Abstract
Persistent activation of the Hedgehog (HH)/GLI signaling pathway has been implicated in the development of a number of human cancers. The GLI zinc finger transcription factors act at the end of the HH signaling cascade to control gene expression, and recent studies have shown that the activity of GLI proteins can be additionally modified by integration of distinct signals, such as the MEK/extracellular signal-regulated kinase (ERK) and phosphinositide-3 kinase (PI3K)/AKT pathway. However, little is known about the identity of the upstream activators of these HH/GLI interacting signaling pathways in cancer. Here, we provide evidence that integration of the HH/GLI and epidermal growth factor receptor (EGFR) pathway synergistically induces oncogenic transformation, which depends on EGFR-mediated activation of the RAS/RAF/MEK/ERK but not of the PI3K/AKT pathway. EGFR/MEK/ERK signaling induces JUN/activator protein 1 activation, which is essential for oncogenic transformation, in combination with the GLI activator forms GLI1 and GLI2. Furthermore, pharmacologic inhibition of EGFR and HH/GLI efficiently reduces growth of basal cell carcinoma (BCC) cell lines derived from mice with activated HH/GLI signaling. The results identify the synergistic integration of GLI activator function and EGFR signaling as a critical step in oncogenic transformation and provide a molecular basis for therapeutic opportunities relying on combined inhibition of the HH/GLI and EGFR/MEK/ERK/JUN pathway in BCC.
Introduction
Hedgehog (HH)/GLI signaling plays a critical role in the initiation and growth of a number of human malignancies (reviewed in refs. 1-3). HH signaling is initiated by the binding of HH protein to its receptor Patched (PTCH), a transmembrane domain protein that represses signal transduction in the absence of ligand by inhibiting the transmembrane domain protein Smoothened (SMO). Binding of HH protein to its receptor blocks PTCH function and unleashes SMO activity. Activation of SMO eventually results in an increase in activator forms of the GLI zinc finger transcription factors GLI2 and GLI1, which regulate target gene expression in response to pathway activation (reviewed in refs. 4, 5).
In cancer, failure to terminate HH/GLI signaling leads to a persistent increase in GLI1 and GLI2 activity, which has been shown to account for the initiation and growth of HH-associated tumors (reviewed in ref. 6). The biological activity of GLI proteins is controlled at the transcriptional and posttranslational level, the latter involving protein phosphorylation, processing, degradation, and interaction with distinct cofactors (reviewed in ref. 7-9). A growing body of evidence suggests that activation of GLI proteins is not controlled exclusively by HH signaling itself but also by other pathways frequently activated in human malignancies. GLI activity can be modulated by phosphinositide-3 kinase (PI3K)/AKT, MEK/extracellular signal-regulated kinase (ERK), protein kinase Cδ, and transforming growth factor β/SMAD, which affect stability, subcellular localization, or expression of GLI proteins (reviewed in refs. 6, 10, 11). In vivo HH/GLI and Ras have been shown to cooperate in the progression of pancreatic cancer lesions and in melanoma growth, although the molecular details of this interaction are still not well understood (reviewed in ref. 6). The complexity of HH/GLI modulation is further highlighted by the unexpected finding that, in brain cancer cells, fibroblast growth factor–mediated activation of MEK/ERK and c-Jun NH2 terminal kinases (JNK) negatively regulates oncogenic HH/GLI signaling (12), suggesting intricate and cell type–specific regulatory mechanisms of signal integration.
Several recent studies have implicated the epidermal growth factor (EGF) receptor (EGFR) pathway in the modulation of HH/GLI activity. For instance, EGF and Sonic HH cooperate to stimulate neural stem cell proliferation and invasive growth of keratinocytes (13-15), and there is preliminary in vitro evidence that both pathways may interact in prostate cancer cells (16). Our own group has recently shown that EGFR signaling synergizes with GLI1 and GLI2 to selectively activate transcription of a subset of direct GLI target genes via stimulation of RAS/RAF/MEK/ERK signaling (17). On the other hand, a negative effect of GLI1 on EGFR and ERK activation can be observed in keratinocytes when cultured under conditions that allow induction of epidermal stem cell markers by GLI1, further underlining the context-dependent regulation of GLI protein activity (18).
Aberrant activation of EGFR signaling has been implicated in a number of human malignancies, which has made EGFR a prime molecular target in drug-based cancer therapy (reviewed in refs. 19, 20). Under normal physiologic conditions, signaling via EGFR/HER1, a member of the ErbB family of receptor tyrosine kinases, involves ligand binding to the extracellular domain of EGFR, which results in activation of the EGFR tyrosine kinase and phosphorylation of multiple COOH terminal tyrosine residues that serve as binding sites for SRC homology 2 and phosphotyrosine binding domain containing cytosolic signaling proteins (21). The selective docking of these proteins relays the signal toward the nucleus via multiple routes, including RAF/MEK/ERK, PI3K/AKT, SRC, and Janus-activated kinase (JAK)/signal transducers and activators of transcription (STAT). Specific regulation of the transcriptional programs in response to EGFR signaling involves context-dependent activation of transcriptional regulators, such as members of the activator protein-1 (AP-1), ETS, and STAT family (reviewed in refs. 22, 23).
In this study, we addressed whether integration of the HH/GLI and EGFR pathway is a critical step in cancer development. We report that EGFR signaling synergistically interacts with HH/GLI in oncogenic transformation and identify a novel molecular mechanism of HH/GLI and EGFR integration relying on EGFR-activated MEK/ERK and JUN/AP-1 function.
Materials and Methods
Cell culture, chemical treatments, and retroviral transduction
RK3E cells, human HaCaT keratinocytes (24), and mouse Egfr−/− fibroblasts (line 1-1; ref. 25) were routinely grown in DMEM (PAA) supplemented with 10% fetal bovine serum (FBS; PAA), penicillin (62.5 μg/mL), and streptomycin (100 μg/mL), at 37°C in a humidified atmosphere of 5% CO2. For the analysis of EGFR signal transduction, cells were starved overnight in DMEM with 0.1% FBS before the respective treatments with pharmacologic compounds. Wortmannin (Calbiochem) was used at a final concentration of 1 Amol/L, UO126 (Sigma) at 10 μmol/L, gefitinib (IRESSA, AstraZeneca) at 1 μmol/L, JAK inhibitor I at 1 μmol/L (Calbiochem), and SP600125 (Sigma) at 10 μmol/L. Recombinant human EGF (Sigma) was used at concentrations as indicated in the text. Induction of GLI1 or GLI2 expression in doxycycline-controlled HaCaT keratinocytes was done as described previously (26). To stably express GLI1 at physiologic levels, RK3E or EGFR-deficient mouse fibroblasts were transduced with a lentiviral vector encoding EGFP-tagged GLI1 construct (27). For stable expression of dominant-active AKT or MEK1, dominant-negative AKT or PTEN, doxycycline-regulated GLI1 HaCaT lines were transduced with respective retroviral constructs followed by selection in medium containing neomycin (for AKT constructs) or puromycin at a concentration of 1 mg/mL and 1.5 μg/mL, respectively. The same protocol was used for reconstitution of Egfr−/− cells with human wild-type or oncogenic EGFR variants (kindly provided by Dr. Heidi Greulich) or with the GRB2 binding mutant EGFRY1068F. For controls, cells were transduced with inactive AKT or empty vector followed by neomycin and puromycin selection, respectively. Virus production and cell transductions were carried out as described in ref. 27, except that Metafectene Pro (Biontex) was used as transfection reagent according to the manufacturer's instructions.
Expression constructs, validation, and real-time PCR
For lentiviral GLI1 expression in RK3E and Egfr−/− mouse fibroblasts, NH2 terminally EGFP-tagged GLI1 was cloned into pLentiLox3.7 vector (28). For transduction with dominant-active, myristoylated Akt1, the corresponding inactive control myrAkt1K179M or dominant-negative Akt1K179M, pLNCX lentiviral vectors were used (29). PTEN (generous gift from Dr. W Sellers, Addgene plasmid 10787), dominant-active ΔN-MEK-EE (MEK*; generous gift from Dr. Graham Neill, Barts and The London School of Medicine and Dentistry), wild-type human EGFR, the kinase-dead mutant EGFRD837A, and all oncogenic variants (EGFRL747_E749del, EGFRD770_N771insNPG, and EGFRL858R; ref. 30), as well as the GRB2 binding mutant EGFRY1068F, were in retroviral pBabe-puro vector. Commercial nontarget control (SHC002) and validated JUN short hairpin RNA (shRNA) lentiviral vectors were selected from the mission shRNA library (Sigma-Aldrich). The integrity of all constructs was verified by sequencing. Expression of all constructs was validated by quantitative PCR (qPCR) or Western blotting. Comparable surface expression of EGFR variants was further analyzed by flow cytometric analysis using FITC-conjugated anti-EGFR antibody 528 (Santa Cruz Biotechnology).
Real-time quantitative reverse transcription-PCR analysis was done, as described previously (26). Primers for mouse EGFR were forward 5′ AGG GGG AAC CAA GGG AGT TTG TGG 3′ and reverse 5′ TGG CGT GGC ATA GGT GGC AGA 3′ or as published previously (17).
in vitro transformation and xenograft assays
To analyze anchorage-independent growth, cells were seeded in 12-well plates in 0.4% select agar on top of 0.5% bottom select agar (Invitrogen) according to standard protocols. Cells (5 × 103) were seeded in 1.5 mL select agar and cultures grown for 14 d (RK3E and mouse fibroblasts) or 28 d (human keratinocytes) at 37°C in a humidified atmosphere of 5% CO2. Anchorage-independent growth was documented on a stereomicroscope equipped with a Cell^D Image capture system. Colony growth in soft agar cultures was quantified using Colony Counter software (Microtech Nition).
In vivo tumor development was analyzed by s.c. injection of 5 × 106 cells into the flank of nude mice (The Jackson Laboratory). To induce and maintain GLI1 expression in the xenografts, doxycycline (Sigma) was added to a final concentration of 10 mg/mL to drinking water supplemented with 5% sucrose. Tumor growth was measured every other day over a period of 25 d.
Western blot analysis, chromatin immunoprecipitation, and gel shift assays
For the analysis of protein expression and modifications, cells were lyzed in Laemmli buffer supplemented with PhosStop (Roche) and 1 mmol/L sodium orthovanadate (Sigma). The following antibodies were used: anti-GLI1 antibody, anti–phosphorylated JUN antibody, anti–β-actin antibody, anti-EGFR antibody (all Santa Cruz Biotechnology), anti–phosphorylated p44/42 mitogen-activated protein kinase (MAPK) antibody, anti–phosphorylated AKT antibody, anti-p44/42 MAPK, anti-AKT antibody, anti–phosphorylated EGFR (p-EGFR; Tyr1173, all Cell Signaling Technology), and anti-JUN antibody (BD Biosciences). Proteins were visualized with horseradish peroxidase–conjugated secondary antibodies in combination with enhanced chemiluminescence detection system (GE Health Care). EGFR staining on human basal cell carcinoma (BCC) specimen was done as described previously using Food and Drug Administration–approved PharmDx EGFR detection kit (Dako; ref. 17).
Chromatin immunoprecipitation was carried out with SimpleChIP Enzymatic ChIP kit (Cell Signaling Technology) according to the manufacturer's instructions. Chromatin was isolated from FLAG-cJUN transduced GLI1-HaCaT cells. Before harvesting, cells were treated with 10 ng/mL EGF for 30 min. Chromatin was precipitated with anti-FLAG-M2 antibody (Sigma). Specific enrichment of DNA was compared with species and isotype-matched IgG control antibodies (Santa Cruz). Primer sequences used for quantitative qPCR measurement of immunoprecipitated promoter fragments were as follows: IL1R2 forward 5′TGGTATTTGGTGAGATTTTCCTAC3′, IL1R2 reverse 5′GGCTTTTCCCATTATTTTGATGA3′; JAG2 forward 5′GAGGGAGCAGAGTGGAGAGG3′, JAG2 reverse 5′CAGACCTACGGGTTGAGACAG3′; S100A9 forward 5′GAAAGTCCACCTGAAAGTTGAGAG3′, S100A9 reverse 5′AAAAAGCATGACAATGAAGCAG3′; JUN forward 5′TTCAGACTAGGTTTCTAAATGAGCA3′, JUN reverse 5′TGAGTCAGGATGGTTTAGGTTATG3′; PTCH forward 5′GAGGATGCACACACTGGGTTGCCTA3′, PTCH reverse 5′GGGCTGTCAGATGGCTTGGGTTTCT3′. Real-time qPCR was performed on a Rotor-Gene 3000 cycler (Corbett Research).
Electrophoretic mobility gel-shift assays (EMSA) were done with doxycycline-inducible GLI1-HaCaT cells. Before harvesting, cells were treated as indicated in the text [i.e., with 10 ng/mL EGF (Roche) for 16 h to induce AP-1 DNA binding activity, with 1 μg/mL doxycycline to induce GLI1 expression, or with a combination of doxycycline/EGF, EGF/U0126 (10 μmol/L), or doxycycline /EGF/U0126]. Cells were harvested in ice-cold PBS, centrifuged for 15 min at 4°C at 13,000 rpm and stored at −80°C until further use. Preparation of extracts and EMSA analysis were done as described previously (31, 32). The sequence of AP-1 binding oligonucleotides and mutated controls was as described in ref. 31.
Results
Induction of anchorage-independent growth by combined activation of GLI1 and EGFR signaling
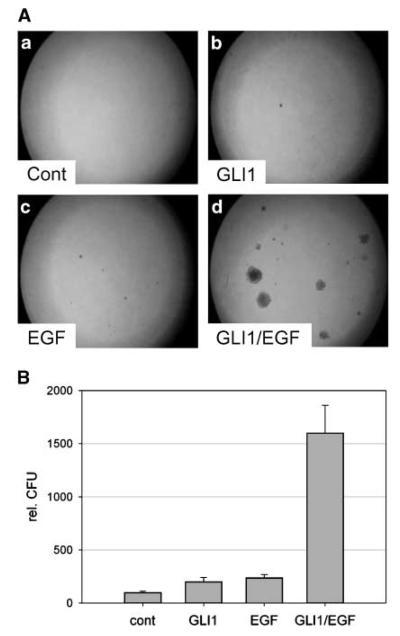
To test a possible role of EGFR and HH/GLI cooperation in oncogenic transformation, we first studied the effect of single and combined GLI1 and EGFR activity on anchorage-independent growth of RK3E rat kidney epithelial cells. As shown in Fig. 1A and B, neither expression of GLI1 nor activation of EGFR signaling alone was sufficient to elicit anchorage-independent growth of RK3E rat kidney epithelial cells. Interestingly, simultaneous activation of GLI1 and EGFR signaling induced anchorage-independent growth, providing first evidence that integration of HH/GLI and EGFR signaling can synergistically promote the emergence of tumorigenic characteristics. The failure of GLI1 to induce transformation on its own differs from results of a previous report and may be due to lower GLI1 expression levels in our study, wherein GLI1 was expressed at levels comparable with or slightly lower than those detected in human BCC (refs. 33, 34; Supplementary Fig. S1).
Figure 1.
Induction of anchorage-independent growth by combined activation of EGFR and GLI1. A, RK3E rat kidney epithelial cells stably expressing GFP (a, c) or GLI1 (b, d). Cells were either left untreated (a, b) or treated with 1 ng/mL EGF (c, d). B, quantification of assays shown in A. rel. CFU, relative number of colony forming units.
Next, we tested whether oncogenic EGFR variants identified in lung cancer patients (35, 36) can also cooperate with GLI1 to enhance anchorage-independent growth. As shown in Supplementary Fig. S2, the constitutively active oncogenic EGFR variants EGFRL747_E749del and EGFRD770_N771insNPG and, to a lesser extent, EGFRL858R (35) induced transformation of RK3E cells but did not synergize with GLI1. As the oncogenic activity of these EGFR mutants has been ascribed to the activation of AKT and STAT (37), we hypothesized that integration of EGFR signaling with GLI1 may be independent of AKT and STAT function and rather involve RAS/MEK/ERK activation, as previously shown for the selective activation of GLI/EGF target genes (17).
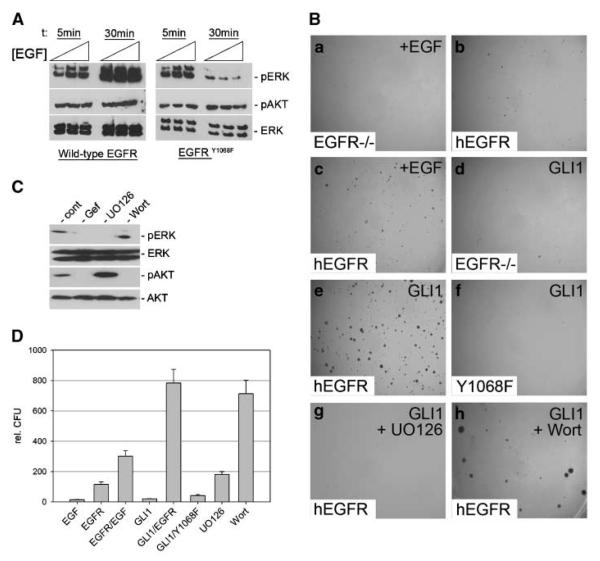
To analyze whether MEK/ERK activation in response to EGFR signaling is required for synergistic GLI1/EGFR-dependent transformation, we first took a genetic approach and reconstituted EGFR-deficient mouse embryonic fibroblasts (25) with either human wild-type EGFR or an EGFR mutant (EGFRY1068F) deficient in GRB2 binding, which is critical for coupling activated EGFR to RAS/RAF/MEK/ERK signaling (38). As shown in Fig. 2A, EGF treatment of Egfr−/− cells reconstituted with EGFRY1068F failed to induce sustained ERK activation, whereas activation of PI3K/AKT signaling was comparable with activation by wild-type EGFR. We found that, like in RK3E cells, transformation of mouse fibroblasts was only observed upon simultaneous expression of human EGFR and GLI1 (Fig. 2B, a–e and D). By contrast, the GRB2-binding mutant EGFRY1068F failed to cooperate with GLI1 (Fig. 2B, f and D), suggesting that sustained activation of MEK/ERK in response to EGFR stimulation is required for synergistic transformation with GLI1. This is further supported by pharmacologic studies showing that inhibition of MEK (by UO126 treatment; Fig. 2B, g and D) but not of PI3K function (by wortmannin treatment; Fig. 2B, h and D) abolished transformation in response to combined GLI1 and EGFR activation.
Figure 2.
Requirement of MEK/ERK function for transformation by synergistic EGFR and GLI1 activity. A, Western blot analysis of Egfr−/− cells reconstituted with wild-type human EGFR or EGFRY1068F and stimulated with different concentrations of EGF (0.5, 1, and 5 ng/mL) for 5 and 30 min, respectively. B, EGFR-deficient mouse embryonic fibroblasts treated with 10 ng/mL EGF (a). b–h, EGFR-deficient mouse embryonic fibroblasts either reconstituted with human wild-type EGFR (b, c, e, g, h) or with the GRB2-binding mutant EGFRY1068F (f) or expressing GLI1 only (d). Reconstitution with functional EGFR was validated by high-dosage EGF treatment (10 ng/mL) known to induce anchorage-independent growth (c). d, no transformation by GLI1 in the absence of EGFR. e, transformation by wild-type EGFR in combination with GLI1 activation. Transformation is lost upon reconstitution of EGFR−/− cells with EGFRY1068F (f)orby pharmacologic inhibition of MEK function (UO126 treatment; g) but not by inhibition of PI3K/AKT by wortmannin treatment (h). C, Western blot analysis of Egfr−/− mouse embryonic fibroblasts reconstituted with wild-type human EGFR and treated with EGF showing efficient inhibition of MEK/ERK (pERK) and PI3K/AKT (pAKT) activation by UO126 and wortmannin treatment, respectively. D, quantitative analysis of assays shown in B. Wort, wortmannin; Gef, gefitinib.
Integration of HH/GLI and EGFR-MEK/ERK signaling induces transformation of human keratinocytes
Ligand-independent activation of HH/GLI signaling in epidermal cells has been identified as the key etiologic factor in BCC, a very common nonmelanoma skin cancer (ref. 3, and references therein). We therefore asked whether integration of GLI and EGFR signaling may also synergize in the transformation of human epidermal cells. Because primary human keratinocytes undergo growth arrest in response to long-term expression of either GLI1 or GLI2, possibly as a result of oncogene-induced senescence,5 we used non-tumorigenic HaCaT keratinocytes, which (a) have retained the capacity to undergo relatively normal epidermal differentiation (24), (b) allow doxycycline-controlled GLI expression, and (c) have proved a valuable model to study GLI-regulated gene expression in the context of BCC (26).
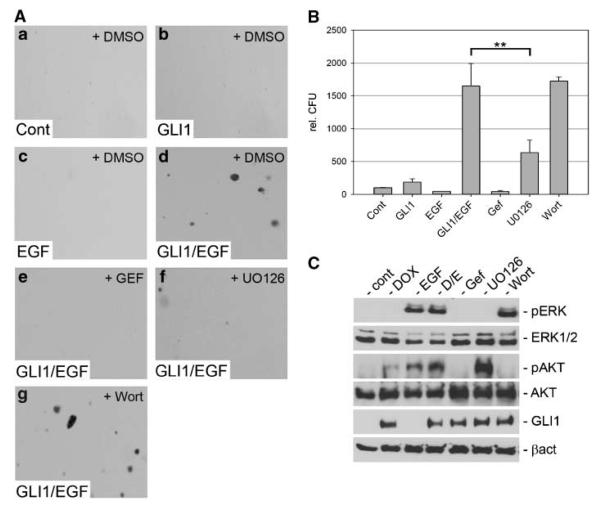
Consistent with our findings in rodent cells, the simultaneous activation of EGFR signaling and GLI1 synergistically induced transformation of human HaCaT keratinocytes (Fig. 3A, a–d and B). Next, we applied pharmacologic and genetic approaches to characterize the downstream pathways regulated by EGFR signaling. In agreement with our studies in mouse fibroblasts, pharmacologic inhibition of MEK/ERK function by administration of UO126 significantly reduced anchorage-independent growth in response to GLI1 and EGFR activation (Fig. 3A, f and B). By contrast, inhibition of PI3K/AKT by wortmannin had no effect on cell transformation (Fig. 3A, g and B). Also, interfering with SRC or JAK/STAT signaling did not affect transformation by EGFR and GLI1 (Supplementary Fig. S3). Similar results were found for GLI2 expressing keratinocytes (Supplementary Fig. S4).
Figure 3.
Anchorage-independent growth of HaCaT keratinocytes by combined activation of GLI1 and EGFR signaling. A, soft agar cultures of control HaCaT keratinocytes (no EGFR, no GLI1; a), keratinocytes expressing GLI1 only (b), EGF (10 ng/mL) treated keratinocytes (c), or HaCaT keratinocytes expressing GLI1 and treated with EGF (d). e-g, HaCaT cells with activated EGFR and GLI1 treated with gefitinib (Gef; e), UO126 (f), or wortmannin (g). B, quantification of assays shown in A. Statistical analysis was done by Student's t test. **, P < 0.005. Data represent the mean value of three independent experiments, each performed in triplicate. C, Western blot analysis of doxycycline-inducible GLI1 HaCaT keratinocytes showing specific activation and inhibition of MEK/ERK and PI3K/AKT function by treatment with the respective compounds. Samples Gef, UO126, and Wort were also treated with doxycycline and EGF. Cont, control; DOX, doxycycline; D/E, doxycycline/EGF treated; Wort, wortmannin.
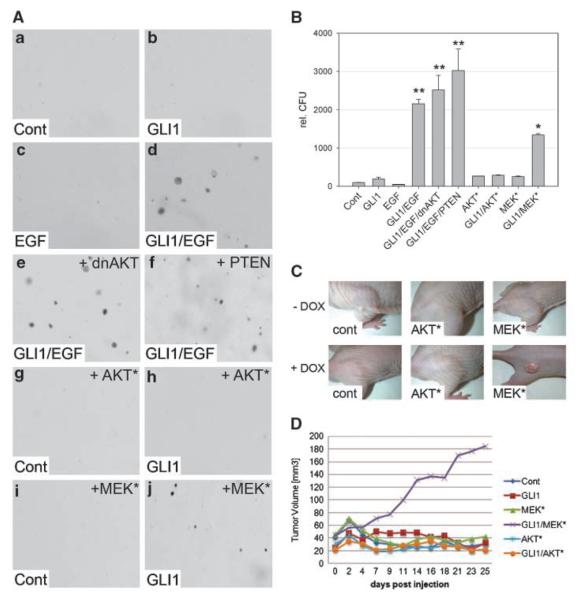
The differential effect of MEK/ERK and PI3K/AKT signaling on the oncogenic activity of GLI1 was further tested by coexpression of GLI1 and negative regulators of AKT and PI3K or dominant-active forms of AKT and MEK. As shown in Fig. 4, neither enforced expression of a dominant-negative AKT (dnAKT) nor of PTEN decreased anchorage-independent growth of keratinocytes induced by simultaneous activation of GLI1 and EGFR signaling. Expression of a dominant-active AKT (AKT*) failed to induce a transformed phenotype irrespective of GLI1 expression. However, expression of constitutively active MEK1 (MEK*) induced clonal soft agar growth but only in combination with GLI1. Efficient transgene expression of dnAKT, AKT*, PTEN, and MEK* is shown in Supplementary Fig. S5.
Figure 4.
MEK but not AKT synergizes with GLI1 in oncogenic transformation. A, HaCaT keratinocytes grown in soft agar either in the absence of both EGF and GLI1 expression (a), in the presence of GLI1 (b), in the presence of EGF (c), or in the presence of both GLI1 and EGF (d). Anchorage-independent growth is not affected by coexpression of dnAKT (e) or PTEN (f). HaCaT cells expressing constitutively active AKT (AKT*) in the absence (g) or presence of GLI1 (h). HaCaT cells expressing dominant-active MEK (MEK*) alone (i) or in combination with GLI1 (j). B, quantitative analysis of soft agar cultures shown in A. Statistical analysis was done by Student's t test. **, P < 0.001; *, P < 0.005. Data represent the mean value of three independent experiments, each performed in triplicate. C, nude mice (n = 8 for each cell line) injected with doxycycline-inducible GLI1 HaCaT keratinocytes expressing either AKT* or MEK*. GLI1 expression was induced by doxycycline in drinking water (+ DOX). D, quantitative analysis of tumor growth in nude mice shown in C.
We also addressed the tumorigenic potential of human HaCaT keratinocytes expressing AKT* or MEK* in combination with GLI1 in xenograft assays. In agreement with results from anchorage-independent growth assays, we found that only the combined expression of GLI1 and MEK* induced tumor growth in nude mice (Fig. 4C and D), further supporting the critical role of MEK/ERK signaling in the modulation of the oncogenic activity of GLI1 downstream of EGFR.
Transformation by integration of HH/GLI and EGFR signaling requires JUN activation
We have previously provided evidence that integration of HH/GLI and EGFR signaling involves convergence at the cis-regulatory region of GLI/EGF target genes (17), although the detailed molecular mechanisms remained unclear. We speculated that regulation of GLI/EGF target genes requires EGFR-dependent activation of transcription factors that cooperate with GLI in selective target gene expression and oncogenic transformation.
Numerous studies have linked EGFR signaling to the activation of the AP-1 transcription factor, a dimeric complex composed of members of the JUN, FOS, ATF, and MAF families (reviewed in ref. 39). As in silico promoter analysis revealed cooccurrence of GLI and consensus AP-1 binding sites in a number of direct GLI/EGF target genes, but not in EGF-independent GLI targets such as PTCH and BCL2 (Supplementary Table S1), we addressed whether EGFR signaling results in activation of the main AP-1 factor JUN, which may then cooperate with GLI to synergistically induce oncogenic transformation and GLI/EGF target gene expression.
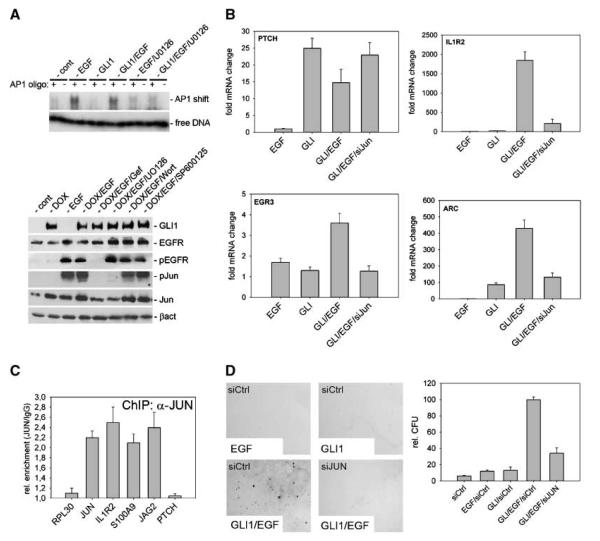
We first analyzed whether EGFR signaling induces activation of AP-1 DNA binding activity and activation of JUN, which is reflected by phosphorylation of the NH2 terminal activation domain at Ser63 and Ser73 mediated by JNK or ERK (40, 41). As shown in Fig. 5A, EGF treatment induced AP-1 DNA binding activity in a MEK/ERK-dependent manner and led to phosphorylation of JUN, which was abolished by inhibition of MEK/ERK but not of JNK or PI3K function. Levels of total EGFR or activated p-EGFR did not significantly change in response to GLI expression. We conclude that EGFR can activate JUN/AP-1 via MEK/ERK signaling.
Figure 5.
EGFR-mediated activation of JUN/AP-1 is essential for synergistic target gene activation and transformation of human keratinocytes by EGFR/GLI. A, top, gel shift assay demonstrating the activation of AP-1 DNA binding activity by EGF treatment of HaCaT keratinocytes. Inhibition of MEK/ERK function by U0126 treatment abolished EGF-induced activation of AP-1. As negative control, a mutated (− lanes) instead of wild-type (+ lanes) AP-1 binding oligonucleotide was used. Bottom, Western blot analysis showing that EGF treatment of HaCaT cells induces phosphorylation of JUN (pJun) via activation of EGFR/MEK/ERK signaling. EGFR was inhibited by gefitinib (Gef), MEK by UO126, AKT by wortmannin (Wort), and JNK by SP600125 treatment. B, siRNA knockdown of JUN expression selectively interferes with the synergistic activation of EGF-dependent GLI target genes, such as IL1R2, ARC, or EGR3. Synergistic activation of GLI/EGF targets was achieved by GLI1 expression and simultaneous EGF treatment (10 ng/mL; ref. 17). C, ChIP analysis showing JUN binding to AP-1 sites in the promoter region of EGF-dependent direct GLI target genes (IL1R2, JAG2, and S100A9). RPL30 and PTCH were used as negative and JUN bound to its own promoter as positive control (48). D, RNAi-mediated knockdown of JUN (shRNA-cJun) inhibits anchorage-independent growth induced by combined activation of EGFR and GLI1.
To address the involvement of JUN in selective GLI/EGF target gene regulation and transformation, we stably knocked down JUN expression by a retroviral shRNA approach. Consistent with a critical role of JUN in GLI/EGF target gene regulation, RNA interference (RNAi) knockdown of JUN inhibited the synergistic transcriptional response of the GLI/EGF target genes EGR3, IL1R2, and ARC to combined GLI/EGFR activation. By contrast, activation of EGF-independent GLI targets, such as PTCH and BCL2, was not affected by RNAi-mediated knockdown of JUN (Fig. 5B; data not shown). To corroborate the involvement of JUN in synergistic GLI/EGF target gene activation, we performed chromatin immunoprecipitation (ChIP) studies using EGF-treated GLI1-HaCaT cells expressing FLAG-tagged JUN. As shown in Fig. 5C, JUN was bound at the predicted AP-1 binding sites in the promoter regions of the EGF-dependent direct GLI target genes IL1R2, JAG2, and S100A9 (also see Table 1, supplementary material), suggesting that EGFR signaling induces JUN binding to promoters of selected direct GLI/EGF target genes.
Consistent with a critical role of JUN downstream of EGFR/MEK/ERK signaling, RNAi against JUN also dramatically reduced transformation in response to synergistic GLI1 (and also GLI2) and EGFR signaling (Fig. 5D and Supplementary Fig. S6). Note that growth of JUN knockdown cells in two-dimensional cultures was comparable with control cells, suggesting that the failure to grow in soft agar cultures is not simply due to a proliferation defect resulting from reduced JUN levels (Supplementary Fig. S7A and B). Also, we did not observe down-regulation of EGFR mRNA levels in response to JUN small interfering RNA (siRNA) expression (data not shown), suggesting that the effect of JUN inhibition on transformation by synergistic GLI and EGFR activity is not due to reduced EGFR expression (42). Based on these data, we propose that EGFR signaling synergizes with GLI activator forms (GLIact) in transformation and selective GLI target gene regulation via activation of MEK/ERK and JUN/AP-1 function.
Combined inhibition of EGFR/MEK/ERK/JUN and HH/GLI signaling efficiently reduces basal cell carcinoma cell growth and viability
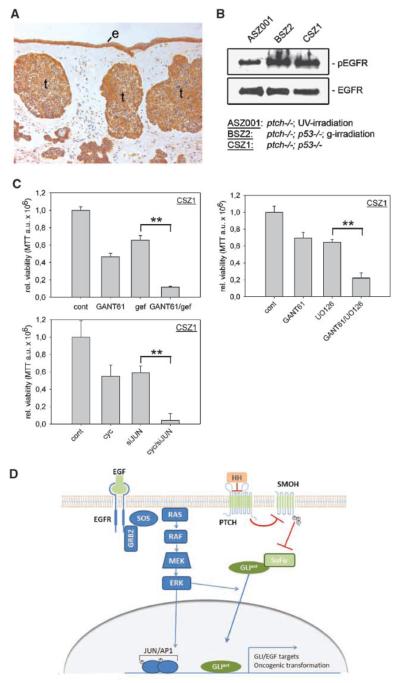
Although it is well established that EGFR signaling plays a critical function in skin cancer development (25), its implication in BCC development and growth has not yet been addressed in detail. By immunohistochemical analysis of a panel of 20 human BCC using a clinically approved diagnostic anti-EGFR antibody, we can show that all BCC tested expressed EGFR. Five BCCs showed EGFR expression at levels slightly lower than normal skin, eight tumors gave signals comparable with normal skin, and seven tumors showed higher EGFR levels than normal skin (Fig. 6A and Supplementary Fig. S8; Supplementary Table S2). This was partially confirmed by qPCR analysis of EGFR mRNA expression in BCC samples compared with normal human keratinocytes or N/TERT-1 keratinocytes (ref. 43; Supplementary Fig. S9A). Similarly, Ptch−/− (ASZ001) and Ptch−/−; p53−/− (BSZ2, CSZ1; ref. 44) mouse BCC cell lines express elevated levels of EGFR mRNA when compared with Ptch+/+ keratinocytes (Supplementary Fig. S9B). In agreement with the mRNA expression data, all BCC cell lines express significant levels of total and activated tyrosine–p-EGFR (Fig. 6B). In light of our findings on synergistic transformation by HH/GLI and EGFR/MEK/ERK/JUN signaling, we asked whether combined targeting of EGFR and HH/GLI may prove an efficient strategy to interfere with BCC growth.
Figure 6.
Combined inhibition of EGFR and HH/GLI efficiently reduces mouse BCC cell growth in vitro. A, immunohistochemical analysis of EGFR expression in a human nodular BCC. B, total EGFR and activated EGFR (p-EGFR) expression in mouse BCC cell lines; the respective genotype (and irradiation) of the cell lines is indicated below. C, single or combined treatment of CSZ1 cells with 5 μmol/L gefitinib (gef) and 10 μmol/L GANT61 (left), 10 μmol/L GANT61, and 10 μmol/L U0126 (right), or 5 μmol/L cyclopamine (cyc) and JUN shRNA (bottom left). Statistical analysis was done by Student's t test. **, P < 0.005. Data represent the mean value of three independent experiments. D, model of integration of EGFR and HH/GLI signaling in the regulation of GLI/EGF target genes and oncogenic transformation. In addition to activation of JUN/AP-1, stimulation of MEK/ERK may also promote the nuclear import of GLIact, thereby enhancing the synergistic effect (46). cyc, cyclopamine; gef, gefitinib; e, epidermis; t, tumor.
As shown in Fig. 6C (left), combined treatment of CSZ1 cells with the GLI antagonist GANT61 (45) and the EGFR inhibitor gefitinib reduced tumor cell proliferation and viability much more efficiently than the single treatments. Strong reduction of cell viability was also observed in response to the combined inhibition of GLI and MEK/ERK or GLI and JUN function (Fig. 6C, right and bottom left). Similar results were obtained for ASZ001 and BSZ2 cells, except that BSZ2 cells did not respond to cyclopamine/gefitinib treatment but were efficiently affected by the combination of GANT61 and gefitinib (Supplementary Fig. S9C and D).
Discussion
Molecular mechanisms of HH/GLI-EGFR integration in oncogenic transformation and GLI target gene regulation
In this study, we show that integration of EGFR and HH/GLI signaling, two prominent pathways with a well-documented etiologic role in a number of human cancers, synergistically promotes oncogenic transformation of various cell types of different species. Our data on EGFR and HH/GLI integration also support the notion that GLI proteins act as an information nexus to integrate multiple signal inputs from different pathways, which modulates their activity in normal and cancer cells (6). Previous studies have provided evidence that PI3K/AKT and MEK/ERK positively regulate the transcriptional activity of GLIact by interfering with the degradation of GLI proteins and/or by promoting the nuclear localization of GLIact (46, 47). However, it was unclear how HH/GLI and EGFR signals are integrated. In our studies, EGFR did not affect the levels of GLIact proteins (see Fig. 3C; data not shown) and the synergistic activation of GLI/EGF target genes by combined GLI1 and EGFR activation was fully pronounced even with a modified form of GLI1, which exclusively localizes to the nucleus (17). Also, PI3K/AKT function is dispensable for GLI/EGF target activation and oncogenic transformation by concurrent GLIact/EGFR signaling. We, therefore, propose that modulation of the oncogenic activity of GLIact by integration of EGFR signaling involves a mechanism that is different from those described previously. Our studies suggest a model wherein EGFR synergizes with GLIact in transformation and cancer development via activation of RAS/RAF/MEK/ERK/JUN signaling. Binding of ligand to EGFR induces activation of the RAS/REF/MEK/ERK cascade, which in turn stimulates JUN/AP-1 DNA binding. Active JUN/AP-1 cooperates with GLIact to induce GLI/EGF target gene expression and oncogenic transformation (Fig. 6D), a scenario that is supported by several findings. First, stimulation of EGFR signaling activates JUN/AP-1 activity in a MEK/ERK-dependent manner and JUN binds to promoters of GLI/EGF target genes. Second, interfering with MEK/ERK function not only prevents activation of JUN but also reduces transformation and GLI/EGF target gene expression in response to parallel EGFR and GLI activity (for reduction of target gene expression by MEK/ERK; see ref. 17). Third, RNAi against JUN dramatically reduces EGFR/GLIact-induced transformation and abrogates synergistic induction of GLI/EGF target genes by combined EGFR/GLI activation.
An intriguing detail of our studies of JUN activation by EGFR signaling is that GLI1 expression led to a moderate increase in the level of total JUN protein (Fig. 5A), consistent with the presence of a functional GLI binding site in the JUN promoter.6 As JUN is activated in response to EGFR signaling and also able to enhance its own expression via an autoregulatory feedback mechanism (48), concurrent activation of EGFR and GLIact is likely to potentiate the level of activated JUN, which may further enforce the synergism between JUN and GLIact in selective target gene activation and transformation.
HH/GLI and EGFR signaling in BCC
Our studies provide evidence that integration of EGFR and HH/GLI signaling may contribute to BCC growth. We found EGFR expression in all BCCs tested, although unlike many other carcinomas, BCCs do not show pronounced overexpression of EGFR compared with nonlesional epidermis (own data; ref. 49). Expression of activated EGFR was evident in Ptch-deficient mouse BCC cell lines. Also, human BCC and murine BCC cell lines express several EGFR ligands (data not shown), pointing to autocrine stimulation of EGFR signaling in BCC cells. Whether EGFR-induced MEK/ERK signaling cooperates with GLIact in BCC remains to be established. We have previously shown that activated ERK is not highly expressed throughout the entire tumor mass of BCC, although it can be readily detected in small subregions of the tumors, such as the peripheral palisading cells with high proliferative activity, as well as in infiltrating cells (18). Because we found that coexpression of activated MEK1 and GLI1 synergistically induces tumor development of human HaCaT keratinocytes in nude mice, it is tempting to speculate that the synergistic interaction of EGFR/MEK/ERK and GLIact in distinct subpopulations of BCC cells is associated with tumor growth and a more aggressive phenotype.
The molecular details of integration of EGFR and HH/GLI signaling presented in this study suggest that combined inhibition of both pathways constitutes a more efficient antitumor strategy than interfering with either signal alone. Indeed, our in vitro studies using mouse BCC cell lines showed that treatment with the EGFR inhibitor gefitinib, in combination with cyclopamine or GANT61, reduced cell proliferation and viability significantly more efficiently than administration of either compound alone. The same may be true of other malignancies with a documented role of both EGFR and HH/GLI signaling. Preliminary evidence supports this assumption as the cytotoxic and antiproliferative effect of gefitinib on metastatic prostate cancer lines has been shown to be augmented by cotreatment with low concentrations of cyclopamine (50). Whether synergistic integration of HH/GLI and EGFR signaling plays an etiologic role in the initiation and/or tumor growth in HH/GLI and EGFR-dependent cancers with high medical need, such as breast and pancreatic cancer, melanoma, and glioblastoma, is well possible and will have to be addressed in future studies using appropriate genetic and pharmacologic in vivo tumor models.
Supplementary Material
Acknowledgments
Grant support: Austrian Science Fund FWF grants P16518 and P20652 (F. Aberger), FWF grants P18421, P18782, SFB-23-B13, DK W1212, the EC program LSHC-CT-2006-037731 (Growthstop), GEN-AU program “Austromouse” (M. Sibilia), FWF grant SFB F028 (R. Moriggl), and the University of Salzburg focus area “Life Sciences and Health” (F. Aberger).
We thank Anna-Maria Frischauf and members of her group for helpful discussions on the project and for critical reading of the manuscript; Drs. Heidi Greulich, Graham Neill, William Sellers, Heidi Hahn, Allan Balmain, and Ervin Epstein for providing plasmid constructs and cell lines; Dr. Rune Toftgard and the Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, NIH for providing GANT61; and Martina Hammer for maintaining the nude mice colony and help in the xenograft assays.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
H. Schnidar and F. Aberger, unpublished observation.
A. Frischauf, personal communication.
References
- 1.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–31. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 2.Wetmore C. Sonic hedgehog in normal and neoplastic proliferation: insight gained from human tumors and animal models. Curr Opin Genet Dev. 2003;13:34–42. doi: 10.1016/s0959-437x(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 3.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–33. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 4.Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat Cell Biol. 2007;9:1005–9. doi: 10.1038/ncb435. [DOI] [PubMed] [Google Scholar]
- 5.Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer. 2006;42:437–45. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–47. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang J. Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle. 2006;5:2457–63. doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- 8.Callahan CA, Ofstad T, Horng L, et al. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 2004;18:2724–9. doi: 10.1101/gad.1221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varjosalo M, Bjorklund M, Cheng F, et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133:537–48. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 10.Lauth M, Toftgard R. Non-canonical activation of GLI transcription factors: implications for targeted anti-cancer therapy. Cell Cycle. 2007;6:2458–63. doi: 10.4161/cc.6.20.4808. [DOI] [PubMed] [Google Scholar]
- 11.Riobo NA, Lu K, Emerson CP., Jr Hedgehog signal transduction: signal integration and cross talk in development and cancer. Cell Cycle. 2006;5:1612–5. doi: 10.4161/cc.5.15.3130. [DOI] [PubMed] [Google Scholar]
- 12.Fogarty MP, Emmenegger BA, Grasfeder LL, Oliver TG, Wechsler-Reya RJ. Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells. Proc Natl Acad Sci U S A. 2007;104:2973–8. doi: 10.1073/pnas.0605770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palma V, Lim DA, Dahmane N, et al. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132:335–44. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palma V, Ruiz i Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–45. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- 15.Bigelow RL, Jen EY, Delehedde M, Chari NS, McDonnell TJ. Sonic hedgehog induces epidermal growth factor dependent matrix infiltration in HaCaT keratinocytes. J Invest Dermatol. 2005;124:457–65. doi: 10.1111/j.0022-202X.2004.23590.x. [DOI] [PubMed] [Google Scholar]
- 16.Mimeault M, Johansson SL, Vankatraman G, et al. Combined targeting of epidermal growth factor receptor and hedgehog signaling by gefitinib and cyclopamine cooperatively improves the cytotoxic effects of docetaxel on metastatic prostate cancer cells. Mol Cancer Ther. 2007;6:967–78. doi: 10.1158/1535-7163.MCT-06-0648. [DOI] [PubMed] [Google Scholar]
- 17.Kasper M, Schnidar H, Neill GW, et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol Cell Biol. 2006;26:6283–98. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neill GW, Harrison WJ, Ikram MS, et al. GLI1 repression of ERK activity correlates with colony formation and impaired migration in human epidermal keratinocytes. Carcinogenesis. 2008;29:738–46. doi: 10.1093/carcin/bgn037. [DOI] [PubMed] [Google Scholar]
- 19.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 20.Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, Holcmann M. The epidermal growth factor receptor: from development to tumorigenesis. Differentiation. 2007;75:770–87. doi: 10.1111/j.1432-0436.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- 21.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–25. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 22.Hackel PO, Zwick E, Prenzel N, Ullrich A. Epidermal growth factor receptors: critical mediators of multiple receptor pathways. Curr Opin Cell Biol. 1999;11:184–9. doi: 10.1016/s0955-0674(99)80024-6. [DOI] [PubMed] [Google Scholar]
- 23.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 24.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–71. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sibilia M, Fleischmann A, Behrens A, et al. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell. 2000;102:211–20. doi: 10.1016/s0092-8674(00)00026-x. [DOI] [PubMed] [Google Scholar]
- 26.Regl G, Kasper M, Schnidar H, et al. The zinc-finger transcription factor GLI2 antagonizes contact inhibition and differentiation of human epidermal cells. Oncogene. 2004;23:1263–74. doi: 10.1038/sj.onc.1207240. [DOI] [PubMed] [Google Scholar]
- 27.Kasper M, Regl G, Eichberger T, Frischauf AM, Aberger F. Efficient manipulation of hedgehog/GLI signaling using retroviral expression systems. Methods Mol Biol. 2007;397:67–78. doi: 10.1007/978-1-59745-516-9_6. [DOI] [PubMed] [Google Scholar]
- 28.Rubinson DA, Dillon CP, Kwiatkowski AV, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–6. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 29.Ramaswamy S, Nakamura N, Vazquez F, et al. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1999;96:2110–5. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kralova J, Liss AS, Bargmann W, Bose HR., Jr AP-1 factors play an important role in transformation induced by the v-rel oncogene. Mol Cell Biol. 1998;18:2997–3009. doi: 10.1128/mcb.18.5.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornfeld JW, Grebien F, Kerenyi MA, et al. The different functions of Stat5 and chromatin alteration through Stat5 proteins. Front Biosci. 2008;13:6237–54. doi: 10.2741/3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regl G, Neill GW, Eichberger T, et al. Human GLI2 and GLI1 are part of a positive feedback mechanism in basal cell carcinoma. Oncogene. 2002;21:5529–39. doi: 10.1038/sj.onc.1205748. [DOI] [PubMed] [Google Scholar]
- 34.Ruppert JM, Vogelstein B, Kinzler KW. The zinc finger protein GLI transforms primary cells in cooperation with adenovirus E1A. Mol Cell Biol. 1991;11:1724–8. doi: 10.1128/mcb.11.3.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 36.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 37.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 38.Batzer AG, Rotin D, Urena JM, Skolnik EY, Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol Cell Biol. 1994;14:5192–201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–68. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 40.Leppa S, Saffrich R, Ansorge W, Bohmann D. Differential regulation of c-Jun by ERK and JNK during PC12 cell differentiation. EMBO J. 1998;17:4404–13. doi: 10.1093/emboj/17.15.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pulverer BJ, Kyriakis JM, Avruch J, Nikolakaki E, Woodgett JR. Phosphorylation of c-jun mediated by MAP kinases. Nature. 1991;353:670–4. doi: 10.1038/353670a0. [DOI] [PubMed] [Google Scholar]
- 42.Zenz R, Scheuch H, Martin P, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4:879–89. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 43.Dickson MA, Hahn WC, Ino Y, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–47. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.So PL, Langston AW, Daniallinia N, et al. Long-term establishment, characterization and manipulation of cell lines from mouse basal cell carcinoma tumors. Exp Dermatol. 2006;15:742–50. doi: 10.1111/j.1600-0625.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 45.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A. 2007;104:8455–60. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stecca B, Mas C, Clement V, et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci U S A. 2007;104:5895–900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2006;103:4505–10. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angel P, Hattori K, Smeal T, Karin M. The jun protooncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–85. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 49.Rittie L, Kansra S, Stoll SW, et al. Differential ErbB1 signaling in squamous cell versus basal cell carcinoma of the skin. Am J Pathol. 2007;170:2089–99. doi: 10.2353/ajpath.2007.060537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mimeault M, Moore E, Moniaux N, et al. Cytotoxic effects induced by a combination of cyclopamine and gefitinib, the selective hedgehog and epidermal growth factor receptor signaling inhibitors, in prostate cancer cells. Int J Cancer. 2006;118:1022–31. doi: 10.1002/ijc.21440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.