Abstract
The use of the Protemist XE, an automated discontinuous-batch protein synthesis robot, in cell-free translation is reported. The soluble Galdieria sulphuraria protein DCN1 was obtained in greater than 2 mg total synthesis yield per mL of reaction mixture from the Protemist XE, and the structure was subsequently solved by X-ray crystallography using material from one 10 mL synthesis (PDB ID: 3KEV). The Protemist XE was also capable of membrane protein translation. Thus human sigma-1 receptor was translated in the presence of unilamellar liposomes and bacteriorhodopsin was translated directly into detergent micelles in the presence of all-trans-retinal. The versatility, ease of use, and compact size of the Protemist XE robot demonstrate its suitability for large-scale synthesis of many classes of proteins.
Introduction
Recent advances, including optimization of the extract preparation and innovations in automation, have greatly improved the productivity of cell-free translation as a tool for protein studies (1-5). Cell-free translation can efficiently support uniform (6) and selective (7) labeling strategies needed for NMR structure determination. This approach also allows the high-fidelity incorporation of SeMet needed for phase determination by X-ray crystallography. Consequently, the ability of cell-free translation to generate sufficient protein for structure determination by NMR spectroscopy or X-ray crystallography has been advancing progressively. Applications of this approach to facilitate functional discovery are also emerging (8,9).
Membrane protein production represents another significant challenge for the biology community. Recent advances in cell-free translation have facilitated the preparation of membrane proteins, and several reviews are available (10-12). One significant advantage of cell-free translation is that stabilizing detergents and lipids can be added directly to the translation reactions to encourage proper folding and solubilization (13). However the breadth of membrane protein types investigated and the number of solved structures are still limited. Therefore, additional methods and better approaches to link small-scale discovery and optimization with large-scale production are needed.
Here we compare the performance of the Protemist XE, a discontinuous-batch robot, with the Protemist10 and the Protemist100 robots (2,14), the latter of which are repeat-batch automated translation systems. In the following, we will use the term ‘Protemist10/100’, to describe both of these robots, as their translation protocols are the same. The Protemist10/100 are large, multi-reaction, completely contained floor-standing robots (W 1.1 m × D 0.9 m × H 1.9 m), while the Protemist XE is a bench-top, single reaction robot. The two robotic platforms were compared for their ability to synthesize protein with yield adequate for structure determination in a single operation. The ability of the Protemist XE to synthesize membrane proteins in the presence of liposomes and/or detergents was also examined. In summary, the Protemist XE robot provides versatility and ease of use, making it suitable for the large-scale synthesis of many classes of proteins. It also represents a useful platform for studying the co-translational stabilization of membrane proteins by exogenously added lipids or detergents.
Materials and Methods
Target proteins
The proteins studied are listed in Table 1. The soluble DCN1 protein from the unicellular red algae Galdieria sulphuraria was chosen for structure determination as a homolog to the yeast DCN1-like 2 neddylation protein involved in protein ubiquitination (15). Two additional proteins were the human sigma-1 receptor (S1R), a 2-transmembrane domain protein involved in calcium signaling known to bind several classes of ligands (16-18), and the Halobacterium salinarium bacteriorhodopsin (19), a 7-transmembrane domain protein with a well-characterized structure and other biophysical properties (20). H. salinarium bacteriorhodopsin was purchased from Sigma-Aldrich (St. Louis, MO) for use as a control.
Table 1.
Expression and purification yields of proteins prepared by cell-free translation
| Protein | Species | Robot | WEPRO Extract | Reaction conditionsa | Labeling | Reaction scale (mL) | Extract volume (mL) | Synthesis yield (mg) | Purified yieldd (mg) |
|---|---|---|---|---|---|---|---|---|---|
| DCN1 | Galdieria sulphuraria | Protemist10/100 | 2240 | A | [13C,15N] | 24 | 6 | 14 | 3.8 |
| DCN1 | Galdieria sulphuraria | Protemist XE | 8240 | B | SeMet | 10 | 5 | 24 | 4.3 |
| S1R | Homo sapiens | Protemist XE | 8240H | C | none | 10 | 5 | 50b | n/d |
| bR | Halobacterium salinarum | Protemist10/100 | 8240H | D | none | 4 | 1 | 1b | n/d |
| bR | Halobacterium salinarum | Protemist10/100 | 8240H | E | none | 4 | 1 | 1b | n/d |
| bR | Halobacterium salinarum | Protemist10/100 | 8240H | F | none | 4 | 1 | 1b | n/d |
| bR | Halobacterium salinarum | Protemist XE | 8240H | G | SeMet | 10 | 5 | 5c | 0.4 |
| bR | Halobacterium salinarum | Protemist XE | 8240 | G | SeMet | 10 | 5 | 4c | 0.9 |
| bR | Halobacterium salinarum | Protemist XE | 2240H | H | SeMet | 10 | 5 | 4c | 1.3 |
A, standard conditions for the Protemist10/100 described in Materials and Methods. B, standard conditions for the Protemist XE described in Materials and Methods. C, conditions from B with 2.4 mg/mL soybean lecithin unilamelar liposomes added to translation reaction. D, 0.1 mM all-trans retinal, 0.4% CHAPS, and 0.2% FC-12 in reaction, 0.1 mM all-trans retinal and 0.04 % CHAPS in feed buffer. E, 0.1 mM all-trans retinal, 0.4% CHAPS, and 0.2% FC-12 in reaction, 0.1 mM all-trans retinal, 0.04% CHAPS, and 0.02% FC-12 in feed buffer. F, 0.1 mM all-trans retinal, 0.4% CHAPS, and 0.2% FC-12 in reaction; 0.1 mM all-trans retinal in feed buffer; G, 0.1 mM all-trans retinal, 0.4% CHAPS, and 0.2% FC-12 in reaction, 0.1 mM all-trans retinal and 0.02% CHAPS in feed buffer. H, 0.1 mM all-trans retinal and 0.2% CHAPS in reaction and feed buffer.
Total synthesis yields were estimated by comparing band intensity on Coomassie-stained SDS-PAGE gels to that of the known quantity of a 31-kDa band in molecular weight standards.
Total synthesis yields were estimated by comparing tryptophan fluorescence to that of quantified marker proteins.
Purification yields were determined by BCA.
Plasmid preparation
All genes were cloned into pEU-His-Flexi adapted for wheat germ cell-free expression (21) using FlexiVector cloning (22). All three proteins were translated from this vector to include an N-terminal His6 tag. Standard FlexiVector cloning reagents were from Promega (Madison, WI). pEU-His-Flexi is available through the Protein Structure Initiative Material Repository (PSI-MR, http://psimr.asu.edu/). Large-scale plasmid preparations were performed using Marligen Biosciences Maxiprep kits (Rockville, MD) followed by treatment of purified plasmid with proteinase K (Part No. P2308 Sigma-Aldrich) to remove residual Rnase (14). Protease-treated plasmids were extracted twice with phenol:chloroform (1:1), precipitated in ethanol, and reconstituted in deionized water. Plasmid concentrations were determined by UV spectrometry.
mRNA synthesis
For small-scale translation reactions, transcription was performed by combining 20μg of ∼0.4μg/μL plasmid with transcription buffer TM (80 mM HEPES-KOH, pH 7.8, containing 20mM magnesium acetate, 2mM spermidine hydrochloride, 10mM dithiothreitol, 4mM NTPs, 1.6U/μL SP6 RNA polymerase (Promega), and 0.8U/μL RNase inhibitor (Promega) in a final volume of 5μL for each small-scale reaction. For large-scale translation reactions using the Protemist10/100 robots (CellFree Sciences, Yokohama, Japan), the amount of plasmid was increased to 200μg of a 1μg/μL solution and other reagent volumes were adjusted accordingly to give a final reaction volume of 4 mL. For a 10mL translation reaction using the Protemist XE (CellFree Sciences), 19.5mL of transcription reaction was required, and the amount of plasmid needed was increased to 750μg of a 1μg/μL solution.
Transcription reactions were incubated for 4h at 37°C. The large-scale reactions were clarified by centrifugation to remove precipitated magnesium pyrophosphate before they were added to the translation reactions. RNA prepared for use with the WEPRO8240 or WEPRO8240H extracts utilized low magnesium 5× transcription buffer from CellFree Sciences (LM buffer, Part #CFS-SUB-SGC-NAA, CellFree Sciences). Transcriptions in the LM buffer were performed for 6h at 37°C.
Small-scale translation
Small-scale translation reactions for comparison among the buffers and extracts were performed in 20μL, using dialysis cups with 12,000 MWCO membranes (Cosmo Bio, Tokyo, Japan) and 2mL receptacle tubes (CellFree Sciences). The reaction mixtures consisted of a half volume of a wheat germ extract, a quarter volume of RNA, 0.05 mg/mL creatine kinase, and the rest of the volume was adjusted with a 1× translation buffer with 0.3mM unlabeled amino acids (either DB or SUB-AMIX SG, defined below). The 1× DB translation buffer (14) consists of 24mM HEPES-KOH, pH 7.8, containing 100mM potassium acetate, 2.5mM magnesium acetate, 0.4mM spermidine-HCl, 4mM dithiothreitol, 1.2mM ATP, 0.25mM GTP, 16mM creatine phosphate, and 0.005% (w/v) sodium azide. The 1× SUB-AMIX SG translation buffer was prepared from 40× concentrates, S1, S2, S3, and S4 (Part #CFS-SUB-SGC-NAA, CellFree Sciences), and amended to contain 0.005% (w/v) sodium azide, and 0.025mg/mL chloramphenicol.
The 2mL receptacle tubes contained 800μL of DB or SUB-AMIX SB buffer with 0.3mM non-labeled amino acids. Translation reactions were conducted at ambient temperature for 41h; the mRNA was supplemented and the translation buffer was replaced at 17h from the start in order to slightly mimic the robotic synthesis. For bacteriorhodopsin expression, detergents and 0.1mM all-trans-retinal were also added to the feed buffer and reaction mixture. All-trans-retinal was prepared as a 100mM stock in ethanol. Translation reactions were conducted in 40μL at ambient temperature for 20h without supplementation of mRNA during synthesis.
Large-scale translation
Translation reactions using the Protemist10/100 are described elsewhere (14). Briefly, 4mL reaction mixtures were prepared in 10,000 MWCO 15mL capacity ultrafiltration concentrators consisting of 1mL wheat germ extract (a 1:4 dilution of the commercial preparation), 1.5mL mRNA solution, and 0.6mg/mL creatine kinase. The remainder of the reaction volume was provided by 1× DB translation buffer containing amino acids. A 50mL aliquot of feed buffer with 2.5mL RNA was prepared for repeated additions over the course of 18h.
The set-up for translation reactions using the Protemist XE was similar to that of the Protemist10/100, but in all cases, the SUB-AMIX SG translation buffer was used. In the Protemist XE, a 10mL reaction consisted of 5mL wheat germ extract, 2.5mL RNA, 0.1mg/mL creatine kinase, and 2.5mL 1× SUB-AMIX SG translation buffer with amino acids. A 770mL aliquot of 1× SUB-AMIX SG feed buffer and a separate tube of 15mL of additional mRNA were prepared for use during the translation cycle. The Protemist XE uses a disposible ultrafiltration cassette (VIVAFLOW 50, MWCO 5000, Part # VF05P1, Sartorius Stedim Biotech, Goettingen, Germany) for every 10mL reaction volume.
For SeMet labeling, 0.3mM each of 19 amino acids other than methionine and 0.6mM selenomethionine were used. For [13C,15N]-labeling, a uniform labeled 20 amino acid mixture (Cambridge Isotope Laboratories, Andover, MA) was used as 0.1% (w/v) concentration.
For translation of DCN1, the above conditions were used. For translation of sigma-1 receptor, 1.6mL of a 15mg/mL preparation of unilamelar liposomes was added to the reaction mixture. For translation of bacteriorhodopsin, detergents and 0.1mM all-trans-retinal were added to the feed buffer and reaction mixture.
Purification of DCN1
Small-scale purification was performed in 96-well format (14). For large-scale purification, the translation reaction was clarified by centrifugation at 9,300×g for 10 min at 4°C. The supernatant was loaded onto a 1mL His-Trap HP column (GE Healthcare, Piscataway, NJ) equilibrated in 50mM NaH2PO4, pH8.0, 50mM imidazole, 300mM NaCl, and 2mM dithiothreitiol at room temperature (RT). An ÄKTAprime (GE Healthcare) was used for the chromatography. The fusion protein was eluted in a 2mL aliquot of the buffer containing 500mM imidazole. The fusion protein was concentrated to 300μL, buffer-exchanged to gel filtration buffer (20 mM Bis-Tris, pH 7.0, containing 100mM NaCl and 5mM dithiothreitol) and treated at room temperature overnight with 0.2mg of His-tagged TEV protease (23). The TEV protease-treated fusion protein was subjected to subtractive IMAC, and the flow-through was applied to a Superdex-75 gel filtration column, equilibrated in the gel filtration buffer. The pooled fractions from the gel filtration were concentrated to ∼0.5mM. The [13C,15N]- and SeMet-labeled samples were purified in the same manner, except that an additional buffer exchange step into a crystallization screening buffer consisting of 5mM HEPES, pH 7.0, containing 50mM NaCl, and 0.3mM tris(2-carboxyethyl)phosphine hydrochloride was included before the final concentration step for the SeMet-labeled sample.
Purification of bacteriorhodopsin
Precipitated bR from a 10mL Protemist XE translation reaction was pelleted by centrifugation at 7,000×g for 15min at 10°C in an Allegra X-22R (Beckman-Coulter, Brea, CA) centrifuge. The pellet was resuspended in 10mL of binding buffer (20mM NaH2PO4, pH8.0, containing 50mM imidazole, 300mM NaCl, 0.5% FC-12) and incubated at room temperature for 2h. During this time, the majority of the bR was solubilized. The suspension was centrifuged again and the soluble fraction was applied to a 5-mL HisTrap-HP column equilibrated in binding buffer containing 0.05% FC-12 and 0.3mM tris(2-carboxyethyl)phosphine hydrochloride on an ÄKTAprime. The bound bR was washed with 10 column volumes of binding buffer containing 0.05% DDM, and eluted in 20mM NaH2PO4, pH8.0, containing 0.3mM tris(2-carboxyethyl)phosphine hydrochloride, and 0.05% DDM with increasing imidazole and decreasing NaCl gradients to 500 mM and 0 mM, respectively.
The eluted purple fractions were immediately pooled and exchanged into gel filtration buffer (10mM MES, pH 5.5, 100mM NaCl, 0.3mM tris(2-carboxyethyl)phosphine hydrochloride, 0.025% DDM) using a 50,000 MWCO Amicon Ultra-15 spin concentrator (Millipore, Billerica MA). This material was concentrated to 500μL and applied to a Superdex-200 10/30-GL 24mL gel filtration column in the same buffer and filtered at 4°C on an ÄKTAFPLC (GE Healthcare). Purple fractions were pooled and quantified by absorbance at 280 and 555nm. The purified bR was concentrated to 10mg/mL using a 50,000 MWCO spin concentrator and stored either at 4°C or frozen as 10μL droplets in liquid N2 and stored at -80°C.
The following detergents were included in a screening panel: Anzergent 3-10; Brij35; CHAPS; DDM, FOS-CHOLINE-12; HEGA-9; LDAO; MEGA-8, Nonidet P40; nonyl-thiogluclopyranoside; octyl-glucopyranoside; octy-thioglucopyranoside; SDS; Triton X-100; Tween 40; Tween 80.
SDS-PAGE and protein quantification
SDS-PAGE analysis was performed using Criterion 10-20% Tris-HCl polyacrylamide gels or stain-free 4-20% gels (BioRad, Hercules, CA). Detection was by Coomassie-Blue staining or tryptophan-fluorescence imaging (24) using a Criterion Stain Free Imager (BioRad). The DCN1 translation was monitored using Coomassie-Blue staining in 10-20% gels. For S1R, soluble fractions and liposome pellets were resuspended in 10mM Bis-Tris, pH7.0, 100mM NaCl, and 0.02% sodium azide before addition of the denaturing PAGE sample buffer. Protein yields were estimated based on comparison of Coomassie-Blue stained band intensities with a 31-kDa band in molecular weight standards (Mark12, Invitrogen, Carlsbad, CA).
Stain-free gels were used in the buffer and extract optimization experiments, and in studies of bR translation time-course experiments. Protein yields were quantified by relative fluorescence intensity as a function of the percentage of Trp residues versus a standard curve of marker proteins of known Trp content (Precision Plus unstained molecular weight markers, BioRad). Final protein concentrations of purified samples were also determined by bicinchoninic acid assay (25).
Detergent quantification
Detection of detergent was performed as described (26). Aliquots of reaction buffer, detergent standards, or reaction unknowns were spotted onto silica gel plates and subjected to thin layer chromatography using chloroform:methanol:ammonium hydroxide (63:35:5) as the mobile phase. The detergents were visualized by incubating the plates with iodine vapor until spots were clearly visible. The plates were imaged using a flatbed scanner.
Absorbance spectroscopy
Purified bR in gel filtration buffer was diluted to 0.5mg/mL in a volume of 200μL and dialyzed against 5mM MES, pH5.5, containing 0.025% DDM overnight at 4°C using a 10,000 MWCO Slide-A-Lyzer cassette (Thermo Scientific, Rockford, IL). The protein concentration and optical purity was measured using a SoloVPE variable path length spectrophotometer (C Technologies, Inc., Bridgewater, NJ), by scanning from 850 to 225nm at a 5mm path length. Optical purity was measured as the ratio of absorbance at 280 to 555nm. The bR control was prepared by dissolving 1mg of lyophilized H. salinarium bR (Sigma-Aldrich) in 2mL of gel filtration buffer, and followed by overnight dialysis of a 200μL aliquot in the buffer described above. The optical purity was measured as described above, except the absorbance maximum of 565nm was used.
Circular dichroism spectroscopy
The H. salinarium and cell-free generated bacteriorhodopsin samples were diluted to concentrations of 0.2 and 0.3mg/mL, respectively, in 5mM MES, pH5.5, containing 0.025% DDM. CD spectra were measured at 22 °C using an Aviv 202SF stopped flow circular dichroism spectrometer with a 0.1cm pathlength quartz cuvette from 300 to 195nm with a 1nm spectral bandwidth. The spectra were normalized to protein concentration and the mean residue ellipticity ([θ]mrw) was plotted as a function of wavelength.
Results
Automated platforms under study
Protemist10/100 are automated in vitro protein synthesizers that employ a repeat-batch strategy (2). These robots contain an internal processor, centrifuge, incubator, and liquid handling system required to perform translation reactions using centrifugal ultrafiltration concentrators. The standard translation protocol is based on a 4mL reaction containing 1mL of wheat germ extract in each concentrator (14). To initiate each translation cycle, 2.5mL of feed buffer is added and the reaction is concentrated to restore the initial 4mL volume. The robot repeats this for 18 cycles including incubation time, which is adjusted so that each cycle takes ∼1h. Using this protocol, these robots can simultaneously accommodate six separate reactions.
The newer the Protemist XE robot (Figure 1) is designed for large-scale production of a single protein employing a discontinuous-batch strategy. It consists of 4 chambers: a feed buffer tank (Fig. 1A); an mRNA reservoir (Fig, 1B), a reaction chamber (Fig. 1C); and a tangential flow ultrafiltration cassette (Fig. 1D). The chambers are connected by a pumping system that is externally controlled by a PC, and two sensors regulate the liquid level of the reaction and detect air bubbles in the flow path. In our experience this simple architecture, with fewer moving parts than the Protemist10/100, improves stability of operations and shortens the user training time. A standard operation protocol consists of repeated cycling through 3 steps: concentration into half of the initial volume; buffer addition to restore the reaction volume; and incubation for 15 min. These 3 steps are repeated 20 times, and then the fourth step of mRNA addition step is executed. The cycle of 4 steps is repeated 6 times during the complete translation protocol. The pumping rate is automatically controlled to adjust for variability during the concentration step so that a run is completed in 48h. The reaction chamber holds 10-40mL of translation reaction (Fig. 1C), with half of the volume accounted for by wheat germ extract.
Figure 1.
The Protemist XE, a cell-free translation robot that uses a discontinuous-batch method. The feed buffer reservoir (A) and mRNA reservoir (B) are used to refill the reaction chamber (C) after 50% reduction in volume by ultrafiltration using a VIVFLOW 50 tangential flow ultrafiltration cassette (D). A stand-alone PC is used to control the pumping system.
Effect of buffer composition and extract on yield
The yield of protein from cell-free translation is influenced by a number of factors including buffers, extract and mRNA. In this study, two transcription buffers and two translation buffer formulations were compared for efficacy. These studies also included comparison of WEPRO8240H, an extract recently developed for higher production yield, with previously available WEPRO2240H. The ‘H’ suffix refers to extracts that have been pretreated with IMAC resin to remove endogenous wheat germ proteins that co-purify with His-tagged proteins.
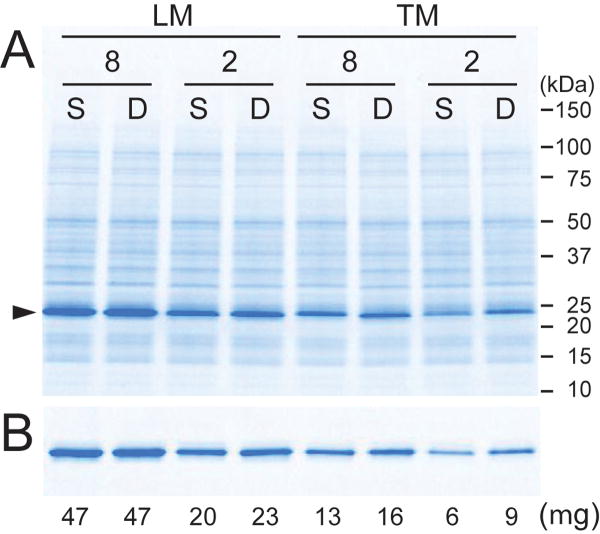
A side-by-side comparison of DCN1 translation in small-scale dialysis reactions showed: 1) the LM transcription buffer performed better than TM transcription buffer with both extracts; 2) the SUB-AMIX and DB translation buffers were overall equivalent with both extracts; and 3) the WEPRO8240H extract gave superior yield relative to WEPRO2240H. These conclusions derive from Figure 2A, and are also supported by the results of IMAC purifications of the reaction product (Figure 2B and also Table 1).
Figure 2.
Comparison of translation and purification results associated with different transcription buffers, wheat germ extracts, and translation buffers. Transcription was performed in either LM or TM buffer. The small-scale translation of DCN1 was performed in dialysis cups with WEPRO8240H (8) or WEPRO2240H (2) and either SUB-AMIX SG (S) or DB (D) translation buffer. A. SDS-PAGE of reaction mixtures performed in different conditions. B. Samples from A after small-scale batch IMAC purification. The triangle marks the migration position of His6-DCN1.
Comparison of the Protemist10/100 and the ProtemistXE translation of DCN1
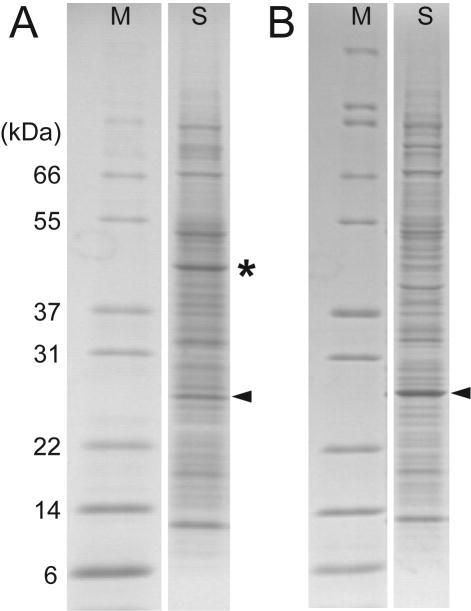
Table 1 shows that use of TM transcription buffer, DB translation buffer, and WEPRO2240 in the Protemist10/100 gave a total synthesis yield of 14mg of DCN1 from six simultaneous 4mL reactions using [13C,15N]-labeled amino acids (also Figure 3A, lane S, marked with a triangle). A total of 6mL of WEPRO2240 was used in this experiment. After purification, including IMAC, TEV protease treatment and subtractive IMAC, and gel filtration, a total of 3.8mg of purified DCN1 was obtained (27% yield).
Figure 3.
Comparison of DCN1 translation using the Protemist100 and the Protemist XE. A. translation of [13C,15N]-labeled DCN1 synthesized with WEPRO2240 using the Protemist100. The yield of purified protein per mL of extract present in the reaction was 2.3mg (Table 1). The asterisk denotes the position of creatine kinase. B. Translation of SeMet-labeled DCN1 synthesized with WEPRO8240 using the Protemist XE in a 10mL reaction. The yield of purified protein per mL of extract present in the reaction was 4.8mg (Table 1). Triangles show the position of DCN1. The two robots yielded samples that gave comparable purity in the final samples (>95%).
Table 1 show results of translation of SeMet-labeled DCN1 in a 10mL reaction using the Protemist XE, LM transcription buffer, 770 mL of SUB-AMIX SG translation buffer and 5mL of WEPRO8240. This reaction gave a total synthesis yield of 24 mg (also Figure 3B, lane S, marked with a triangle). After IMAC, TEV protease treatment and subtractive IMAC, and gel filtration, a total of 4.3mg of purified DCN1 was obtained (18% yield). Although the total synthesis yield was higher with the Protemist XE, the purified yield of SeMet-labeled protein from the purification of this material was less than the [13C,15N]-labeled protein. Thus the SeMet substitution may have adversely affected the solubility or stability of DCN1 relative to the less intrusive isotopic substitutions. The purified DCN1 was concentrated to 10 mg/mL in a final volume of 432 μL using ultrafiltration with a 10,000 MWCO filter. The DCN1 purified from the Protemist XE synthesis was crystallized and the structure was solved (27) using multi-wavelength anomalous diffraction phasing [PDB ID: 3KEV, 1.3 Å resolution, Rfree 0.178]. By use of the Center for Eukaryotic Structural Genomics miniaturized crystallization screening platform, 360μL of the prepared sample was used for the structure determination. In this case, automated protein production and improved crystallization screening are well matched for complementary application to structural biology problems.
Protemist XE translation of S1R in liposomes
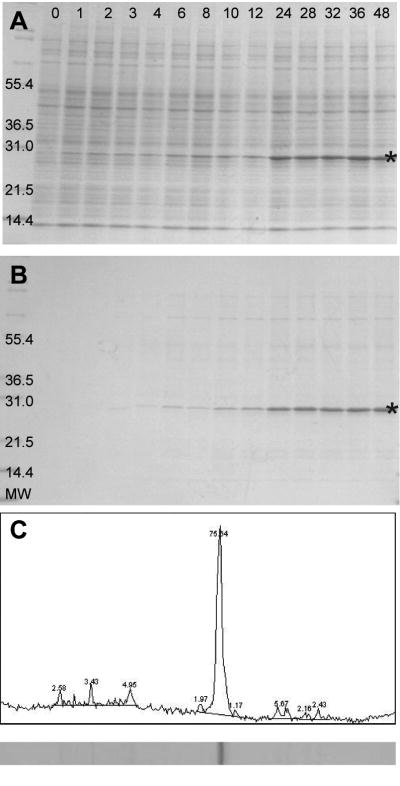
In small-scale trials, human S1R could be translated in the presence of liposomes, and efficiently captured into the liposomes using a previously established approach (11). The proteoliposomes were easily purified by centrifugation. With these properties established by small-scale screening, the ability of the Protemist XE to carry out large-scale liposome-facilitated translation of a membrane protein was examined. For these translation reactions, unilamellar liposomes were added only to the reaction chamber, while SUB-AMIX SG translation buffer was used in the feed buffer. Figure 4A shows a reducing SDS-PAGE analysis of the time course of S1R translation. Accumulation of S1R could be detected by visual inspection after 1h. The amount of synthesized S1R increased most rapidly up to 24h, but continued to accumulate up to ∼36h, indicating that WEPRO8240H maintained translational activity during this time period.
Figure 4.
Cell-free translation of human integral membrane S1R using WEPRO8240H and the Protemist XE in reaction mixtures supplemented with 2.4mg/mL soybean lecithin unilamellar liposomes. A. Time course of the translation reaction analyzed by SDS-PAGE. The star indicates the migration position of His6-S1R. B. SDS-PAGE of S1R proteoliposomes pelleted from the translation reaction and re-suspended in buffer. C ImageJ (42) analysis of the 48h sample from B. S1R is the most abundant protein in these fractions, representing >70% purification in a single step.
At the indicated times, proteoliposomes could be recovered from the reaction mixture by centrifugation (Figure 4B). These proteoliposomes could be easily resuspended in buffer along with all of the S1R. This indicated that S1R was stoichiometrically associated with proteoliposomes. The coupling of automated cell-free translation with the addition of liposomes provides a rapid and easy method for preparation of membrane proteins (11,28). Moreover, simple centrifugation and resuspension gave S1R with purity >70%; comparable levels of purity have been obtained with other membrane proteins (8,9). The total yield of S1R from the Protemist XE reaction was 50mg, representing a volumetric productivity of 10mg/mL relative to the volume of WEPRO8240H used. For this membrane protein, the productivity is comparable to well-behaved soluble proteins such as GFP(29,30) and dihydrofolate reductase (30).
Synthesis of bacteriorhodopsin directly into detergents
bR is a well-studied integral membrane protein (20). It is often used as a model for production and handling of G-protein coupled receptors. Recent studies have shown that bR can be produced by cell-free translation using Escherichia coli extracts and refolded (31) or obtained in a folded state in the presence of liposomes (28), detergents (32) or nanodiscs (12). Here, detergent-mediated translation of bR was evaluated using the Protemist10/100 and the Protemist XE.
To identify detergents suitable for cofactor incorporation with the wheat germ extract, bR was translated using small-scale dialysis in the presence of various combinations of CHAPS, FC-12, all-trans-retinal, and liposomes. The suitability of the different combinations was monitored by examination of each reaction for the appearance of purple color, which indicated that the newly translated bR was properly folded and bound with all-trans-retinal. Purple coloration was observed in reactions containing CHAPS (0.4%), and when CHAPS was supplemented with FC-12. Less intense purple color was observed when CHAPS and liposomes were combined. The purple coloration was not observed in reactions containing either FC-12 (0.2%) or liposomes, and no color was observed in reactions lacking all-trans-retinal. From these results, CHAPS and FC-12 were evaluated for further use.
Partitioning of detergents upon concentration cycling
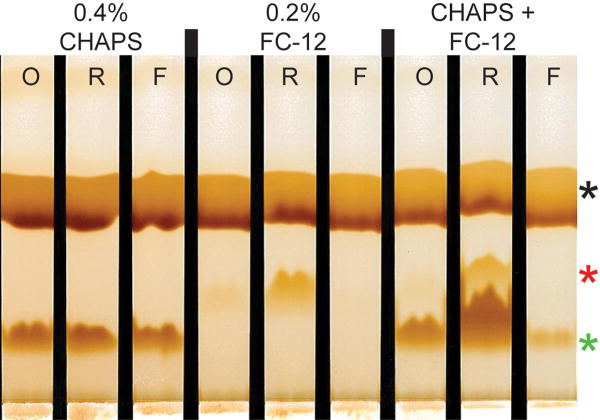
The partitioning of CHAPS and FC-12 between the retained and eluted fractions from the 10,000 MWCO ultrafiltration membrane was investigated and Figure 5 shows the comparative distribution of the detergents between the retained and eluted fractions. In this experiment, solutions of individual detergents and their combinations were prepared in a mock feed buffer consisting of 24mM HEPES-KOH, pH7.8, containing 100mM potassium acetate, 2.5mM magnesium acetate, 1.2mM ATP, and 0.005% sodium azide. The operating cycle was completed three times and detergent concentrations were determined before and after each reaction cycle using thin layer chromatography (26).
Figure 5.
Detergent retention in 10,000 MWCO ultrafiltration concentrators. Buffers containing the indicated detergents were centrifuged in the same concentrators used by the Protemist10/100 robots. After centrifugation at 3000×g for 10 min, thin layer chromatography was performed on the original sample (O), retentate (R), and flow-through (F) and visualized by iodine vapor staining. Black asterisk, non-detergent buffer components; red asterisk, FC-12; green asterisk, CHAPS.
CHAPS at 0.4% w/v (slightly below the critical micelle concentration) passed freely through a 10,000 MWCO ultrafiltration membrane. The same result was obtained from multiple concentration cycles when buffer without detergent was added. When FC-12 was tested alone at 0.2% w/v (4× critical micelle concentration), it was quantitatively retained with the 10,000 MWCO ultrafiltration membrane. When the mixed detergent solution of 0.4% CHAPS and 0.2% FC-12 was tested, the majority of CHAPS in the reaction chamber was retained through repeated reaction cycles along with all of the FC-12 (Figure 5).
These experiments showed that detergent mixtures might change during the course of a multi-step translation reaction, depending on their identity. From this example, FC-12 should not be included in the feed buffer, because it would be retained during concentration. Thus new additions would cause it to accumulate and potentially contribute to extract or protein destabilization during the course of many cycles of the translation reaction. In contrast, CHAPS alone could be added to the feed buffer and a steady-state concentration of detergent might be maintained in the reaction chamber during the duration of the translation reaction. Mixed micelles behaved more like FC-12 alone, indicating no further additions in the feed buffer (or minor adjustments) would be required. The possibility that detergent mixture composition can be controlled during the course of the reaction to customize the translation of different membrane proteins is an obvious extension of these finding.
Large-scale translation of bR in mixed detergents
The detergent screening results from above were investigated in the Protemist10/100. CHAPS (0.4%), FC-12 (0.2%), and all-trans-retinal (0.1 mM) were included in the reaction chambers, and the feed buffer compositions were varied in 4mL reactions (Table 1). The feed buffers contained either 0.04% CHAPS, 0.04% CHAPS plus 0.02% FC-12, or no detergent, reflecting understanding from Figure 5 that detergents may accumulate during the course of the reaction. After completion of the Protemist10/100 run, reactions containing 0.04% CHAPS in the feeding buffer had the darkest purple color. In contrast, feed buffers containing FC-12 plus either CHAPS or no additional detergent had a less intense (or not noticeable) purple color. After overnight incubation of the Protemist10/100 reactions at 4°C, no obvious precipitation or change in the purple color was observed for the reaction with CHAPS alone in the feed buffer, whereas a yellow precipitate formed in the reactions containing FC-12 plus CHAPS or with no additional detergent in the feed buffer. This indicated successful, high-level incorporation of all-trans-retinal in reactions containing CHAPS and FC-12 with cyclic addition of low levels of CHAPS.
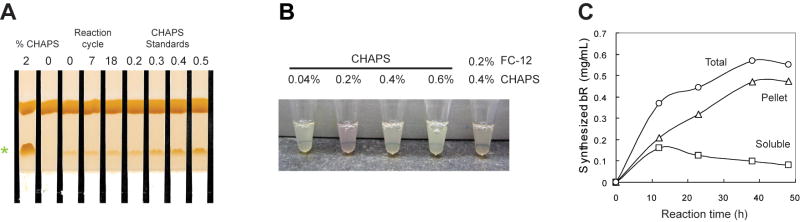
These results were used to perform initial scale-up translation in the Protemist XE (Table 1, two reactions with condition G). The reaction chamber contained 0.4% CHAPS and 0.2% FC-12 as in the Protemist10/100. The CHAPS concentration in the feed buffer was dropped to 0.02%, to compensate for the increased volume of feed buffer used by the Protemist XE and partial retention of CHAPS in the presence of FC-12 (Figure 5). This condition gave a lower yield than anticipated, which was attributed to an imbalance of detergents in the mixed CHAPS/FC-12 micelles. To further test whether CHAPS alone partitioned differently in the Protemist XE with a 5,000 kDa MWCO ultrafiltration cassette, 0.2% CHAPS was added to a feed buffer reaction and a mock synthesis was carried out using the Protemist XE (Figure 6A). In this case, the operating cycle was completed 18 times in order to mimic better the duration of the protein translation reaction, and detergent concentrations were determined before and after each reaction cycle using thin layer chromatography (26). As expected, 0.2% CHAPS remained at constant concentration throughout the operation cycle. This constant detergent level was considered appropriate for scale-up of bR translation.
Figure 6.
Detergent-mediated expression of the bR in the Protemist XE. A. Results of small-scale translation in dialysis cups with the indicated detergent additives. B. CHAPS partitioning in the Protemist XE with VIVAFLOW 50 ultrafiltration membrane. 0.2% CHAPS was added to both the reaction and feed buffers in a mock translation buffer with no extract in the reaction buffer. Samples were removed before start (0) and after 7 and 18 cycles of operation and analyzed by thin layer chromatography and iodine vapor staining. Standards of known concentrations of CHAPS are shown. C. Time course of bR synthesis with 0.2% CHAPS and 0.1 mM all-trans-retinal included in reaction and feed buffers. Synthesized bR was determined by tryptophan fluorescence (24) using Stain-Free SDS-PAGE (BioRad).
A further small-scale optimization showed that CHAPS concentration could be decreased from 0.4% to 0.2% while improving on chromophore formation (Figure 6B). With this revised condition, bR syntheses were performed again using the Protemist XE (Table 1, condition H). The reaction chamber and the feed buffer contained 0.2% CHAPS and 1 mM all-trans-retinal.
Figure 6C shows the time course of bR translated in the Protemist XE. SDS-PAGE analysis indicated that the synthesized bR was initially present as equal portions of solubilized and precipitated material. As the amount of bR synthesized increased (after ∼10 h of reaction), the total translated bR increasingly partitioned into the precipitate fraction. This partitioning was assigned to development of an imbalance between total detergent available and the amount of total protein produced by the ongoing translation reaction (see Purification, below). The total synthesis yield of bR produced in the presence of 0.2% CHAPS was ∼4 mg (Table 1). This was somewhat lower than other proteins tested with the Protemist XE, but within the range expected for a broad range of many other soluble proteins studied by wheat germ cell-free translation (33).
Purification of bR from cell-free translation
In the detergent-limited reactions from the Protemist XE synthesis, bR partitioned between the soluble and precipitate fractions. The precipitate had an orange color, suggesting cofactor incorporation. As the majority of the bR precipitated after the run was completed, a series of solubilization and purification experiments were performed to determine its status. Small aliquots of the precipitate were resuspended in a panel of different detergents (listed in Materials and Methods) that were added to IMAC binding buffer at different concentrations. The samples were incubated for 1h, centrifuged, and then the soluble and pellet fractions were analyzed by SDS-PAGE. These experiments revealed that the best detergents for solubilization of bR were FC-12 and LDAO; 0.5% FC-12 was sufficient to solubilize >90% of the bR synthesized in 2 h and to form the purple chromophore. The FC-12 solubilized bR was purified by IMAC in pH 8.0 buffer also containing 0.5% FC-12. The solubilized bR bound to the column and was eluted with 500mM imidazole in the same detergent conditions, but rapidly turned yellow upon elution, indicating that bR was not stable in this buffer.
By using on-column exchange into DDM, the stability of bR was improved. Moreover, the elution conditions were also modified to include an increasing concentration gradient of imidazole and a decreasing concentration gradient of NaCl. The incorporation of DDM and new elution conditions yielded 35% recovery of purified bR from the IMAC step. After IMAC elution, the pooled bR was immediately exchanged into 10mM MES, pH 5.5, 100mM NaCl, 0.3mM tris(2-carboxyethyl)phosphine hydrochloride, and 0.025% DDM. The drop in pH in the presence of DDM increased the stability of the bR, and storage for several days at 4°C gave no change in the solubility or absorbance spectrum.
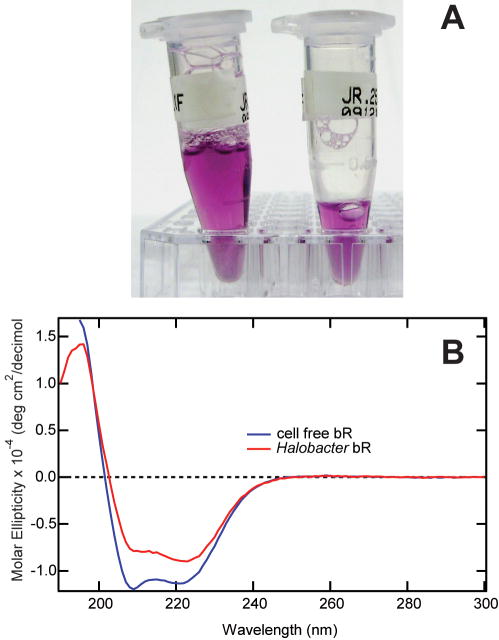
Gel filtration was used to complete the purification and determine the aggregation state of bR in the DDM micelles. The colored fractions containing bR eluted with an Mr of 134kDa, corresponding to either a dimeric or trimeric quaternary structure in the DDM micelles. The purified bR obtained from cell-free translation was concentrated to 10mg/mL by ultrafiltration, and could be drop-frozen and thawed without loss of chromophore or solubilization status. Moreover, the purple chromophore in the purified protein was stable for several weeks at 4°C (Figure 7A). Absorbance spectroscopy showed that purified bR had an absorption maximum at 555nm, and the optical purity ratio (A280/A555) was 2.2. For comparison, a control sample prepared from lyophilized and reconstituted bR had a visible peak of 565nm and an optical purity (A280/A565) of 1.9. Previous studies have shown the bR absorption maximum is sensitive to detergent and lipid composition (32,34). Circular dichroism spectroscopy indicated the Protemist XE-generated bR was largely α-helical, as predicted for folded bacteriorhodopsin (Figure 7B). The commercial preparation of H. salinarium bR was similarly α-helical, but showed peak flattening indicative of the presence of residual membrane lipids (35).
Figure 7.
Results from bR purified from a Protemist XE translation. A. CHAPS partitioning in the Protemist XE with VIVAFLOW 50 ultrafiltration membrane. 0.2% CHAPS was added to both the reaction and feed buffers in a mock translation buffer with no extract in the reaction buffer. Samples were removed before start (0) and after 7 and 18 cycles of operation and analyzed by thin layer chromatography and iodine vapor staining. Standards of known concentrations of CHAPS are shown. B. Results of small-scale translation in dialysis cups with the indicated detergent additives. C. Time course of bR synthesis with 0.2% CHAPS and 0.1 mM all-trans-retinal included in reaction and feed buffers. Synthesized bR was quantified by tryptophan fluorescence24 using Stain-Free SDS-PAGE (BioRad).
Discussion
Robots, buffers and extracts
In batch operation, cell-free protein translation can be inhibited by imbalanced changes in substrates and products that occur as the reaction proceeds. In order to alleviate this inhibition, diffusion-based methods (6,29) have been developed to continue replenishment of essential reagents and reduce the accumulation of by-products.
Here, we evaluated a new discontinuous ultrafiltration-type robot for cell-free translation. Overall, the Protemist XE provided comparable or higher total yield of protein per mL of extract than the earlier Protemist10/100 robots (Table 1). The compact, accessible design and simplicity of operation are advantages of the Protemist XE, which also used less wheat germ extract than the Protemist10/100 to carry out a comparable reaction (5mL versus 6mL for the Protemist10). The Protemist XE requires specialized, consumable peristaltic tubing assemblies and utrafiltration membrane cassette, uses 13-fold more feed buffer for operation, and has a standard operation cycle that is 2.6 time longer.
Side-by-side comparison of transcription buffers, translation buffers and a new preparation of wheat germ extract indicate that optimal performance will be obtained by combination of the LM transcription buffer and WEPRO8240 extract. The WEPRO8240 extract was clearly superior to the WEPRO2240 extract for soluble proteins, but further investigations are needed to make a conclusive comparison with membrane proteins (Table 1). The DB and SUB-AMIX SG translation buffers were overall equivalent in our small-scale dialysis reactions. The extensive buffer exchange enabled by the robots may account for this. There was no noticeable difference in translation performance between the WEPRO8240 and WEPRO8240H extracts. The latter extract can simplify purification due to the removal of endogenous wheat germ proteins that bind and elute from IMAC resins along with His-tagged proteins. WEPRO8240H is also more costly, so consideration of purity needs and ease of separation of the endogenous proteins by additional chromatography steps should be considered.
Translation of soluble protein DCN1
Wheat germ cell-free translation has been previously used to solve NMR structures of soluble eukaryotic proteins (36). Others and we have now established that this approach can be used to determine X-ray crystal structures. The first X-ray structure (37) was the cytotoxic restriction enzyme PabI [3.0 Å, Rfree 0.318, PDB ID 2DVY]. More recently, we determined X-ray structures of agmatine deiminase (1.50 Å, Rfree 0.174, PDB ID 3H7C), human replication protein A complex (1.98 Å, Rfree 0.206, PDB ID 3KDF), and DCN1 (1.3 Å, PDB ID 3KEV). The present study of DCN1 showed that the Protemist XE was able to generate a sufficient amount of highly purified protein from a single run to support the X-ray structure determination. Decreases in material required for crystallization screening and the improved performance of WEPRO8240H extract and the Protemist XE robotics are well matched to enable protein structure determination in an automated manner.
Translation of integral membrane proteins
Cell-free translation is an emerging technology for production of membrane proteins. Although membrane proteins are largely insoluble in cell-free reactions unless additives such as liposomes or detergents are included (38), the open nature of the reaction allows addition of these materials. This work has described translation of human S1R and bR using two adaptations of cell-free translation. With S1R, the translation reaction included unilamellar liposomes, while with bR, the translation reaction included detergents. S1R has been previously produced in E. coli as an MBP fusion, and purified by detergent extraction from bacterial membranes (39). The cell-free translation approach provides a new method for preparation of this interesting protein.
The production of bR using cell-free translation has been previously described, as this is a model membrane protein. bR was first synthesized in wheat germ extract as a precipitate and refolded in organic solvents and anionic detergent to produce a spectrally active protein (31). A related proteorhodopsin was reconstituted as a pigmented protein by solubilizing a colorless cell-free precipitate with n-octyl-β-D-glucopyranoside (40). Shimono and coworkers showed that chromophoric bR could be synthesized in the presence of unilamellar vesicles using an E. coli-based cell-free system and extracted using DDM (32). Moreover, chromophoric bR could be obtained by co-translation in the presence of nanodiscs (12).
Here we have found that chromophoric bR could be produced in the presence of detergents without added lipids. This promising result suggests possibilities for enhanced translation based on knowledge of compatible detergents and additives needed for folding and stability of other membrane proteins. The wheat germ extract may also contribute some lipids suitable to maintain bR solubility. Indeed, wheat germ extract WEPRO2240H has ∼725 mM phospholipid (Jacob Ludington, personal communication). The potential contributions of endogenous wheat germ substances warrant further consideration as protocols for co-translational solubilization of membrane proteins are developed.
Use of automation in cell-free translation of membrane proteins
Automation offers significant advantages for screening conditions needed for successful cell-free translation of membrane proteins. Under optimal circumstances, screening reactions would be coupled to determination of enzyme function, such as we have recently demonstrated for human stearoyl-CoA desaturase (8) and trypanosomal sphingolipid synthases (9). In the present study, we have begun to explore possibilities and methods to enhance co-translational solubilization of membrane proteins in the presence of detergents. Significant challenges still remain to be addressed to achieve the promise of these early indications. One important consideration is the identification of detergents and other additives that are compatible with the translation reaction (13). For bR, certain combinations of CHAPS and FC-12 fulfill this requirement with wheat germ extracts.
Protemist XE and the Protemist10/100 robots use ultrafiltration as an essential step of their operation cycles. Consequently, understanding the behavior of detergents during ultrafiltration is essential. As shown in Figure 5, detergents may be retained depending on critical micelle concentration (41) and thus increase in concentration during operation. This would be the case for FC-12, while not so for CHAPS. Knowledge of these properties can be used to make experimental choices for whether detergents should be included in the feed buffer, reaction chamber or both. More sophisticated applications may consider partition of detergents between retained and flow-through, and devise customized feeding strategies to accommodate loss of detergent in the flow-through.
Figure 6 suggests that membrane protein translation may exceed the capacity of detergent needed for solubilization. With bR, this apparently led to the formation of a folded precipitate that could be otherwise easily recovered and purified as a folded protein upon solubilization with additional detergent. We do not anticipate this will be a universal strategy for production of membrane proteins, so further consideration of how to experimentally balance detergent addition relative to the amount of newly synthesized membrane protein is needed.
Conclusions
These studies examined the efficacy of a newly developed cell-free protein translation robot, the Protemist XE. It was found to be useful in the automated production of proteins in a short time period and with minimal supporting labor. The discontinuous-batch approach used by this robot was compatible with liposome-mediated production of a membrane protein, and also with translation and direct solubilization of another membrane protein in the presence of translation-compatible detergents. Our previously established use of small-scale automation (14) is clearly amenable to functional studies and screening for optimal conditions as a prerequisite to scale-up for structural biology studies using the Protemist XE. Improvements in robots and reagents such as those described here contribute to a continually improving prospect for the use of cell-free translation in production of valuable proteins for structure determination and other large-scale applications.
Acknowledgments
This work was supported by NIH/NIGMS Protein Structure Initiative grant U54 GM074901 (JL Markley, PI; GN Phillips, Jr. and BG Fox, Co-Investigators). We thank Dr. Jorge Escalante for generously supplying the bacteriorhodopsin gene, We thank Masaki Madono, Tomo Sato, and Miwako Denda from CellFree Science for their insight and responsiveness and also thank members of the Center for Eukaryotic Structural Genomics and the Department of Biochemistry for technical assistance and many helpful discussions.
Abbreviations
- bR
bacteriorhodopsin
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propansulfonate
- DB
dialysis buffer
- DCN1
DCUN1-domain containing protein isoform 1
- DDM
n-dodecyl-β -D-maltopyranoside
- FC-12
FOS-CHOLINE-12
- IMAC
immobilized metal affinity chromatography
- MWCO
molecular weight cut-off
- TEV
tobacco etch virus
- SeMet
selenomethionine
- S1R
sigma-1 receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klammt C, Lohr F, Schafer B, et al. High level cell-free expression and specific labeling of integral membrane proteins. Eur J Biochem. 2004;271:568–80. doi: 10.1111/j.1432-1033.2003.03959.x. [DOI] [PubMed] [Google Scholar]
- 2.Vinarov DA, Loushin Newman CL, Markley JL. Wheat germ cell-free platform for eukaryotic protein production. FEBS J. 2006;273:4160–9. doi: 10.1111/j.1742-4658.2006.05434.x. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz D, Klammt C, Koglin A, et al. Preparative scale cell-free expression systems: New tools for the large scale preparation of integral membrane proteins for functional and structural studies. Methods. 2007;41:355–69. doi: 10.1016/j.ymeth.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Takai K, Sawasaki T, Endo Y. Practical cell-free protein synthesis system using purified wheat embryos. Nature Prot. 2010;5:227–38. doi: 10.1038/nprot.2009.207. [DOI] [PubMed] [Google Scholar]
- 5.Vinarov DA, Newman CL, Tyler EM, et al. Wheat germ cell-free expression system for protein production. Curr Protoc Protein Sci. 2006;Chapter 5(Unit 5 18) doi: 10.1002/0471140864.ps0518s44. [DOI] [PubMed] [Google Scholar]
- 6.Kigawa T, Yabuki T, Yoshida Y, et al. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999;442:15–9. doi: 10.1016/s0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- 7.Kainosho M, Torizawa T, Iwashita Y, et al. Optimal isotope labelling for NMR protein structure determinations. Nature. 2006;440:52–7. doi: 10.1038/nature04525. [DOI] [PubMed] [Google Scholar]
- 8.Goren MA, Fox BG. Wheat germ cell-free translation, purification, and assembly of a functional human stearoyl-CoA desaturase complex. Protein Expr Purif. 2008;62:171–8. doi: 10.1016/j.pep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sevova ES, Goren MA, Schwartz KJ, et al. Cell-free synthesis and functional characterization of sphingolipid synthases from parasitic trypanosomatid protozoa. J Biol Chem. 2010 doi: 10.1074/jbc.M110.127662. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klammt C, Schwarz D, Lohr F, et al. Cell-free expression as an emerging technique for the large scale production of integral membrane protein. FEBS J. 2006;273:4141–53. doi: 10.1111/j.1742-4658.2006.05432.x. [DOI] [PubMed] [Google Scholar]
- 11.Goren MA, Nozawa A, Makino S, Wrobel RL, Fox BG. Cell-free translation of integral membrane proteins into unilamelar liposomes. Methods Enzymol. 2009;463:647–73. doi: 10.1016/S0076-6879(09)63037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cappuccio JA, Hinz AK, Kuhn EA, et al. Cell-free expression for nanolipoprotein particles: building a high-throughput membrane protein solubility platform. Methods Mol Biol. 2009;498:273–96. doi: 10.1007/978-1-59745-196-3_18. [DOI] [PubMed] [Google Scholar]
- 13.Berrier C, Park KH, Abes S, et al. Cell-free synthesis of a functional ion channel in the absence of a membrane and in the presence of detergent. Biochemistry. 2004;43:12585–91. doi: 10.1021/bi049049y. [DOI] [PubMed] [Google Scholar]
- 14.Makino S, Goren MA, Fox BG, Markley JL. Cell-free protein synthesis technology in NMR high-throughput structure determination. Methods Mol Biol. 2010;607:127–47. doi: 10.1007/978-1-60327-331-2_12. [DOI] [PubMed] [Google Scholar]
- 15.Kurz T, Chou YC, Willems AR, et al. DCN1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 17.Lee IT, Chen S, Schetz JA. An unambiguous assay for the cloned human sigma1 receptor reveals high affinity interactions with dopamine D4 receptor selective compounds and a distinct structure-affinity relationship for butyrophenones. Eur J Pharmacol. 2008;578:123–36. doi: 10.1016/j.ejphar.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontanilla D, Johannessen M, Hajipour AR, et al. The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science. 2009;323:934–7. doi: 10.1126/science.1166127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oesterhelt D, Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–78. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 20.Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665–88. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 21.Blommel PG, Martin PA, Wrobel RL, Steffen E, Fox BG. High efficiency single step production of expression plasmids from cDNA clones using the Flexi Vector cloning system. Protein Express Purif. 2006;47:562–70. doi: 10.1016/j.pep.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Blommel PG, Martin PA, Seder KD, Wrobel RL, Fox BG. Flexi Vector cloning in high throughput protein expression and purification. In: Doyle S, editor. Methods in Molecular Biology. Totowa, N.J.: The Humana Press Inc; 2007. [Google Scholar]
- 23.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Express Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazmin D, Edwards RA, Turner RJ, Larson E, Starkey J. Visualization of proteins in acrylamide gels using ultraviolet illumination. Anal Biochem. 2002;301:91–6. doi: 10.1006/abio.2001.5488. [DOI] [PubMed] [Google Scholar]
- 25.Smith PK, Krohn RI, Hermanson GT, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 26.Eriks LR, Mayor JA, Kaplan RS. A strategy for identification and quantification of detergents frequently used in the purification of membrane proteins. Anal Biochem. 2003;323:234–41. doi: 10.1016/j.ab.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Burgie, S., Makino, S-i, Bingman, C.A., Fox, B.G. and Phillips, G.N.. (2010) submitted,
- 28.Kalmbach R, Chizhov I, Schumacher MC, et al. Functional cell-free synthesis of a seven helix membrane protein: In situ insertion of bacteriorhodopsin into liposomes. J Mol Biol. 2007;371:639–48. doi: 10.1016/j.jmb.2007.05.087. [DOI] [PubMed] [Google Scholar]
- 29.Sawasaki T, Hasegawa Y, Tsuchimochi M, et al. A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 2002;514:102–5. doi: 10.1016/s0014-5793(02)02329-3. [DOI] [PubMed] [Google Scholar]
- 30.Madin K, Sawasaki T, Ogasawara T, Endo Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:559–64. doi: 10.1073/pnas.97.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonar S, Patel N, Fischer W, Rothschild KJ. Cell-free synthesis, functional refolding, and spectroscopic characterization of bacteriorhodopsin, an integral membrane protein. Biochemistry. 1993;32:13777–81. doi: 10.1021/bi00213a004. [DOI] [PubMed] [Google Scholar]
- 32.Shimono K, Goto M, Kikukawa T, et al. Production of functional bacteriorhodopsin by an Escherichia coli cell-free protein synthesis system supplemented with steroid detergent and lipid. Prot Sci. 2009;18:2160–71. doi: 10.1002/pro.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyler RC, Aceti DJ, Bingman CA, et al. Comparison of cell-based and cell-free protocols for producing target proteins from the Arabidopsis thaliana genome for structural studies. Proteins. 2005;59:633–43. doi: 10.1002/prot.20436. [DOI] [PubMed] [Google Scholar]
- 34.Lopez F, Lobasso S, Colella M, Agostiano A, Corcelli A. Light-dependent and biochemical properties of two different bands of bacteriorhodopsin isolated on phenyl-sehparose CL-4B. Photochem Photobiol. 1999;69:599–604. [Google Scholar]
- 35.Wu SG, Elsayed MA. CD spectrum of bacteriorhodopsin - Best evidence against exciton model. Biophys J. 1991;60:190–7. doi: 10.1016/S0006-3495(91)82042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markley JL, Aceti DJ, Bingman CA, Fox BG, et al. The Center for Eukaryotic Structural Genomics. J Struct Funct Genomics. 2009;10:165–79. doi: 10.1007/s10969-008-9057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyazono K, Watanabe M, Kosinski J, et al. Novel protein fold discovered in the PabI family of restriction enzymes. Nucleic Acids Res. 2007;35:1908–18. doi: 10.1093/nar/gkm091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz D, Dotsch V, Bernhard F. Production of membrane proteins using cell-free expression systems. Proteomics. 2008;8:3933–46. doi: 10.1002/pmic.200800171. [DOI] [PubMed] [Google Scholar]
- 39.Ramachandran S, Lu H, Prabhu U, Ruoho AE. Purification and characterization of the guinea pig sigma-1 receptor functionally expressed in Escherichia coli. Protein Expr Purif. 2007;51:283–92. doi: 10.1016/j.pep.2006.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gourdon P, Alfredsson A, Pedersen A, et al. Optimized in vitro and in vivo expression of proteorhodopsin: A seven-transmembrane proton pump. Prot Express Purif. 2008;58:103–13. doi: 10.1016/j.pep.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Prive GG. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41:388–97. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]