal muscle and brown fat but PGC-1β is not induced in brown fat in response to cold exposure [70,71]. PGC-1β also exhibits a marked preference for promoting the ligand-dependent activity of ERα while having a minimal effect on ERβ [71]. Although initially implicated in liver gluconeogenesis [70], PGC-1β was subsequently found to be a poor inducer of gluconeogenic genes in liver and cultured hepatocytes because of its failure to interact with hepatic nuclear receptor 4α (HNF4α) and forkhead transcription factor O1 (FOXO1) [73,74]. However, PGC-1β appears identical to PGC-1α in its functional interaction with NRF-1 and in its ability to promote the expression of nuclear respiratory genes and mitochondrial mass when expressed from viral vectors [73,74]. Interestingly, despite the functional similarities between the two family members, PGC-1β promotes a much higher level of coupled respiration than PGC-1α because of differences in proton leak suggesting differences between the two in metabolic efficiency [75].
The functional differences between the two are further reflected by the observation that loss of PGC-1α, while reducing the expression of thermogenic genes, did not block brown fat differentiation [76]. In fact, loss of either coactivator alone had little effect on differentiation-induced mitochondrial biogenesis. However, silencing of PGC-1β in a PGC-1α null background, while leaving basal levels of mitochondria unaffected, completely abolished the increase in mitochondrial number and function upon brown fat differentiation [76]. This is indicative of complementary functions in promoting the differentiated state. The complementary role for these coactivators as differentiation factors is also seen at the tissue level in the specification of fiber types in skeletal muscle. Over expression of PGC-1α in the skeletal muscle of transgenic mice promotes the conversion to slow, oxidative type I and IIA muscle fibers [77]. By contrast, skeletal muscle over expression of PGC-1β induces the specific formation of type IIX fibers which are highly oxidative but have fast twitch characteristics [78]. This was accompanied by the induction of oxidative phosphorylation and fatty acid oxidation genes along with increased mitochondrial mass. Thus, in these gain of function experiments, the two factors can execute distinct programs of fiber type differentiation.
6. In vivo functions of PGC-1α and β revealed by gene knockouts
Massive increases in mitochondrial content both in cultured cells and in mouse tissues upon over expression of either PGC-1 or PGC-1β is thought to occur through coactivator interactions with key transcription factors (NRF-1, NRF-2, ERRα) in promoting the expression of genes required for the biogenesis of mitochondria [31,50,55,56,62,78,79]. Surprisingly, mice with a germ line targeted disruption of PGC-1α are viable and exhibit normal mitochondrial abundance and morphology in liver or brown fat [80]. A subtle mitochondrial phenotype is reflected in diminished oxygen consumption in isolated hepatocytes and lower steady-state levels of several mRNAs required for mitochondrial function. Both the striatum of the brain and brown adipose tissue were morphologically abnormal and the null mice were markedly hyperactive. This likely results from the loss of striatal axons and reduced expression of respiratory and brain-specific genes. Analysis of an independently constructed PGC-1α null mouse confirmed that the coactivator is not required for normal development or for the global biogenesis or maintenance of mitochondria [81]. It is important to note, however, that under physiological stress the PGC-1α knockouts exhibit more severe phenotypes such as an inability to defend body temperature in response to cold and reduced cardiac ATP production and work output in response to physiological stimuli [80–82]. A skeletal muscle-specific PGC-1α knockout mouse exhibited multiple muscle defects including reduced exercise tolerance and abnormalities in the maintenance of normal muscle fiber composition [83].
A similar theme is echoed in the construction and analysis of PGC-1β knockouts. Two PGC-1β homozygous nulls [84,85] and one homozygous hypomorphic variant that retains the partial ability to trans-activate through ERRα [86], were generated independently. In all three cases, the mice were viable and fertile and the homozygous null animals had normal food intake, energy expenditure and respiratory exchange ratios [85,86]. Thus, as with PGC-1α, the gene encoding PGC-1β is nonessential and by itself does not function as an absolute determinant of mitochondrial biogenesis. However, all of the PGC-1β mice exhibited some level of mitochondrial dysfunction especially under stress conditions. Although mitochondrial volume density was only marginally affected in brown fat, mRNAs encoding a number of nuclear respiratory genes were down regulated and the animals exhibited deficiencies in thermoregulation. The fact that elevated levels of PGC-1α mRNA in the brown fat of the PGC-1β knockouts could not compensate for the loss of PGC-1β is consistent with the conclusion that PGC-1β is not functionally redundant. Interestingly, mitochondrial content was somewhat reduced in soleus muscle and heart along with modest reductions in respiratory gene expression. However, where measured, the mitochondria retained normal internal structure and the respiratory function of isolated mitochondria was indistinguishable between the wild type and knockout animals [84–86]. The results support the idea that neither coactivator alone is essential for viability or for the biogenesis of a near normal complement of mitochondria. However, they each serve to optimize mitochondrial function especially under conditions of physiological stress.
Functional overlap between PGC-1α and β was recently revealed by the construction and analysis of PGC-1α/β double knockout mice [87]. These mice were viable at birth indicating that the absence of both coactivators was compatible with a full course of prenatal development. However, the majority died of cardiac failure within 24 hours with none surviving beyond 14 days. In contrast to the individual knockouts, the mitochondrial volume density in the brown fat of the PGC-1α/β double knockout was markedly reduced and the mitochondria exhibited diminished density of cristae coinciding with reduced expression of a number of respiratory genes. In addition, cardiac failure was accompanied by mitochondrial abnormalities in the heart marked by a reduction in mitochondrial number and size as well as the presence of internal vacuoles with reduced cristae. These differences resulted from a defect in mitochondrial maturation which normally occurs during the late fetal and early postnatal stages [10,88]. The expression of fatty acid oxidation and oxidative phosphorylation genes were also diminished including a number of ERRα and ERRγ target genes [87]. Since these factors are targets of the PGC-1 coactivators it is likely that impairment of the PGC-1-ERR pathway contributes to the mitochondrial maturation defect. These results argue that the two coactivators provide complementary functions in supporting postnatal cardiac differentiation.
7. PRC: black sheep of the family?
Because regulated coactivators are relatively rare, it was of interest to identify additional PGC-1 family members that might respond to alternative regulatory signals. A search for similarities to PGC-1α identified a large partial cDNA containing a carboxy-terminal RS domain and RNA recognition motif [89]. Molecular cloning of the full-length cDNA revealed additional conserved sequence motifs imbedded among otherwise dissimilar sequence (Fig. 2). These included an acidic amino-terminal region, an LXXLL signature for nuclear receptor coactivators and a central proline-rich region. Both the sequence and spatial conservation of these motifs was highly suggestive of related function. The encoded protein was thus designated as PGC-1α-related coactivator (PRC) [89].
PRC resembles PGC-1α and β in its binding to nuclear transcription factors associated with mitochondrial function including NRF-1, CREB and ERRα [23,90]. NRF-1 and CREB binding to PRC was mapped to 34 amino acids located between the amino-terminal activation domain and the proline-rich region and also to the R/S domain near the carboxy terminus [23]. The significance of the dual binding sites for transcription factors is not known but may be required for coupling transcription and RNA processing [72]. Immunoprecipitation experiments confirmed that antibodies directed against PRC could precipitate NRF-1 and CREB from cell extracts, indicating that these molecules likely associate in vivo as well [23,89,90].
In addition to the carboxy-terminal R/S and RRM, PRC and PGC-1α share an extremely potent, amino terminal, transcriptional activation domain that is required for NRF-1-dependent trans-activation by PRC [89]. PRC trans-activates a number of nuclear genes required for mitochondrial respiratory function including cytochome c, 5-aminolevulinate synthase, Tfam and the mitochondrial transcription factor B isoforms, TFB1M and TFB2M [89,91]. In each case, the functional NRF-1 recognition sites are required for maximal promoter activation by PRC [89,91]. PRC also utilizes CREB and NRF-2 sites for the activation of cytochrome c and TFB promoters, respectively. In the latter case, a head-to-head comparison demonstrated nearly identical requirements for activation of both TFB promoters by PRC and PGC-1α [91].
Despite these many similarities between PRC and the other PGC-1 family members there are also important differences that point to distinct biological functions. In mouse, PGC-1α mRNA is abundantly expressed in tissues with high mitochondrial content such as heart, kidney and brown fat [53] in contrast to PRC mRNA, whose expression does not appear to track mitochondrial abundance [89]. In addition, PGC-1α mRNA is markedly induced in brown fat in response to cold exposure in keeping with its role in adaptive thermogenesis [53] whereas PRC mRNA shows little induction under these conditions [89]. Thus, unlike PGC-1α, PRC expression is not linked to the variations in mitochondrial content associated with differentiated tissues.
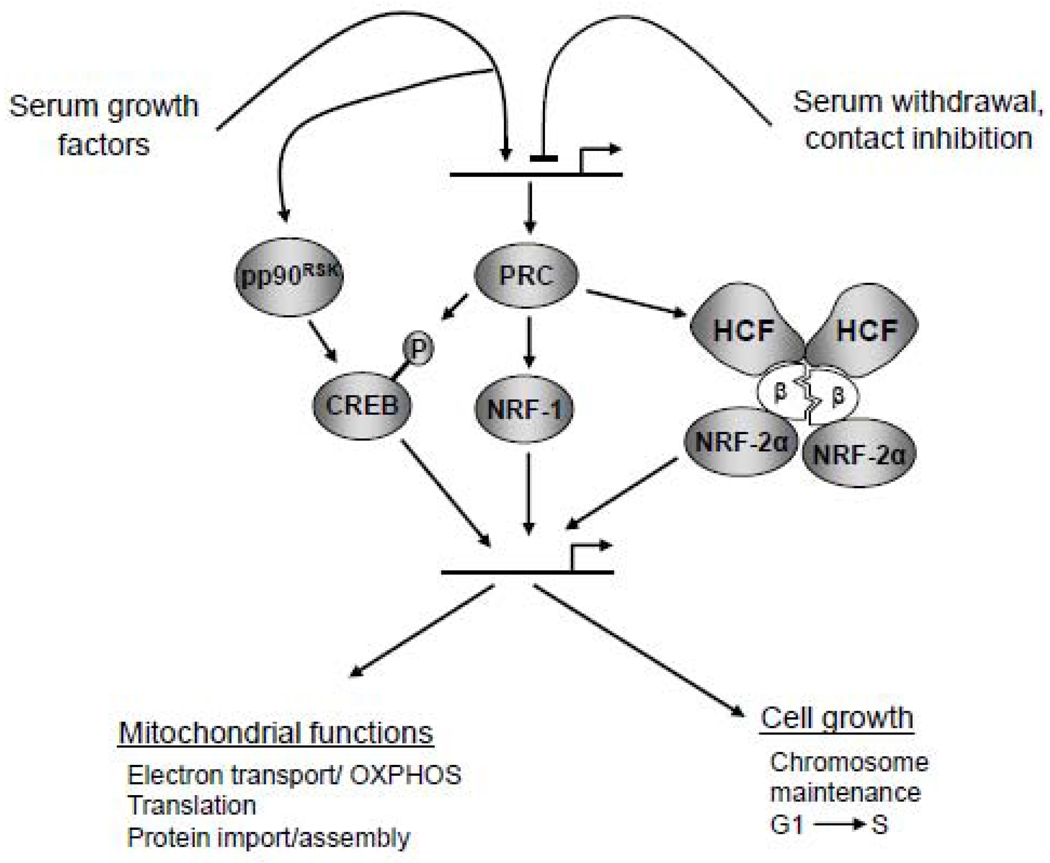
Interestingly, as depicted in Fig. 3, PRC mRNA and protein are expressed at higher levels in proliferating cells compared to those that are growth arrested because of serum withdrawal or contact inhibition [23,89]. PRC is also markedly induced upon serum stimulation of quiescent cells in the absence of de novo protein synthesis, demonstrating that its rapid induction occurs through the use of preexisting factors [23]. This property along with its relatively short mRNA half life is shared by a class of genes known as the immediate early genes. These genes include chemokines, growth factors, proto-oncogenes, serine-threonine kinases, enzymes of nucleic acid metabolism and transcription factors, among others [92,93]. PRC is the only known transcriptional coactivator belonging to this class of growth regulators suggesting a role for PRC in the integration of respiratory chain expression with the cell growth program (Fig. 3).
Fig. 3.
A model depicting the integration of mitochondrial and growth regulatory functions by PRC. PRC is rapidly induced by serum stimulation of quiescent cells and down regulated by serum withdrawal or contact inhibition. It binds both NRF-1 and CREB in stimulating the expression of cytochrome c and other genes required for mitochondrial function. It also engages in an indirect, functional interaction with NRF-2 through its binding of host cell factor (HCF). (Binding stoichiometry between NRF-2 and HCF has not been determined experimentally.) Regulation of PRC expression and interactions among transcriptional regulatory factors are discussed in the text.
Serum induction of PRC is associated with a gene expression profile that is similar to that observed in response to elevated PGC-1α [91,94]. This includes increased expression of Tfam, TFB1M and 2M mRNAs as well as those for both nucleus and mitochondria encoded respiratory subunits [94]. PRC, NRF-1 and Tfam are also elevated in thyroid oncocytomas along with increases in cytochrome oxidase activity and mtDNA content [95]. These thyroid tumors exhibit dense mitochondrial accumulation and are apparently lacking PGC-1α, suggesting that PRC may drive mitochondrial proliferation in these tumors.
8. Host cell factor (HCF): a platform for PGC-1 coactivator-NRF-2(GABP) interactions
Although both PRC and PGC-1α required NRF-2 binding sites for maximal activation of target promoters, there was no direct binding of the NRF-2α or β subunits to either coactivator in vitro. However, antibodies directed against NRF-2α or β subunits could efficiently immunoprecipitate PRC from cell extracts suggesting that PRC and NRF-2 are bound to the same in vivo complex through one or more intermediaries [90]. A candidate for such an intermediary is HCF (host cell factor), a molecule that binds both NRF-2β(GABPβ) [96] and the PGC-1 coactivators [70]. HCF is an abundant chromatin-associated protein that was identified as a cellular factor required for herpes virus immediate early gene expression [97] and for progression through G1 of the cell cycle [98]. It associates with a number of transcription factors including VP16 and NRF-2β(GABPβ) (Fig. 3) as well as chromatin remodeling activities. Like the other PGC-1 family coactivators [70], PRC binds HCF in vitro through a subfragment that contains the consensus DHDY HCF binding motif [90]. Moreover, antibodies directed against either PRC or HCF immunoprecipitate NRF-2β from cell extracts. In fact, deletion of the DHDY HCF binding site on PRC markedly reduced its ability to activate NRF-2-dependent transcription [90]. Likewise, the same amino acid residues required for HCF binding to the NRF-2β activation domain are also required for NRF-2-dependent promoter activation by PRC. Moreover, all three proteins have also been localized to the TFB1M and TFB2M promoters by chromatin immunoprecipitation confirming their association with biologically relevant promoters. Thus, HCF functions as a common platform for all three PGC-1 family members. The PGC-1 coactivators may be limiting components of a higher order transcription complex where HCF facilitates access to constitutively assembled transcription factors and chromatin modifying activities.
9. PRC silencing supports a role in nucleo-mitochondrial interactions
The similarities and differences between PGC-1α and PRC raise the question of whether PRC is functionally equivalent to PGC-1α, differing only in its responses to environmental signals. This appears not to be the case. PRC displays only weak binding to nuclear hormone receptors and this binding is not enhanced by ligand. In contrast, ligand enhances the binding of PGC-1α to both TRβ and RAR in vitro [53,90]. PRC also exhibits only very weak binding to PPARγ [90], a known target for PGC-1α [53]. Moreover, enhanced respiratory gene expression and mitochondrial biogenesis upon over expression of PRC in cultured cells has not been observed, although combined expression with ERRα does induce citrate synthase and cytochrome oxidase activities [99]. This may reflect the weak binding of PRC to nuclear hormone receptors.
Despite differences in gain of function experiments, PRC loss of function in cultured cells results in defects in respiratory chain expression and mitochondrial biogenesis. A subfragment of PRC that binds both NRF-1 and CREB inhibits the PRC activation of the cytochrome c promoter when present in trans. This same subfragment when stably expressed in lentiviral transductants also inhibits respiratory growth on galactose [23], which requires mitochondrial respiration and ATP production [100]. The results suggested that the interaction between PRC and its target transcription factors may be important for mitochondrial respiratory function in living cells [23].
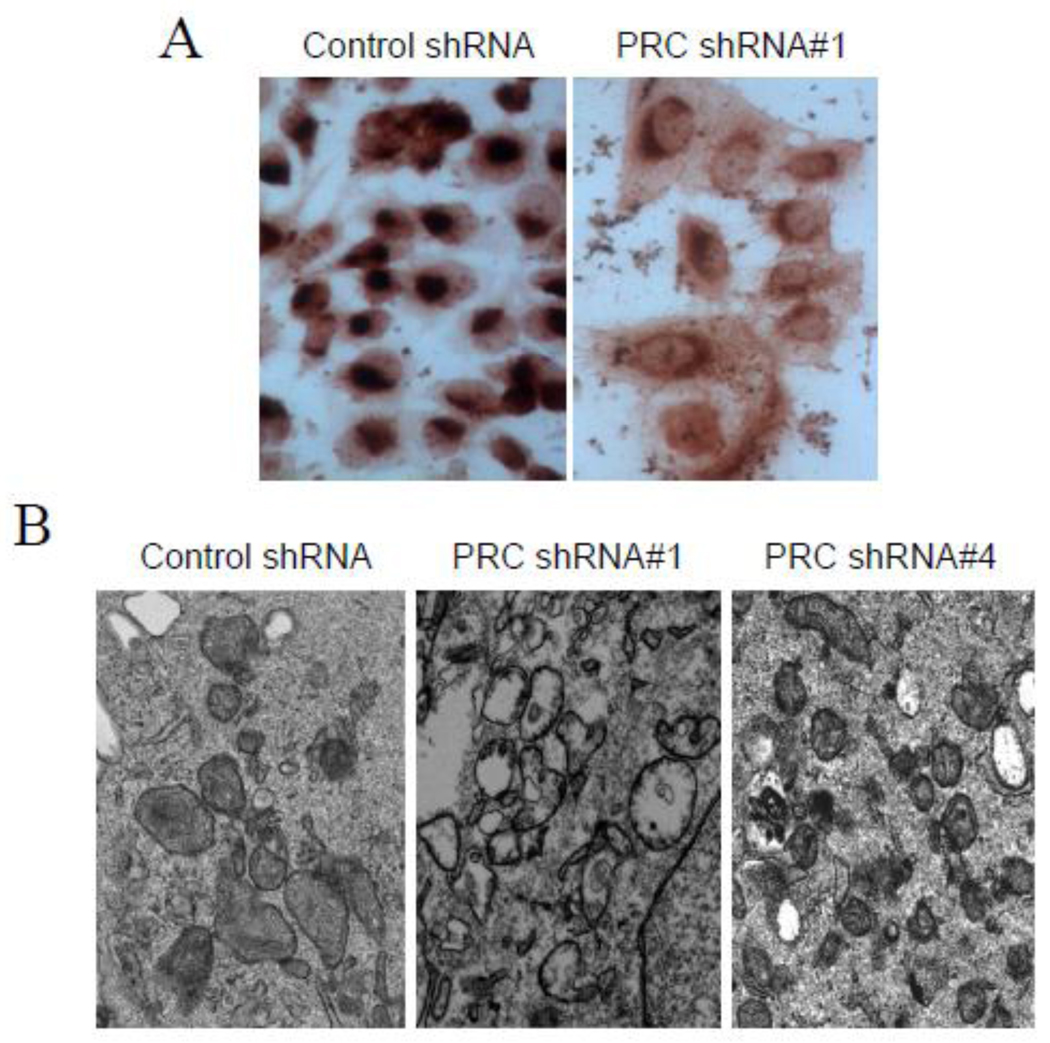
A more definitive link between PRC and mitochondrial biogenesis came from lentiviral transductants where PRC expression was diminished by short hairpin RNAs (shRNAs). Lentiviral expression of two distinct shRNAs, shRNA1 and shRNA4, designed from different PRC sequences, resulted in either complete (shRNA1) or partial (shRNA4) knockdown of PRC protein [101] in U2OS cells, where PRC expression is growth regulated [23]. Although both partial and complete PRC silencing resulted in growth inhibition in the G1/S transition, complete PRC silencing by shRNA1 resulted in nearly complete inhibition of growth on galactose. This was accompanied by diminished expression of both nuclear and mitochondrial respiratory chain subunits, lower levels of respiratory complexes I and IV (Fig. 4A) and reduced production of mitochondrial ATP. Moreover, the shRNA1 transductant had abundant, morphologically defective mitochondria marked by the absence of well defined cristae and a matrix space devoid of internal structure (Fig. 4B). This phenotype is reminiscent of that observed in the tissue-specific disruption of nuclear genes whose products are localized to mitochondria and are required by the mitochondrial genetic system [39,41,42]. The shRNA4 transductants, despite having increased numbers of smaller mitochondria (Fig. 4B) and a milder galactose growth defect, had no apparent deficit in respiratory chain expression and ATP production [101]. This indicates that the reduced level of PRC in these cells (about 15% of wild type) is sufficient to maintain near normal mitochondrial content and respiratory function. This low functional threshold may explain why PRC over expression alone does not enhance mitochondrial content and respiratory gene expression.
Fig. 4.
Effects of PRC silencing on mitochondrial morphology and oxidative function. (Panel A) Cytochrome oxidase staining of lentiviral transductants of U2OS cells expressing a control shRNA or PRC shRNA #1. Control shRNA transductants display a more rounded morphology with intense cytochrome oxidase staining of the cytoplasm. PRC shRNA#1 transductants display a flattened morphology with much weaker cytochrome oxidase staining. (Panel B) Electron micrographs of mitochondria from lentiviral transductants of U2OS cells expressing either a control shRNA or PRC shRNAs associated with either complete (shRNA#1) or partial (shRNA#4) PRC silencing. Complete PRC silencing in the PRC shRNA #1 transductant is associated with abundant, atypical mitochondria lacking well defined cristae.
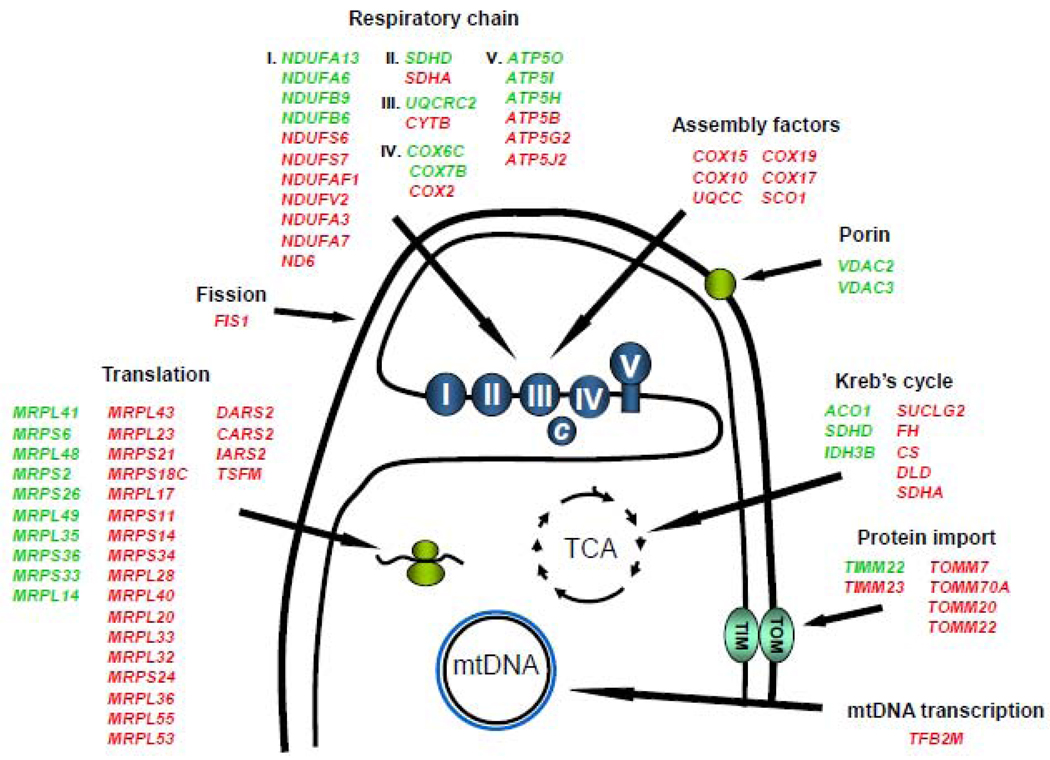
From the severity of the mitochondrial defect one might expect that respiratory gene expression would be affected in the PRC knockdown. Gene arrays revealed 79 mitochondria-related genes whose expression was altered significantly upon complete PRC silencing [101]. These included respiratory chain subunits, mitochondrial protein import and assembly factors, and mitochondrial ribosomal proteins and tRNA synthetases, among others (Fig. 5). Approximately two-thirds of the genes were down regulated. Notably, nearly all of the genes involved in protein import and assembly, which were affected by PRC silencing, showed reduced expression (Fig. 5). The diminished expression of multiple genes in this category could have an adverse effect, not only on the entire respiratory machinery, but also on the import of enzymes and other proteins necessary for the proper assembly of a functional organelle. A small subset of mitochondria-related genes was affected upon partial PRC silencing in keeping with the milder mitochondrial phenotype. Transient PRC silencing by siRNA also affects the expression of the respiratory chain at both the mRNA and protein level and was most pronounced for complexes I, II and IV [102]. This provides independent confirmation that PRC acts as a positive regulator of the mitochondrial respiratory complexes. Thus, blocking PRC function by independent means, dominant negative inhibition [23] and shRNA [101] or siRNA [102] silencing results in a mitochondrial phenotype with features similar to those observed in mouse knockouts of genes whose products function exclusively within the mitochondria. It is important to note that the severity of the mitochondrial phenotypes associated with PRC silencing may result from the absence of detectable PGC-1α and β expression in many cultured cell lines. It remains to be determined whether the PGC-1 coactivators can compensate for the loss of PRC in other physiological settings. Conversely, it has been suggested that PRC may support embryogenesis in the PGC-1α/β double knockout mouse [87].
Fig. 5.
Summary of the effects of PRC silencing on the expression of genes required for mitochondrial biogenesis and metabolic function. Genes whose expression is altered significantly (p < 0.01, FDR p < 0.05) upon complete PRC silencing in lentiviral transductants are listed under each functional category. Those up regulated are shown in green while those down regulated are shown in red. Arrows point to a schematic representation of the mitochondrial functions affected.
10. Summary and perspective
The PGC-1 family coactivators are key components in the molecular network that regulates the expression of the respiratory chain and the biogenesis of mitochondria. They engage in multiple interactions with a complex array of transcriptional regulators and are modified post-translationally in response to energy sensing pathways. Much work has focused on these coactivators in the expression of nucleus encoded respiratory subunits or proteins governing the mitochondrial genetic system. However, very little is known of their potential contributions to the synthesis and assembly of the membrane architecture comprising the mitochondrial reticulum itself. In addition, mouse knockouts have revealed important distinctions among the various regulatory factors. Homozygous knockouts of genes that function exclusively within the mitochondria exhibit phenotypic similarities including respiratory chain dysfunction and abundant abnormal mitochondria as is also seen in a number of human mitochondrial diseases. This suggests that the nuclear response to mitochondrial impairment at a number of sites within mitochondria can be fairly uniform. By contrast, knockout of individual components in the PGC-1 family regulatory network can result in diverse phenotypes ranging from early embryonic lethality to complete viability with very mild effects on global mitochondrial content and function. This indicates that some factors provide a unique and essential role that is not exclusive to mitochondria whereas others engage in compensatory interactions, possibly among family members, as illustrated by the PGC-1α/β double knockout. In either event, there appears to be no single regulator whose sole function is to exert exclusive control over mitochondrial content. Nevertheless, the PGC-1 family coactivators play a key role in modulating mitochondrial function and energy homeostasis in a number of physiological settings. Further understanding of the specific regulatory contributions of the three family members will continue to provide important insights into the control of mitochondrial energy metabolism during growth, differentiation and disease.
Acknowledgements
Work in the author’s laboratory was supported by National Institute of General Medical Sciences Grant GM 32525-27.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hatefi Y. The mitochondrial electron transport chain and oxidative phosphorylation system. Annu. Rev. Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 2.Balaban RS. Regulation of oxidative phosphorylation in the mammalian cell. Am. J. Physiol. Cell Physiol. 1990;258:C377–C389. doi: 10.1152/ajpcell.1990.258.3.C377. [DOI] [PubMed] [Google Scholar]
- 3.Gray MW, Burgess G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 4.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta Bio-Energetics. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC. A Mitochondrial Paradigm of Metabolic and Degenerative Diseases, Aging, and Cancer: A Dawn for Evolutionary Medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Scarpulla RC. Transcriptional paradigms in Mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 8.Becker T, Gebert M, Pfanner N, van der LM. Biogenesis of mitochondrial membrane proteins. Curr. Opin. Cell Biol. 2009;21:484–493. doi: 10.1016/j.ceb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Vizarra E, Tiranti V, Zeviani M. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta. 2009;1793:200–211. doi: 10.1016/j.bbamcr.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 10.Valcarce C, Navarrete RM, Encabo P, Loeches E, Satrustegui J, Cuezva JM. Postnatal development of rat liver mitochondrial functions. J. Biol. Chem. 1988;263:7767–7775. [PubMed] [Google Scholar]
- 11.Holloszy JO. Biochemical adaptations in muscle. Effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. J. Biol. Chem. 1967;242:2278–2282. [PubMed] [Google Scholar]
- 12.Williams RS, Garcia-Moll M, Mellor J, Salmons S, Harlan W. Adaptation of Skeletal Muscle to Increased Contractile Activity. J. Biol. Chem. 1987;262:2764–2767. [PubMed] [Google Scholar]
- 13.Tata JR, Ernster L, Lindberg O, Arrhenius E, Pederson S, Hedman R. The action of thyroid hormones at the cell level. Biochem. J. 1963;86:408–428. doi: 10.1042/bj0860408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booth FW, Holloszy JO. Effect of thyroid hormone administration on synthesis and degradation of cytochrome c in rat liver. Arch. Biochem. Biophys. 1975;167:674–677. doi: 10.1016/0003-9861(75)90511-1. [DOI] [PubMed] [Google Scholar]
- 15.Ricquier D, Bouillaud F. Mitochondrial uncoupling proteins: from mitochondria to the regulation of energy balance. J. Physio. 2000;529:3–10. doi: 10.1111/j.1469-7793.2000.00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 17.Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J. Cell Biochem. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- 18.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 19.Ongwijitwat S, Wong-Riley MT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J. Biol. Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gopalakrishnan L, Scarpulla RC. Differential regulation of respiratory chain subunits by a CREB-dependent signal transduction pathway. Role of cyclic AMP in cytochrome c and COXIV gene expression. J. Biol. Chem. 1994;269:105–113. [PubMed] [Google Scholar]
- 22.Herzig RP, Scacco S, Scarpulla RC. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J. Biol. Chem. 2000;275:13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- 23.Vercauteren K, Pasko RA, Gleyzer N, Marino VM, Scarpulla RC. PGC-1-related coactivator (PRC): immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol. Cell Biol. 2006;26:7409–7419. doi: 10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basu A, Lenka N, Mullick J, Avadhani NG. Regulation of murine cytochrome oxidase Vb gene expression in different tissues and during myogenesis - Role of a YY-1 factor- binding negative enhancer. J. Biol. Chem. 1997;272:5899–5908. doi: 10.1074/jbc.272.9.5899. [DOI] [PubMed] [Google Scholar]
- 25.Seelan RS, Grossman LI. Structural organization and promoter analysis of the bovine cytochrome c oxidase subunit VIIc gene - A functional role for YY1. J. Biol. Chem. 1997;272:10175–10181. doi: 10.1074/jbc.272.15.10175. [DOI] [PubMed] [Google Scholar]
- 26.Wan B, Moreadith RW. Structural characterization and regulatory element analysis of the heart isoform of cytochrome c oxidase VIa. J. Biol. Chem. 1995;270:26433–26440. doi: 10.1074/jbc.270.44.26433. [DOI] [PubMed] [Google Scholar]
- 27.Ramachandran B, Yu G, Gulick T. Nuclear respiratory factor 1 controls myocyte enhancer factor 2A transcription to provide a mechanism for coordinate expression of respiratory chain subunits. J. Biol. Chem. 2008;283:11935–11946. doi: 10.1074/jbc.M707389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ. Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor α (ERRα) functions in PPARgamma coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mootha VK, Handschin C, Arlow D, Xie XH, St Pierre J, Sihag S, Yang WL, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 32.Morrish F, Giedt C, Hockenbery D. C-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17:240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Kuzmin I, Duh FM, Latif F, Geil L, Zbar B, Lerman MI. Identification of the promoter of the human von Hippel-Lindau disease tumor suppressor gene. Oncogene. 1995;10:2185–2194. [PubMed] [Google Scholar]
- 36.Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, Wallace DC. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nature Genetics. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 37.Murdock DG, Boone BE, Esposito LA, Wallace DC. Up-regulation of nuclear and mitochondrial genes in the skeletal muscle of mice lacking the heart muscle isoform of the adenine nucleotide translocator. J. Biol. Chem. 1999;274:14429–14433. doi: 10.1074/jbc.274.20.14429. [DOI] [PubMed] [Google Scholar]
- 38.Torraco A, Diaz F, Vempati UD, Moraes CT. Mouse models of oxidative phosphorylation defects: powerful tools to study the pathobiology of mitochondrial diseases. Biochim. Biophys. Acta. 2009;1793:171–180. doi: 10.1016/j.bbamcr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsson NG, Wang JM, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genetics. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 40.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase γ is essential for mammalian embryogenesis. Hum. Mol. Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 41.Park CB, sin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, Falkenberg M, Gustafsson CM, Larsson NG. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 42.Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson N-G. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nature Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 44.Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol. Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ristevski S, O'Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. The ETS transcription factor GABPα is essential for early embryogenesis. Mol. Cell. Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse yin yang 1 transcription factor results in peri-implantation lethality. Mol. Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo J, Sladek R, Carrier J, Bader J, Richard D, Giguere V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor α. Mol. Bio. Cell. 2003;23:7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villena JA, Hock MB, Chang WY, Barcas JE, Giguere V, Kralli V. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1418–1423. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol. Metab. 2008;19:269–276. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 52.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–1066. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 54.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin JD. Minireview: the PGC-1 coactivator networks: chromatin-remodeling and mitochondrial energy metabolism. Mol. Endocrinol. 2009;23:2–10. doi: 10.1210/me.2008-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and function through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 57.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 58.White R, Morganstein D, Christian M, Seth A, Herzog B, Parker MG. Role of RIP140 in metabolic tissues: connections to disease. FEBS Lett. 2008;582:39–45. doi: 10.1016/j.febslet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 59.Nichol D, Christian M, Steel JH, White R, Parker MG. RIP140 expression is stimulated by estrogen-related receptor alpha during adipogenesis. J. Biol. Chem. 2006;281:32140–32147. doi: 10.1074/jbc.M604803200. [DOI] [PubMed] [Google Scholar]
- 60.Powelka AM, Seth A, Virbasius JV, Kiskinis E, Nicoloro SM, Guilherme A, Tang X, Straubhaar J, Cherniack AD, Parker MG, Czech MP. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J. Clin. Invest. 2006;116:125–136. doi: 10.1172/JCI26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hallberg M, Morganstein DL, Kiskinis E, Shah K, Kralli A, Dilworth SM, White R, Parker MG, Christian M. A functional interaction between RIP140 and PGC-1alpha regulates the expression of the lipid droplet protein CIDEA. Mol. Cell Biol. 2008;28:6785–6795. doi: 10.1128/MCB.00504-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu. Rev. Physiol. 2000;62:439–466. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- 64.Wenz T, Diaz F, Spiegelman BM, Moraes CT. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–256. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiol Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 67.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, Ogata H, Kubota N, Takamoto I, Hayashi YK, Yamauchi N, Waki H, Fukayama M, Nishino I, Tokuyama K, Ueki K, Oike Y, Ishii S, Hirose K, Shimizu T, Touhara K, Kadowaki T. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 70.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. PGC-1β: A novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 71.Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor α. J. Biol. Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 72.Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 73.Meirhaeghe A, Crowley V, Lenaghan C, Lelliott C, Green K, Stewart A, Hart K, Schinner S, Sethi JK, Yeo G, Brand MD, Cortright RN, O'Rahilly S, Montague C, Vidal-Puig AJ. Characterization of the human, mouse and rat PGC1β (peroxisome-proliferator-activated receptor-gamma co-activator 1β) gene in vitro and in vivo. Biochem. J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin JD, Tarr PT, Yang RJ, Rhee J, Puigserver P, Newgard CB, Spiegelman BM. PGC-1β in the regulation of hepatic glucose and energy metabolism. J. Biol. Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 75.St Pierre J, Lin J, Krauss S, Tarr PT, Yang RJ, Newgard CB, Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J. Biol. Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 76.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 77.Lin J, Wu H, Tarr PT, Zhang CY, Wu ZD, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional coactivator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 78.Arany Z, Lebrasseur N, Morris C, Smith E, Yang W, Ma Y, Chin S, Spiegelman BM. The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 79.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin J, Wu P, Tarr PT, Lindenberg KS, St-Pierre J, Zhang C, Mootha VK, Jäger S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan ML, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 81.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS. Biol. 2005;3:0672–0687. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 83.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance and myopathy in PGC-1alpha muscle-specific knockout animals. J. Biol. Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 84.Lelliott CJ, Medina-Gomez G, Petrovic N, Kis A, Feldmann HM, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, Zhang D, Litwin SE, Zaha VG, Fountain KT, Boudina S, Jimenez-Linan M, Blount M, Lopez M, Meirhaeghe A, Bohlooly Y, Storlien L, Stromstedt M, Snaith M, Oresic M, Abel ED, Cannon B, Vidal-Puig A. Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS. Biol. 2006;4:2042–2056. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vianna CR, Huntgeburth M, Coppari R, Choi CS, Lin J, Krauss S, Barbatelli G, Tzameli I, Kim YB, Cinti S, Shulman GI, Spiegelman BM, Lowell BB. Hypomorphic mutation of PGC-1beta causes mitochondrial dysfunction and liver insulin resistance. Cell Metab. 2006;4:453–464. doi: 10.1016/j.cmet.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2010;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marin-Garcia J, Ananthakrishnan R, Goldenthal MJ. Heart mitochondrial DNA and enzyme changes during early human development. Mol. Cell Biochem. 2000;210:47–52. doi: 10.1023/a:1007031919298. [DOI] [PubMed] [Google Scholar]
- 89.Andersson U, Scarpulla RC. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell. Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vercauteren K, Gleyzer N, Scarpulla RC. PGC-1-related Coactivator Complexes with HCF-1 and NRF-2{beta} in Mediating NRF-2(GABP)-dependent Respiratory Gene Expression. J. Biol. Chem. 2008;283:12102–12111. doi: 10.1074/jbc.M710150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu. Rev. Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 93.Winkles JA. Serum- and polypeptide growth factor-inducible gene expression in mouse fibroblasts. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:41–78. doi: 10.1016/s0079-6603(08)60033-1. [DOI] [PubMed] [Google Scholar]
- 94.Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann. N. Y. Acad. Sci. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Savagner F, Mirebeau D, Jacques C, Guyetant S, Morgan C, Franc B, Reynier P, Malthièry Y. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem. Biophys. Res. Commun. 2003;310:779–784. doi: 10.1016/j.bbrc.2003.09.076. [DOI] [PubMed] [Google Scholar]
- 96.Vogel JL, Kristie TM. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 2000;19:683–690. doi: 10.1093/emboj/19.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiao P, Capone JP. A cellular factor binds to the herpes simplex virus type 1 transactivator Vmw65 and is required for Vmw65-dependent protein-DNA complex assembly with Oct-1. Mol. Cell Biol. 1990;10:4974–4977. doi: 10.1128/mcb.10.9.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima M, Herr W, Nishimoto T. A single point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 99.Mirebeau-Prunier D, Le PS, Jacques C, Gueguen N, Poirier J, Malthiery Y, Savagner F. Estrogen-related receptor alpha and PGC-1-related coactivator constitute a novel complex mediating the biogenesis of functional mitochondria. FEBS J. 2010;277:713–725. doi: 10.1111/j.1742-4658.2009.07516.x. [DOI] [PubMed] [Google Scholar]
- 100.Robinson BH. Use of fibroblast and lymphoblast cultures for detection of respiratory chain defects. Methods Enzymol. 1996;264:454–464. doi: 10.1016/s0076-6879(96)64041-5. [DOI] [PubMed] [Google Scholar]
- 101.Vercauteren K, Gleyzer N, Scarpulla RC. Short hairpin RNA-mediated silencing of PRC (PGC-1-related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. J. Biol. Chem. 2009;284:2307–2319. doi: 10.1074/jbc.M806434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Raharijaona M, Le PS, Poirier J, Mirebeau-Prunier D, Rouxel C, Jacques C, Fontaine JF, Malthiery Y, Houlgatte R, Savagner F. PGC-1-related coactivator modulates mitochondrial-nuclear crosstalk through endogenous nitric oxide in a cellular model of oncocytic thyroid tumours. PLoS. One. 2009;4:e7964. doi: 10.1371/journal.pone.0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]