Abstract
Summary: The Sample avAILability system—SAIL—is a web based application for searching, browsing and annotating biological sample collections or biobank entries. By providing individual-level information on the availability of specific data types (phenotypes, genetic or genomic data) and samples within a collection, rather than the actual measurement data, resource integration can be facilitated. A flexible data structure enables the collection owners to provide descriptive information on their samples using existing or custom vocabularies. Users can query for the available samples by various parameters combining them via logical expressions. The system can be scaled to hold data from millions of samples with thousands of variables.
Availability: SAIL is available under Aferro-GPL open source license: https://github.com/sail.
Contact: gostev@ebi.ac.uk, support@simbioms.org
Supplementary information: Supplementary data are available at Bioinformatics online and from http://www.simbioms.org.
1 INTRODUCTION
For many years biobanks have been collecting biological samples enriching them with annotations of phenotype, familial and environmental data stored in diverse computational systems. With some notable exceptions, such as the UK DNA Banking Network (Yuille et al., 2010), biobank data are usually restricted to in-house use (Hirtzlin et al., 2003; Kauffman and Cambon-Thomsen, 2008; Yuille et al., 2007). With an increasing need for improved statistical power in genome-wide epidemiological studies, accessibility to samples and their annotation from many collections is essential (McCarthy et al., 2008). However, with considerable differences in how sample collections describe their content, and access to individual data restricted by ethical and legal regulations this may be a daunting task. Even when access to summary data for individual phenotypes is available, it is difficult to estimate how many samples have measurements for a combination of phenotypes. For instance, how many samples in a given cohort have the combination of genome-wide association results, metabolomic data and information on a given clinical phenotype? There is a need for an efficient means of finding which biobanks contain samples relevant to a particular study annotated with the necessary metadata (Founti et al., 2009; Helgesson et al. 2007).
By sample availability data we understand the information describing which meta-data and measurements exist for each sample in a collection without necessarily revealing the actual content. The Sample avAILability system (SAIL) is a platform that uses this paradigm to help researchers to integrate their resources and search sample availability across any number of sources. The system can either be used by individual biobanks or by consortia of sample collections to facilitate their research.
2 BASIC CONCEPTS AND DATA STRUCTURES
A sample in SAIL is a general concept which can for example represent a human individual, a biopsy or a derived sample preparation. Biobank samples are typically annotated with relevant phenotypic information, such as the type of sample (e.g. blood), the age and disease state of the person the sample was taken from and possibly with various measurement data such as glucose level and blood pressure at the time the sample was taken. When the same biosample has been used to collect measurements at different time points, these are given separate sample identifiers to simplify searching and maximize the visibility of each individual measurement. The situation where more than one sample has been taken from a person can be represented in various ways (a record per sample or a record per person) which is down to the user; however in the data sets currently loaded in SAIL this situation is not typical.
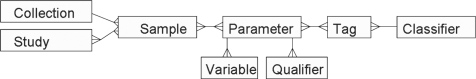
Samples are grouped in collections (e.g. cohorts); collections may be annotated with information common to the collection (e.g. descriptions of the origin of the samples and the collection protocols). While a sample can only belong to one collection, it can participate in many studies. Moreover studies may contain samples from multiple collections (see Fig. 1).
Fig. 1.
A high level data structure in SAIL. (See Supplementary Materials for a complete database schema).
The basic functional descriptor unit in SAIL is a parameter. Parameters are described by variables, which are mandatory fields, and qualifiers, which are optional additional descriptors. For instance, we can define ‘Glucose’ as a parameter with a variable ‘concentration’. We can refine the Glucose parameter by adding a qualifier ‘timing’ to specify whether the measurement was taken while ‘fasting’ or ‘non fasting’.
Parameters can be grouped using tags. Tags are assigned values to identify the parameter group. They are a general concept in SAIL that provides the flexibility to group parameters without being bound by the semantics of the specific type of group. For example, tags can be used to group parameters that come from a specific vocabulary, parameters that are used in the definition of a specific disease or parameters that are synonymous with each other. Tags can themselves be grouped into classifiers that can be used for more general parameter classification. For example, tags for different vocabularies can be grouped into the classifier ‘Vocabulary’. Similarly, tags for specific types of parameter relationships are grouped into the classifier ‘Relation’. When a tag is added to a parameter, the user indicates the value of the tag (such as the specific vocabulary) and the classifier to which the tag belongs (‘Vocabulary’). Tags can be used to map external taxonomic structures to data structures in SAIL, for example data schemas produced by DataSHaPER (Fortier et al., 2010), the supplier of standardized data schemas for biobanks, can be easily represented in SAIL.
Using such a flexible semantic structure SAIL can accommodate parameters from several vocabularies and store relations between them enabling searches that span across more than one data collection as well as for samples only partially matching the search term. Relations can be defined by the users or data providers and can vary from generic forms (i.e. synonym or partial match) to more detailed forms to express associations between specific vocabularies (or even specific for a group of terms).
3 QUERIES
Users can form queries by selecting parameters listed in the interface and combining them via logical expressions. Parameters can be filtered by a selection of classification tags, such as parameters associated with a particular disease. Query results can be limited by defining value ranges for specific parameters.
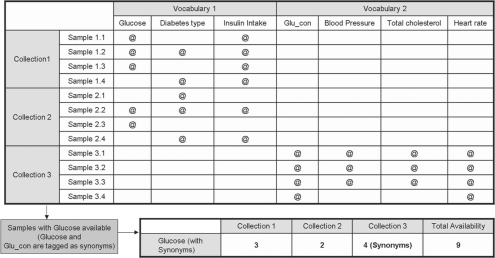
When data from more than one collection is available, users can select which collections to query. Users can also select what type of relation between parameters they allow; for example to retrieve samples annotated using particular vocabularies where only synonymous terms are considered (see Fig. 2).
Fig. 2.
Sample availability matrix for three collections, where collection 1 and 2 are annotated with one vocabulary and collection 3 with a different vocabulary. As Glucose and Glu_con are tagged as synonymous, the result of a query for Glucose will show samples available from the three collections.
The result of a query is presented as a report showing how many samples with the specified parameters are available from each of the collections. SAIL allows for the creation of predefined queries by the systems administrator as a part of the systems customization process. Predefined queries can be used for work with rather complex combinations of variables.
4 IMPLEMENTATION
SAIL is designed as a client-server system where the sample availability data and vocabularies are stored at a server instance, with tools for browsing, searching and editing content accessible through a web application interface using common web browsers.
Parameters can be introduced in SAIL by batch import of vocabularies (see Supplementary Material) or manually using the parameter creation tool. Availability data for a group of samples, annotated using one of the preloaded vocabularies, is uploaded using a tab-separated spreadsheet file with one row per sample and one column per parameter. Sample data may contain the actual values or the symbol @ representing the availability of data for this parameter without disclosing the actual value. It is up to the data provider to decide for which parameters to provide the actual values or only the availability. The actual values will not be presented at any time (except for values specified for presentation), but they allow for finer data filtering during querying. Thus SAIL has been designed to minimize (even eliminate) the storage of identifiable human data, allowing the browsing and sharing of data without the need for access restrictions that need to be imposed for ethical reasons. Nevertheless, depending on how the system is used, access control may still be useful, and SAIL allows for three types of users: (i) system administrator with full access; (ii) data manager with vocabulary and data upload rights; and (iii) basic user with search engine access.
The web interface of SAIL was developed in Java using the Google Web Toolkit and Ext-JS widget libraries. The Java servlet specifications were followed. The system is run as a Tomcat web application. In our implementation, all data are stored in a relational database on the server and the part selected for queries are loaded into memory for highest performance. This allows for fast execution and flexible query formulation. The execution of queries is not dependant on the relational database, and can be formulated using linked data solutions such as RDF, OWL and SPARQL. However, we opted for a simple customized semantic data structure instead, in order to maximize performance and to be able to flexibly contain annotation structures from different sources.
5 DISCUSSION
SAIL has been developed as a tool that can be installed and used independently by any biobank or research group in need for a system for keeping track of their samples. However, we believe that the main use of SAIL is in a centralized instance holding data from many providers and used to identify samples across multiple data sources.
For SAIL to function effectively across multiple data sources, it is important that their vocabularies are compatible and that the correct relations are made between data elements in different vocabularies. There are two possible strategies to achieve this: (i) harmonizing the vocabularies in advance of loading them into the system or (ii) by using SAIL to create mappings between terms of vocabularies already loaded into the system. In each case this is not a trivial task and, although it is central to the system, describing this is outside the scope of this application note.
A particular use case is the design of meta-analysis studies based on genome-wide genotype data availability across many collections. We give two examples on how SAIL can be used in this context.
Example 1. —
Meta-analysis of genome-wide association studies for glucose levels in plasma.
A consortium wants to conduct a meta-analysis of genome-wide association studies for fasting plasma glucose levels. It is of importance to know diabetes status for the study, and age, gender and BMI are to be used as study covariates. For the study design, an estimate is sought for how many samples can be included in the study, and from which cohorts.
In the SAIL report constructor a query is constructed by selecting the parameters of interest, and adding them one by one to the query: glucose concentration (GLU), diabetes status (DB), age (AGE), gender (SEX) and BMI (BMI). We add the requirement for genome-wide genotyping data (GW_GT) and retrieve a report with detailed information about the availability in each collection for samples supporting the query (Supplementary Figure S1). The report tells us that 12 487 samples in three different cohorts may be eligible for the study. A further query restricted to these cohorts show the exact genome-wide genotyping arrays these samples have been measured on, and whether or not genome-wide imputed genotypes are available (Supplementary Figure S2). Based on the results from SAIL, it is now easy to contact the administrators of the different cohorts to ask for specific information and coordinate the sharing of data for the meta-analysis.
Example 2. —
Metabolic Syndrome.
Metabolic Syndrome is a term for a combination of phenotypes that affect the risk for diseases involving the metabolic system and diseases that may follow as the conditions progress. The definition of Metabolic Syndrome is complex and can be done in different ways. Three commonly used definitions are those of the International Diabetes Federation (IDF), the US National Cholesterol Education Program (NCEP) and the World Health Organization (WHO). For example, WHO defines Metabolic Syndrome as a combination of impaired glucose regulation and two out of four additional risk factors. Impaired glucose regulation in itself is determined by the presence of either type 2 diabetes, impaired fasting glucose, impaired glucose tolerance or insulin resistance. The four additional risk factors are: (i) central obesity (threshold is gender dependent and determined by waist-to-hip ratio or BMI); (ii) raised plasma triglyceride levels and raised HDL cholesterol level (where the threshold depends on gender); (iii) raised blood pressure; and (iv) raised urinary albumin secretion ratio or raised albumin : creatinine ratio in serum. SAIL supports queries for such a complex combination of phenotypes, with operations such as ‘two out of four’, but encodes them as pre-defined queries.
To query SAIL for Metabolic Syndrome by the WHO definition, we select the eligible collections and simply add the predefined MetS_WHO query and submit the request. In the current installation of SAIL at the EBI, 16 903 out of 85 979 samples across 13 collections have sufficient measurements available to determine Metabolic Syndrome status according to WHO (Supplementary Figure S3).
Although SAIL has been developed with a focus on biobanks and biological sample collections, its design allows for the integration of data from other sources where information can be arranged into annotated records. The largest of the four SAIL instances which are currently run for various projects is accessible at http://www.ebi.ac.uk/Tools/sail/ and http://sail.simbioms.org with data from approximately 189 000 samples from 14 collections. Technically SAIL software can scale to any number of cohorts though the system may slow down as more cohorts are added. According to our current assessment, on the existing hardware (Intel Xeon 2.66 GHz, 4 GB RAM) the system can scale up to millions of samples.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank members of the ENGAGE consortium for their support and feedback, as well as the data providers, especially HUNT Biobank for the largest dataset deposited in SAIL and for the valuable discussions and continuous support.
Funding: Functional Genomics budget of the European Molecular Biology Laboratory and the ENGAGE grant HEALTH-F4-2007-201413 of the FP7 programme by the European Commission.
Conflict of Interest: none declared.
REFERENCES
- Fortier I., et al. Quality, quantity and harmony: the DataSHaPER approach to integrating data across bioclinical studies. Int. J. Epidemiol. 2010;39:1383–1393. doi: 10.1093/ije/dyq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Founti P., et al. Biobanks and the importance of detailed phenotyping: a case study-the European Glaucome Society GlaucoGENE project. Br. J. Ophthalmol. 2009;93:577–581. doi: 10.1136/bjo.2008.156273. [DOI] [PubMed] [Google Scholar]
- Helgesson G., et al. Ethical framework for previously collected biobank samples. Nat. Biotechnol. 2007;25:973–976. doi: 10.1038/nbt0907-973b. [DOI] [PubMed] [Google Scholar]
- Hirtzlin I., et al. An empirical survey on biobanking of human genetic material and data in six EU countries. Eur. J. Hum. Genet. 2003;11:475–488. doi: 10.1038/sj.ejhg.5201007. [DOI] [PubMed] [Google Scholar]
- Kauffmann F., Cambon-Thomsen A. Tracing biological collections: between books and clinical trials. JAMA. 2008;299:2316–2318. doi: 10.1001/jama.299.19.2316. [DOI] [PubMed] [Google Scholar]
- McCarthy M.I., et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Yuille M., et al. Biobanking for Europe. Brief. Bioinform. 2007;9:14–24. doi: 10.1093/bib/bbm050. [DOI] [PubMed] [Google Scholar]
- Yuille M., et al. The UK DNA Banking Network: a “fair access” biobank. Cell Tissue Bank. 2010;11:241–251. doi: 10.1007/s10561-009-9150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.