Abstract
The topology of metabolic networks can provide insight not only into the metabolic processes that occur within each species, but also into interactions between different species. Here, we introduce a novel pair-wise, topology-based measure of biosynthetic support, reflecting the extent to which the nutritional requirements of one species could be satisfied by the biosynthetic capacity of another. To evaluate the biosynthetic support for a given pair of species, we use a graph-based algorithm to identify the set of exogenously acquired compounds in the metabolic network of the first species, and calculate the fraction of this set that occurs in the metabolic network of the second species. Reconstructing the metabolic network of 569 bacterial species and several eukaryotes, and calculating the biosynthetic support score for all bacterial-eukaryotic pairs, we show that this measure indeed reflects host-parasite interactions and facilitates a successful prediction of such interactions on a large-scale. Integrating this method with phylogenetic analysis and calculating the biosynthetic support of ancestral species in the Firmicutes division (as well as other bacterial divisions) further reveals a large-scale evolutionary trend of biosynthetic capacity loss in parasites. The inference of ecological features from genomic-based data presented here lays the foundations for an exciting “reverse ecology” framework for studying the complex web of interactions characterizing various ecosystems.
Key words: biosynthetic support, host-parasite, metabolic networks, reverse ecology, seed set species interaction
1. Introduction
In recent years, researchers have come to realize that a network-based representation of various biological systems encapsulates many of the essential properties of these systems. Such networks provide valuable insights into the system's function and dynamics and often lend themselves to well-established tools and algorithms borrowed from graph theory (Alon, 2003), which, interestingly, has deep roots in the analysis of chemical reactions (Bonchev and Rouvray, 1991). In particular, a wide range of computational approaches have been applied to study topological characteristics of metabolic networks and their bearings on various functional properties, including scaling (Jeong et al., 2000), regulation (Stelling et al., 2002), universality (Smith and Morowitz, 2004), robustness (Deutscher et al., 2006), and adaptation (Borenstein et al., 2008; Kreimer et al., 2008). Further analyses have also considered the environmental context in which each network functions and examined the effect of the biochemical environment on various metabolic processes (Almaas et al., 2004; Ibarra et al., 2002).
The analysis of metabolic networks, however, can provide valuable insight not only into the function and dynamics of the metabolic process, but also into the biochemical environment in which each species evolved. This is because interactions with the environment throughout the evolutionary process and selection pressures that the environment exerts on the evolving metabolic network affect the structure of the network and often produce a detectable topological “signature.” Following this ‘reverse ecology’ approach (Parter et al., 2007), we previously developed a computational framework designed to extract large-scale ecological insights from the analysis of the topology of metabolic networks (Borenstein et al., 2008). In this framework, the topology of a given network is analyzed using a novel graph theory-based algorithm, to infer the set of compounds that are exogenously acquired. This set, termed the ‘seed set’ of the network (Raymond and Segre, 2006), reflects the metabolic interface between the organism and its surroundings, approximating its effective biochemical environment. We constructed and analyzed the metabolic networks of 478 species and identified their seed sets, presenting a comprehensive large-scale reconstruction of such metabolic environments. We further showed that the seed sets' composition strongly correlates with several properties characterizing the species' habitats and agrees with observations concerning major adaptations. Phylogenetic analysis of the seeds also revealed the complex dynamics governing the evolution of metabolic networks and the gain and loss of biosynthetic capacity. This framework thus enables tracing the evolutionary history of both metabolic networks and the interaction of these networks with their environments.
In reality, however, the biochemical environment of one species is often affected (or even fully determined) by the metabolic context of another species (Brown et al., 2001). This is clearly the case in obligate intracellular parasites, where environments are fully determined by the internal biochemical state of their hosts. Such host-parasite relationships, as well as other forms of species interactions, clearly shape the selection pressures governing the evolution of each species (Dybdahl and Lively, 1998), and could markedly affect the structure of their metabolic networks. Therefore, these interactions are likely to have a unique signature in the joint topology of the species' metabolic networks, providing a computational, large-scale method for characterizing and detecting ecological patterns. For example, many obligate intracellular parasites have lost a considerable number of biosynthetic genes, and rely on their host for nutrients they cannot synthesize (McCutcheon and Moran, 2007; Shigenobu et al., 2000; Stephens et al., 1998). This will be manifested as an exceptionally high number of externally acquired compounds in the parasite's network that can be synthesized by the network of the host.
In this paper, we build upon the computational framework described above and formulate a novel pair-wise, topology-based measure of biosynthetic support, reflecting the complementarity between the nutritional requirements of one species and the biosynthetic capacity of another. We demonstrate that this measure correlates with host-parasite interactions on a large-scale and provides valuable insights into their evolution.
2. Results
We constructed the metabolic networks of 569 bacterial species, based on a large-scale metabolic database (Kanehisa et al., 2006). Using a computational “reverse ecology” algorithm developed previously (Borenstein et al., 2008), we identified in each network the set of compounds that are exogenously acquired (i.e., the seed set of the network). We also retrieved data describing the natural habitat of each bacterial species and labeled each species as either a parasite or a free living bacteria. We further collected new data and labeled each parasitic species as being a parasite of either mammals, insects, or plants.
We first compared the overall size of the networks (a measure of biosynthetic capacity) and the size of the seed sets (a measure of environmental variability) (Parter et al., 2007) in parasites versus free-living bacteria. We find that generally parasitic bacteria not only have significantly smaller networks than free-living bacteria (as expected), but that they also extract fewer compounds from their environments (Table 1).This is in agreement with our findings in a previous analysis (Borenstein et al., 2008) for a substantially smaller set of species. These findings suggest that intracellular bacteria that live in highly predictable and fixed environments, and potentially could rely on these environments for the supply of numerous required metabolites, take up a relatively small number of metabolites in comparison to bacteria that have to cope with a wide range of environmental niches. Interestingly, focusing on parasites of specific hosts, we find that this trend does not hold for plant parasites, which appear to possess markedly larger networks and seed sets than other organisms. This accords with biological observations concerning at least two major families of plant parasites: The nitrogen fixing Rhizobia species, which are characterized by extremely complex and diverse metabolism (and remain, at least part of the time, exterior to the plant cell, interacting directly with a soil environment) (Lodwig and Poole, 2003), and the xylem-inhabiting bacteria, such as Xylella fastidiosa, which exhibit extensive biosynthetic capabilities in an environment of the nutrient-poor xylem sap (Simpson et al., 2000).
Table 1.
Size of Networks and Seed Sets in Free-Living versus Parasitic Bacteria
| Network size | Seed set size | |
|---|---|---|
| Free-living | 819.6 (242.9) | 188.6 (56.4) |
| Parasites (all) | 701.5 (312.6)b | 163.8 (70.8)b |
| Mammal parasites | 677.8 (287.0)b | 158.0 (61.7)b |
| Insect parasites | 448.8 (180.0)c | 112.7 (40.2)c |
| Plant parasites | 993.4 (432.5)a | 239.6 (106.5)a |
The size of a network is measured by the number of compounds in the network. The size of the seed set is estimated by the number of source components. Each entry in this table denotes the average size and the standard deviation (in parentheses). Footnote symbols indicate the p-value obtained in a Wilcoxon rank sum test, comparing the focal values to those associated with free-living bacteria.
p < 0.01.
p < 10−6.
p < 10−13.
Next, we introduce a new pair-wise, topology-based measure of biosynthetic support, which aims to capture the extent to which the nutritional requirements of a potential parasite are met by the biosynthetic capacity of a potential host. Formally, given a pair of species b and e, the biosynthetic support of b in e is defined as the fraction of exogenously acquired compounds required by b (i.e., its seed set) that occur in the metabolic network of e. As obligatory parasites often lose the ability to synthesize certain compounds, and instead rely on their hosts for the supply of these compounds, biosynthetic support values obtained for true host-parasite pairs are expected to be higher than those obtained for a random pair of species. Here, we wish to utilize this novel measure, which accounts not only for the interactions between organisms and their environments (as in Borenstein et al., 2008), but also for interactions between pairs of species, and to examine whether this large-scale computational approach can indeed provide an informative characterization of host-parasite interactions.
To this end, we consider three representative eukaryotic hosts: human, fruit fly, and Arabidopsis. We constructed the metabolic networks of these eukaryotic species, and calculated the biosynthetic support of all the bacterial species included in our analysis in these organisms. We first compared the biosynthetic support values obtained for parasites versus free-living bacteria. We find that, while the average biosynthetic support of free-living bacteria in the metabolic network of human is 0.694, the average biosynthetic support of parasites is significantly higher (0.751; p < 10−13; Wilcoxon rank sum test). A similar trend is observed for biosynthetic support values in the metabolic network of fruit fly; the average biosynthetic support of parasitic bacteria is significantly higher that that of free-living bacteria (0.701 and 0.640, respectively; p < 10−11; Wilcoxon rank sum test), and in Arabidopsis (0.725 for parasites vs. 0.687 for free-living; p < 10−6; Wilcoxon rank sum test). This difference between parasites and free-living bacteria goes beyond the difference in the size of networks and seed sets reported above, as demonstrated by the markedly lower p-values obtained by the Wilcoxon rank sum test for the biosynthetic support score (and compare with Table 1; see also the improved prediction accuracy below). Moreover, it should be noted that the biosynthetic support scores are normalized for the size of the seed set, and therefore, in an important sense, represent novel information.
To further examine the overlap between the characteristics encapsulated in the biosynthetic support scores and other, global features of the network, we calculated the correlation between these scores and other network based measures. We find a significant negative correlation between the biosynthetic support score (in human) and the number of reactions in the network, as well as with the size of the seed set (−0.637, and −0.7 respectively; p < 10−300 for both; Spearman rank correlation). These moderate correlation coefficients suggest that while biosynthetic support is linked to other global properties of the network, it may still encapsulate fundamentally different and independent information. Furthermore, these correlations are markedly different in parasites versus free-living bacteria: Specifically, the coefficients of the correlation with the number of reactions and with the size of the seed set in parasites are significantly higher than those obtained for free-living bacteria (−0.788 and −0.807 vs. −0.494 and −0.578; p < 10−300 for both; Spearman rank correlation).
It is also instructive to examine the distribution of biosynthetic support scores in human and in fruit fly obtained for parasites and for free-living bacteria. Although these distributions are relatively broad, the biosynthetic support scores obtained for parasites clearly have a propensity toward higher values (Figs. 1A and 1B). Remarkably, this trend becomes even more prominent when we consider only those parasites that are labeled as infecting hosts that are phylogenetically close to the host species we used to calculate the biosynthetic support scores; mammal parasites display a slightly more skewed distribution in human, and insect parasites display a markedly more skewed distribution in fruit fly (the relatively weaker effect in mammal parasites may be attributed to the fact that most of the parasites in our set—179 out of 212—are labeled as mammal parasites). As before, plant parasites exhibit a fundamentally different trend, with many species having very low biosynthetic support scores, indicating that a large group of these parasites do not rely strongly on their hosts for the supply of the various nutrients they require (Fig. 1C). A scatter plot of biosynthetic support scores in human versus fruit fly (Fig. 1D) further demonstrates the overall similarities between the scores obtained in these two hosts (although, as also demonstrated below, host specificity can still be predicted to a certain extant). It also appears that the highest biosynthetic support scores are often obtained for bacterial species that infect both mammals and insects (e.g., insect-borne human pathogens).
FIG. 1.

The distribution of biosynthetic support scores for various bacterial species. (A) The biosynthetic support of free-living bacteria, parasites, and mammal parasites alone, in the metabolic network of human. (B) The biosynthetic support of free-living bacteria, parasites, and insect parasites alone, in the metabolic network of fruit fly. (C) The biosynthetic support of free-living bacteria, parasites, and plant parasites alone, in the metabolic network of Arabidopsis. (D) Scatter plot of biosynthetic support scores in human versus fruit fly. To highlight the similarities and differences between the scores in these hosts, the scores in each host were ordered and the ranks were used instead of the actual values.
This increased biosynthetic support of parasites in various hosts can be used to predict parasitic species. This is achieved by defining a biosynthetic support threshold, above which a bacterial species would be classified as a parasite. To evaluate the performance of this method, we tested its predictive power in classifying the set of 569 bacterial species included in our analysis. For comparison, we also predicted parasites versus free-living bacteria using the size of the networks and the size of the seed sets (i.e., defining a minimal size, below which a bacterial species is classified as a parasitic species). Figure 2 demonstrates the quality of each of these predictors, using precision versus recall and receiver operating characteristic (ROC) curves. Evidently, biosynthetic support-based predictions are superior to predictions which are based on the size of the network or on the size of the seed set. Moreover, considering the simple network model used in our analysis, and the incompleteness of the data, the resulting biosynthetic support-based predictions are remarkably accurate (especially for insect parasites). Figure 2B further demonstrates the success of host-specific parasite prediction, based on the biosynthetic support scores obtained in different potential hosts (considering the results shown above for plant parasites, we focus here only on human and fruit fly as hosts).
FIG. 2.
Accuracy of parasite prediction. (A) Precision versus recall of parasite prediction based on network size, seed set size, and biosynthetic support scores in human and in fruit fly. (B) Receiver operating characteristic (ROC) curves for parasite prediction. In addition to predicting all parasites, host-specific parasite prediction is also examined. Mammal parasites are predicted according to their biosynthetic support in human. Insect parasites are predicted according to their biosynthetic support in fruit fly. For comparison, ROC curves are also presented for prediction of all parasites based on either the size of the network or the size of the seed set. The areas under the ROC curves are 0.623, 0.632, 0.691, 0.678, 0.699, and 0.898, respectively (following the order in the legend).
Finally, we wish to examine the changes in biosynthetic support scores throughout the evolutionary process. We first focus on the bacterial division of Firmicutes, which includes many host-associated species. Using a well-established, sequence-based phylogenetic tree (Ciccarelli et al., 2006) to relate the various Firmicutes species in our dataset and to reconstruct the metabolic networks of ancestral species across this tree, we can calculate the biosynthetic support of both extant and ancestral species and compare it with their phylogeny (Fig. 3). We find a significantly and markedly high correlation between the biosynthetic support scores of the various species (extant and ancestral) and their distances from the common ancestor of Firmicutes (0.795, p < 10−300; Spearman rank correlation). Limiting this analysis only to ancestral species (to control for potential differences between extant networks and ancestral reconstructed networks), a comparable correlation is still obtained (0.788, p < 10−8; Spearman rank correlation). Repeating this analysis in other divisions that include many parasitic species (and for which enough species are included in our analysis) reveals a similar trend. Specifically, we find a marked correlation between the biosynthetic support scores of extant and ancestral species and their distances from their common ancestor also in Gamma-proteobacteria, Alpha-proteobacteria, and Actinobacteria (0.812, p < 10−300; 0.67, p < 10−3; 0.686, p < 10−3, respectively). This finding suggests that overall (at least in these large divisions of parasitic species) biosynthetic support tends to increase from ancestral species to descendants. This is in agreement with a gradual adaptation of parasites to their host environments, and the loss of biosynthetic capacity (McCutcheon and Moran, 2007; Moran and Mira, 2001).
FIG. 3.
Biosynthetic support scores of 39 extant and 38 ancestral Firmicutes species. Extant species are labeled according to KEGG organism codes. The gray scale corresponds to biosynthetic support scores in current species.
3. Discussion
This study introduces a novel measure of biosynthetic support and demonstrates how this measure relates to host-parasite interactions. Specifically, parasitic species are shown to have higher biosynthetic support scores in several hosts than free-living bacteria. Integrating this method with phylogenetic data further reveals an evolutionary trend of biosynthetic capacity loss across numerous parasites (McCutcheon and Moran, 2007; Moran and Mira, 2001), providing a first computational, large-scale characterization of this trend.
It should be noted that our measure is based on network topology alone, ignoring several other quantitative properties of metabolic reactions (e.g., stoichiometry, metabolic rates, and regulation). We further assume a static model, where all pathways are active, ignoring dynamic regulation and dependency on environmental conditions. In this sense, the seed set and biosynthetic support score of a given species can be conceived as representing its overall metabolic potential. More involved models (such as constraint-based stoichiometric models) (Edwards and Palsson, 2000; Ibarra et al., 2002), may provide additional insights into the metabolic interaction between species (Rokhlenko et al., 2007). A recent study, for example, constructed a stoichiometric model of a simple, two-species mutualistic system (Desulfovibrio vulgaris and Methanococcus maripaludis), and successfully predicted metabolic fluxes and growth rates (Stolyar et al., 2007). Yet, these complex, manually curated models are available for only a handful of species. In contrast, topology-based models can be readily reconstructed for hundreds of species, and analyzed using a plethora of tools borrowed from graph theory and complex network analysis. Such studies therefore facilitate the detection of large-scale patterns across numerous species.
Another potential caveat of any study that is based on large-scale metabolic data (such as those obtained from KEGG) stems from missing and inaccurate annotation. These data are often based on automated comparison-based methods of genome annotation (Kanehisa et al., 2006) and are associated with high levels of noise and incompleteness (Green and Karp, 2006). This can affect the prediction of seed sets and consequently the calculation of biosynthetic support scores. We previously examined the effect of missing or erroneous data on seed prediction and showed that the composition of the seed set is fairly robust to perturbations of the raw metabolic data (Borenstein et al., 2008). Moreover, as biosynthetic support scores reflect the complementarity between two networks, a consistent bias in annotation (such as a missing annotation of an entire metabolic pathway), may in fact still allow comparison between the scores of various species. Still, considering the inherent incompleteness associated with these data, our main goal in this paper is to demonstrate large-scale patterns, such as the complementarity between hosts and parasites and the loss of biosynthetic capacity across evolutionary timescales, rather than to provide specific predictions that concern specific species. As such, the findings presented above for various prediction methods (e.g., using ROC curves) serve to facilitate a comparison between the information content of various measures, and to shed light on the way in which the co-evolution of hosts and parasites, as well as the continuing adaptation of parasites to their hosts, are manifested in the topology of metabolic networks.
The method presented in this paper can assist in preliminary characterization of newly detected parasites and can be utilized to identify patterns of potential variation in hosts or in transmission vectors. Further phylogenetic analysis has the potential to provide important insights into the evolutionary dynamics that govern the continuing race between parasites and hosts (Dybdahl and Storfer, 2003; Maynard Smith, 1976; McCutcheon and Moran, 2007; Sole et al., 1999). Of special interest is the use of network topology-based analysis for the characterization of the complex web of interactions within diverse microbial communities (Schloss and Handelsman, 2005), ranging from those living on sunken whale skeletons (Tringe et al., 2005) to those inhabiting the human gut (Eckburg et al., 2005). Advances in metagenomics—the analysis of genetic material recovered from environmental samples—are expected to give rise to an explosion of data in coming years and call for system-level computational analyses of that sort. As such, the “reverse ecology” approach presented here, inferring ecological insights from genomic-based data, is a promising vehicle for addressing key ecological questions in the post-genomic era (Borenstein et al., 2008; Parter et al., 2007).
4. Methods
4.1. Metabolic networks construction and relevant data
Data concerning the metabolic reaction of a large array of species were retrieved from the KEGG database (Kanehisa et al., 2006), release 45.0 (January 1, 2008). A list of the main reactions in the database was retrieved from the file reaction\mapformula.lst in the KEGG LIGAND database. This file also lists for each reaction its definition (i.e., the substrates and product compounds) and its directionality (if known) in each pathway in which it participates. The chemical compounds are limited to main reactants. The list of reactions present in each species (and the pathways in which they are found) was retrieved from the rn files in the PATHWAY database. Using these data, the metabolic network of each species was reconstructed. We represent each network as a directed graph where nodes denote compounds and edges denote reactions. Formally, a directed edge from compound a to compound b indicates that compound a is a substrate in some reaction which produces compound b (i.e., for each given reaction, all the nodes that represent its substrates are connected by directed edges to all the nodes that represent its products). In total, the metabolic networks of 782 species, of which 595 are bacterial species, were reconstructed. We further discarded species that have less than 100 reactions, leaving a total of 569 bacterial species. We also constructed merged networks for mammals, insects, and plants, representing the union of the metabolic networks of all the species in each of these phyla.
Data concerning various environmental properties of Prokaryotes were obtained from the prokaryotic attributes table of the NCBI Genome Project (www.ncbi.nlm.nih.gov/genomes/lproks.cgi). This table specifies for each species the basic habitat in which the organism is commonly found and specifically whether it is host-associated (i.e., the organism is often or obligately associated with a host organism), aquatic, terrestrial, specialized, or multiple (i.e., the organism can be found in more than one of the above environments). For the purpose of this study we considered all species that were defined in this dataset as host-associated as parasites, and all others as free-living bacteria. We also conducted an extensive literature search to identify the potential host of each parasitic species. Specifically, we defined species that infect humans or other mammals as mammal parasites, those that infect various insects or other arthropods as insect parasites, and those that infect plants as plant parasites. It should be noted that a parasitic species can be, for example, both a mammal-parasite and an insect-parasite, as is the case for insect-borne human pathogens.
4.2. Identifying seed compounds
Seed compounds are the set of compounds that, based on the network topology, are exogenously acquired. Formally, we define the seed set of a metabolic network (Raymond and Segre, 2006) as the minimal subset of the network's compounds that cannot be synthesized from other compounds in the network and whose existence permits the production of all other compounds in the network (Borenstein et al., 2008). Such seed sets form ecological “interfaces” between metabolic networks and their surroundings, and can serve as a proxy for the biochemical environment of each species. The seed set of a given network can be identified as follows: (i) The network is decomposed into its Strongly Connected Components (Aho et al., 1974) (a strongly connected component is a maximal set of nodes such that for every pair of nodes u and v there is a path from u to v and a path from v to u). (ii) The strongly connected components form a Directed Acyclic Graph (DAG) whose nodes are the components and whose edges are the original edges in the graph that connect nodes in two different components. (iii) Components without incoming edges and at least one outgoing edge are identified and defined as source components. (iiii) Each source component forms a collection of candidate seed compounds, where a viable seed set must include exactly one compound from each source component (and no other compounds). Each possible selection of one compound out of each source component yields a seed set solution that satisfies the above definition. Further details concerning the seed compounds detection algorithm and its validation can be found elsewhere (Borenstein et al., 2008).
4.3. Calculating biosynthetic support scores
The biosynthetic support score represents the extent to which the metabolic requirements of a potential parasitic organism can be supported by the biosynthetic capacity of a potential host. Formally, we define 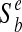 —the biosynthetic support of a bacterial species b in a eukaryotic species e—as the fraction of the seed set of b that can be found in the metabolic network of e. As seed sets cannot be fully determined in cases where a source component comprises more than one compound (see Section 4.2), we define the biosynthetic support as the maximal support that can be obtained across all viable seed sets. This is measured by calculating the fraction of the source components of b, in which at least one of the compounds can be found in the network of e. Additionally, we examined a more complex version of this measure, where highly prevalent seeds (those that are exogenously acquired by many organisms and that therefore may not carry significant information on the effective biosynthetic support value) were initially discarded form the analysis. This omission, however, did not qualitatively affect the results reported in this paper.
—the biosynthetic support of a bacterial species b in a eukaryotic species e—as the fraction of the seed set of b that can be found in the metabolic network of e. As seed sets cannot be fully determined in cases where a source component comprises more than one compound (see Section 4.2), we define the biosynthetic support as the maximal support that can be obtained across all viable seed sets. This is measured by calculating the fraction of the source components of b, in which at least one of the compounds can be found in the network of e. Additionally, we examined a more complex version of this measure, where highly prevalent seeds (those that are exogenously acquired by many organisms and that therefore may not carry significant information on the effective biosynthetic support value) were initially discarded form the analysis. This omission, however, did not qualitatively affect the results reported in this paper.
We calculated the biosynthetic support of each bacterial species in several eukaryotic species. In this study we focus only on the support obtained in H. sapiens (as a representative mammalian host), in D. melanogaster (as a representative insect host), and in A. thaliana (as a representative plant host). Using other representative hosts from each class did not qualitatively change the results presented in this paper (except for differences that stem from annotation level). This suggests that metabolic data alone (and specifically, metabolic network topology) are not sufficient to distinguish parasites of different mammals (or different insects), without potentially integrating other data (e.g., immune response). We also calculated the support of each bacterial species in the merged networks of mammals, insects, and plants. In general, the results obtained with these merged networks (not shown here) demonstrated qualitatively similar trends, but were markedly more noisy.
4.4. Phylogenetic analysis and reconstruction of ancestral metabolic networks
We consider a well-established, sequence similarity-based tree (Ciccarelli et al., 2006), to identify the phylogenetic relations between the species included in our analysis. Specifically, this tree covers 39 Firmicutes species. We followed Borenstein et al. (2008), using the presence/absence pattern of each reaction across extant species and applying Fitch's small-parsimony algorithm (Fitch, 1971), to determine the presence/absence of each reaction in the internal nodes of the tree and to reconstruct the ancestral metabolic networks. As ancestral networks are reconstructed based on reactions that were detected in extant species, this analysis is not affected by bias in homology detection that may stem from different distances of various ancestral species from well annotated species. It should also be noted that the biosynthetic support score is normalized by the size of the seed set, and is therefore not likely to be affected by variation in gene detection probabilities. We also used this tree to measure the distance of each extant and ancestral species from the last common ancestors of Firmicutes. As most Firmicutes infect mammals, biosynthetic support scores for these species were calculated in relation to the human metabolic network. A similar analysis was also performed for all major bacterial divisions. To obtain sufficient statistics, this analysis was limited to divisions that included at least 10 species in our dataset.
Acknowledgments
We thank the anonymous reviewers for their helpful comments. The research of E.B. is supported by the Morrison Institute for Population and Resource Studies, and by a grant to the Santa Fe Institute from the James S. McDonnell Foundation 21st Century Collaborative Award Studying Complex Systems. Research is supported in part by the NIH grant GM28016 to M.W.F.
Disclosure Statement
No competing financial interests exist.
References
- Aho A. Hopcroft J. Ullman J. The Design and Analysis of Computer Algorithms. Addison-Wesley; Reading, MA: 1974. [Google Scholar]
- Almaas E. Kovacs B. Vicsek T., et al. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427:839–843. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- Alon U. Biological networks: The tinkerer as an engineer. Science. 2003;301:1866–1867. doi: 10.1126/science.1089072. [DOI] [PubMed] [Google Scholar]
- Bonchev D. Rouvray D. Chemical Graph Theory: Introduction and Fundamentals. Taylor & Francis; London: 1991. [Google Scholar]
- Borenstein E. Kupiec M. Feldman M., et al. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc. Natl. Acad. Sci. USA. 2008;105:14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. Whitham T. Ernest S., et al. Complex species interactions and the dynamics of ecological systems: long-term experiments. Science. 2001;293:643–650. doi: 10.1126/science.293.5530.643. [DOI] [PubMed] [Google Scholar]
- Ciccarelli F.D. Doerks T. von Mering C., et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Deutscher D. Meilijson I. Kupiec M., et al. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat. Genet. 2006;38:993–998. doi: 10.1038/ng1856. [DOI] [PubMed] [Google Scholar]
- Dybdahl M. Lively C. Host-parasite coevolution: evidence for rare advantage and time-lagged selection in a natural population. Evolution. 1998;52:1057–1066. doi: 10.1111/j.1558-5646.1998.tb01833.x. [DOI] [PubMed] [Google Scholar]
- Dybdahl M. Storfer A. Parasite local adaptation: red queen versus suicide king. Trends Ecol. Evol. 2003;18:523–530. [Google Scholar]
- Eckburg P. Bik E. Bernstein C. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. Palsson B. Robustness analysis of the Escherichia coli metabolic network. Biotechnol. Prog. 2000;16:927–937. doi: 10.1021/bp0000712. [DOI] [PubMed] [Google Scholar]
- Fitch W. Towards defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 1971;20:406–416. [Google Scholar]
- Green M. Karp P. The outcomes of pathway database computations depend on pathway ontology. Nucleic Acids Res. 2006;34:3687–3697. doi: 10.1093/nar/gkl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra R. Edwards B. Palsson J.S. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- Jeong H. Tombor B. Albert R., et al. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. Goto S. Hattori M., et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer A. Borenstein E. Gophna U., et al. The evolution of modularity in bacterial metabolic networks. Proc. Natl. Acad. Sci. USA. 2008;105:6976–6981. doi: 10.1073/pnas.0712149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwig E. Poole P. Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 2003;22:37–78. [Google Scholar]
- Maynard Smith J. A comment on the red queen. Am. Nat. 1976;110:325–330. [Google Scholar]
- McCutcheon J. Moran N. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2:research0054.1–12. doi: 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parter M. Kashtan N. Alon U. Environmental variability and modularity of bacterial metabolic networks. BMC Evol. Biol. 2007;7:169. doi: 10.1186/1471-2148-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J. Segre D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- Rokhlenko O. Shlomi T. Sharan R., et al. Constraint-based functional similarity of metabolic genes: going beyond network topology. Bioinformatics. 2007;23:2139–2146. doi: 10.1093/bioinformatics/btm319. [DOI] [PubMed] [Google Scholar]
- Schloss P. Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 2005;6:229. doi: 10.1186/gb-2005-6-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S. Watanabe H. Hattori M., et al. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Simpson A.J. Reinach F.C. Arruda P., et al. The genome sequence of the plant pathogen Xylella fastidiosa. Nature. 2000;406:151–159. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- Smith E. Morowitz H. Universality in intermediary metabolism. Proc. Natl. Acad. Sci. USA. 2004;101:13168–13173. doi: 10.1073/pnas.0404922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole R. Ferrer R. Gonzalez-Garcia I., et al. Red queen dynamics, competition and critical points in a model of RNA virus quasispecies. J. Theor. Biol. 1999;198:47–59. doi: 10.1006/jtbi.1999.0901. [DOI] [PubMed] [Google Scholar]
- Stelling J. Klamt S. Bettenbrock K., et al. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–193. doi: 10.1038/nature01166. [DOI] [PubMed] [Google Scholar]
- Stephens R. Kalman S. Lammel C., et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Stolyar S. Van Dien S. Linnea Hillesland K. Metabolic modeling of a mutualistic microbial community. Mol. Syst. Biol. 2007;3:92. doi: 10.1038/msb4100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe S. von Mering C. Kobayashi A., et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]




