Abstract
The balance between hematopoietic progenitor commitment and self-renewal versus differentiation is controlled by various transcriptional regulators cooperating with cytokine receptors. Disruption of this balance is increasingly recognized as important in the development of leukemia,by causing enhanced renewal and differentiation arrest. We studied regulation of renewal versus differentiation in primary murine erythroid progenitors that require cooperation of erythropoietin receptor (EpoR),the receptor tyrosine kinase c-Kit and a transcriptional regulator (glucocorticoid receptor (GR)) for sustained renewal. However, mice defective for GR- (GRdim/dim), EpoR- (EpoRH) or STAT5ab function (Stat5ab−/−) show no severe erythropoiesis defects in vivo. Using primary erythroblast cultures from these mutants, we present genetic evidence that functional GR, EpoR, and Stat5 are essential for erythroblast renewal in vitro. Cells from GRdim/dim, EpoRH, and Stat5ab−/− mice showed enhanced differentiation instead of renewal, causing accumulation of mature cells and gradual proliferation arrest. Stat5ab was additionally required for Epo-induced terminal differentiation: differentiating Stat5ab−/− erythroblasts underwent apoptosis instead of erythrocyte maturation, due to absent induction of the antiapoptotic protein Bcl-XL. This defect could be fully rescued by exogenous Bcl-XL. These data suggest that signaling molecules driving leukemic proliferation may also be essential for prolonged self-renewal of normal erythroid progenitors.
Keywords: erythroid progenitor self-renewal, terminal erythropoiesis, Stat5, glucocorticoid receptor, erythropoietin receptor, Bcl-XL
Introduction
Pluripotent hematopoietic stem cells (HSCs) have continuously to decide between commitment, self-renewal and terminal differentiation to maintain proper numbers of all mature blood cell types. Originally, HSCs were thought to represent the only cells with self-renewal ability, and many human leukemias indeed involve mutated HSCs (Shet et al., 2002). Recently, however, multi- or unipotent progenitors were also found to undergo normal or leukemic renewal (Blau et al., 2001; Pardal et al., 2003). This renewal is driven by normal or mutated cytokine receptors and receptor tyrosine kinases (RTKs), which cooperate with transcriptional and chromatin regulators as shown in culture (Schulte et al., 2002; Carotta et al., 2004; Von Lindern et al., 2004) and human leukemia (Stirewalt and Radich, 2003; Tenen, 2003).
Erythropoiesis requires tight control of erythrocyte numbers but red cell production must increase rapidlyin response to blood loss, hypoxia, or anemia. This flexibility is effected by tight regulation of erythroid progenitor renewal versus maturation via an interplay of cytokine- and nuclear receptors essential for stress erythropoiesis: Stem cell-factor (SCF) receptor (c-Kit) and erythropoietin (Epo) receptor cooperate with a nuclear hormone receptor (glucocorticoid receptor (GR)) to induce prolonged expansion of primary avian, murine, and human erythroid progenitors (Bauer et al., 1999; von Lindern et al., 1999; Carotta et al., 2004). These cells resemble proerythroblasts with properties of burst- and colony forming units-erythroid (BFU-E, CFU-Es) (Sawada et al., 1990). In response to differentiation factors, they undergo in vivo-like erythroid maturation (Dolznig et al., 1995, 2001; von Lindern et al., 2001). Owing to their close resemblance to in vivo progenitors, primary murine erythroblasts are particularly useful to analyse the balance between renewal and differentiation and its disturbance in leukemia at the molecular level, and are superior to established erythroid cell lines for such studies (Silva et al., 1996, 1999).
Culture models from avian erythroid and multipotent progenitors have already proven their value in analysing leukemic renewal (Beug et al., 1995; Schulte et al., 2002). Importantly, mutated or amplified RTKs (c-Kit; Flt-3) (Stirewalt and Radich, 2003) cooperating with transcriptional regulators (including mutated nuclear receptors) are increasingly implicated in human leukemias (He et al., 1998; Tenen, 2003).
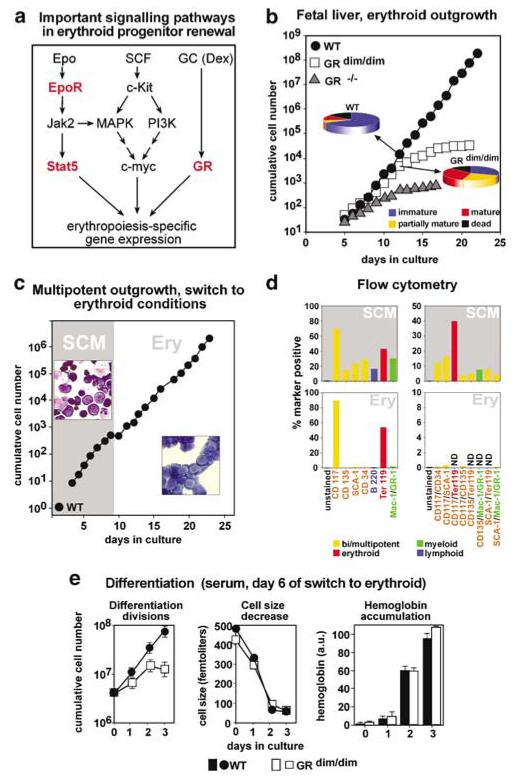
Analysis of signaling pathways for erythroid progenitor renewal (Figure 1a) revealed that EpoR activates Stat5, probably cooperating with the GR (Doppler et al., 2000), whereas c-Kit activates the PI3K-pathway. Whereas PI3K-signaling was indispensable for erythroblast renewal, the requirement for Stat5 activation was only studied in differentiating myeloid progenitors, where it was essential for apoptosis protection via the antiapoptotic protein Bcl-XL (Kieslinger et al., 2000). Interestingly, erythroid maturation also involves protection from apoptosis by Epo-dependent upregulation of Bcl-XL (Motoyama et al., 1999; Dolznig et al., 2002).
Figure 1.
Pre-expansion of proliferation-defective GRdim/dim-erythroblasts as multipotent cells yields erythroblasts defective for renewal but not for differentiation. (a) Signal transduction pathways important in erythropoiesis. Signaling molecules ablated or mutated in cells from genetically modified mice used in this study are indicated in red. (b) E12.5 fetal liver cells from GRdim/dim (white squares), GR−/− (gray triangles) or wild-type mice (WT, black circles) were cultivated in ‘erythroid proliferation medium’ and cumulative cell numbers determined daily. At day 12, aliquots were subjected to cytocentrifugation, stained and quantified for cells of increasing maturity (pie diagrams, blue, yellow, red) and apoptotic cells (black) as described (Kolbus et al., 2002). Cells (≥300) were counted per slide and mean values ±s.d. calculated from at least three independent determinations. Percentages of immature/partially mature/mature/dead cells were 70±6/5±1/9±2/16±3 for wild-type cells and 31±4/27±5/26±4/16±3 for GRdim/dim erythroblasts. (c) Cells from wild-type mouse fetal livers were pre-expanded in ‘stem cell mix’ medium (gray area, ‘SCM’), switched to erythroid conditions (day 9, white area, ‘Ery’) and cumulative cell numbers determined. Insets, cytospins from multipotent progenitors (top left) and cells switched to the erythroid lineage (day17, bottom right). (d) Multipotent cells (top panels, day9) and erythroblasts derived thereof (bottom panels, day 17, see panel (c) were subjected to flow cytometry. Percentages of cells single- (left) or double positive (right) for immature (green bars), erythroid (red bars), myeloid (green bars), and lymphoid surface markers (blue bars) detected by respective antibodies are shown. (e) Fetal liver cells from GRdim/dim- (white symbols) and wild-type (WT) mice (black symbols) were expanded in SCM, switched to the erythroid lineage (day 0, not shown) and analysed for differentiation kinetics in serum-containing differentiation medium. Cumulative cell numbers (left panel; mean values ±s.d., n=3), cell size (middle panel) and hemoglobin content (right panel) were determined at the times indicated.
Several transgenic mouse studies addressed how ablation of the above signal transducers would interfere with erythropoiesis in vivo. Deletion of the GR (GR−/−) or a knocked-in mutation leading to a dimerization-defective protein (GRdim/dim) prevented stress erythropoiesis (Bauer et al., 1999). Loss of c-Kit affected multiple progenitor types, including erythroid ones, leading to severe anemia (Broudy, 1997). Mice lacking EpoR or Jak2 are deficient for definitive erythropoiesis (Wu et al., 1995; Neubauer et al., 1998; Parganas et al., 1998). In contrast, knock-in mice expressing cytoplasmically truncated EpoR variants – binding Jak2 but lacking most (EpoRH) or all (EpoRHM) of signal transducer docking sites (Zang et al., 2001) – show largely normal erythropoiesis. Finally, Stat5ab−/− mice have no obvious defects in steady-state erythropoiesis except mild anemia (Socolovsky et al., 1999). Thus, the above-mentioned mouse models do not reflect the strict requirement of EpoR/Stat5- and GR-signaling for progenitor renewal in culture.
We therefore sought to obtain genetic evidence for essential, cell-autonomous roles of GR-signaling and EpoR-signaling (via Stat5) for renewal and apoptosis protection in cultured erythroid progenitors from genetically modified mice, as multiple, compensatory mechanisms might obscure defects in the respective erythroid progenitors in vivo. We show that primary erythroblasts from GR−/−, GRdim/dim, EpoRH, EpoRHM, and Stat5ab−/− mouse embryos – cultured ex vivo under fully defined conditions – indeed display distinct proliferation defects in vitro. These defects were due to increased terminal differentiation at the expense of renewal rather than to enhanced apoptosis. While differentiation of GR−/−, GRdim/dim, and EpoRH erythroblasts proceeded normally, Bcl-XL induction and survival of Stat5ab−/− cells was regulated in a complex fashion, involving Epo, serum factors, and compensatory upregulation of other Stat-family members.
Results
Impaired expansion of GR-defective erythroblasts
Erythroblasts from mice expressing a dimerization-defective GR (GRdim/dim; Bauer et al., 1999) showed impaired stress erythropoiesis in vivo and could not be expanded in culture (Reichardt et al., 1998). While wild-type cells proliferated for >20 days, GRdim/dim erythroblasts stopped after 5–10 days and GR−/− cells could hardly be expanded at all (Figure 1b). At day 12, cytospin analysis of GRdim/dim cultures showed strongly increased numbers of mature cells and fewer immature cells than wild-type cultures. Dead cells were infrequent in both cultures. Therefore, the proliferation defect of GRdim/dim erythroblasts in vitro was due to increased differentiation at the expense of renewal. A similar, even more rapid increase in mature cells occurred in GR−/− erythroblasts (not shown).
Pre-expansion of proliferation-defective mutant erythroblasts as multipotent progenitors
Owing to their enhanced rate of spontaneous differentiation, GR-deficient erythroblasts could not be analysed for potential maturation defects. We thus tried to obtain homogenous, immature erythroblasts by pre-expansion of GR-defective multipotent progenitors. We expected that these cells would lack erythroid-specific defects and thus should be able to undergo commitment to the erythroid lineage in erythroid-specific proliferation medium (Schulte et al., 2002). In establishing this procedure, wild-type fetal liver-derived cells could be expanded more than 106-fold (not shown) in serum-free ‘stem cell medium’ (SCM). Cytospin analyses after ~ 500-fold expansion (8 days) revealed >50% of cells with immature morphology, whereas the remainder consisted of maturing erythroblasts, monocytes/macrophages, neutrophils, eosinophils, and mast cells (Figure 1c). Flow cytometry of such cell populations revealed >20% of cells with markers typical of multipotent, immature progenitors (CD117/c-Kit, CD135/Flk2/Flt3, CD34, Sca-1), confirmed by coexpression of several immature markers plus the absence of lineage-specific markers on the same cells (Figure 1d). As expected, we also detected committed cells expressing erythroid (Ter119), myeloid (CD11b/Mac-1, GR-1), or lymphoid lineage markers (CD45R/B220).
From these wild-type cells, pure erythroid progenitors were generated by transfer into media containing SCF/Epo/Dex. After continuous proliferation for 5–7 days, populations of >90% pure erythroblasts were obtained as verified by cytospins (Figure 1c) and flow cytometry (CD117/c-Kithigh, Ter119low Mac-1/GR-1low, negative for CD135, CD34, and Sca-1; Figure 1d, lower panels). These erythroblasts could be expanded for ~2 weeks and induced to terminally differentiate at any time throughout their life span.
Applying this procedure to GRdim/dim fetal liver cells yielded similarly homogenous erythroid cultures, suitable for detailed analysis of differentiation kinetics. Purified GRdim/dim erythroblasts still underwent strongly increased spontaneous differentiation at the expense of renewal, showing the expected defect (as in Figure 1b, data not shown). Nevertheless, pre-expansion as multipotent cells yielded enough GRdim/dim erythroblasts with proliferation rate and size identical to wild-type cells to allow analysis of differentiation parameters. During terminal maturation, GRdim/dim erythroblasts showed normal kinetics of size decrease and hemoglobin accumulation as compared to that of wild-type cells, but displayed a reduced proliferation capacity (Figure 1e). Similar results were obtained in serum-containing- and serum-free differentiation media (see below and data not shown).
EpoR intracellular domain: required for renewal but not for differentiation
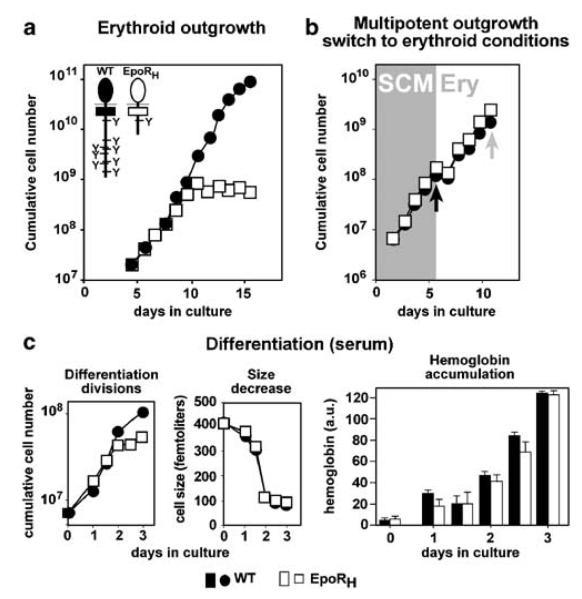
To address whether the EpoR cytoplasmic domain was required for erythroblast renewal, cells from mice lacking most (EpoRH) or all (EpoRHM) (Zang et al., 2001) of this domain (schemes in Figure 2a) were analysed. EpoRH erythroblasts grown in ‘erythroid proliferation medium’ exhibited gradual proliferation arrest after 7–8 days (Figure 2a), caused by massively enhanced spontaneous differentiation under renewal conditions (data not shown). Again, pre-expansion of EpoRH fetal liver cells as multipotent progenitors and switch to erythroid conditions (Figure 2b, black arrow, day 6) yielded homogenous mass cultures of mutant erythroblasts for differentiation analysis (Figure 2b, gray arrow, day 12). In both serum-containing- and serum-free media, EpoRH cells showed the same maturation kinetics as that of wild-type cells, as judged by the number of differentiation divisions, size decrease, and hemoglobin accumulation (Figure 2c). Similar results were obtained with erythroblasts from EpoRHM mice. Thus, cytoplasmic docking sites for signal transducers were required for sustained erythroblast renewal but not for terminal differentiation, in line with EpoRH/EpoRHM mice showing normal steady-state erythropoiesis.
Figure 2.
Erythroblasts expressing cytoplasmically truncated EpoRH: impaired renewal but normal differentiation. (a) Fetal liver-derived cells from EpoRH- (white symbols) and WT-mice (black) were cultivated in serum-free ‘erythroid proliferation medium’ and cumulative cell numbers determined as in Figure 1. Inset, scheme of mutated EpoR's used. (b) Cells as in (a) were pre-expanded in SCM (gray area, ‘SCM’) and switched to erythroid proliferation medium after 6 days (black arrow, white area, ‘Ery’). (c) After expansion for another 6 days in (gray arrow in (b) cells were analysed for differentiation parameters as in Figure 1 (three separate experiments, representative data shown).
Stat5ab: essential for erythroid progenitor renewal and terminal differentiation?
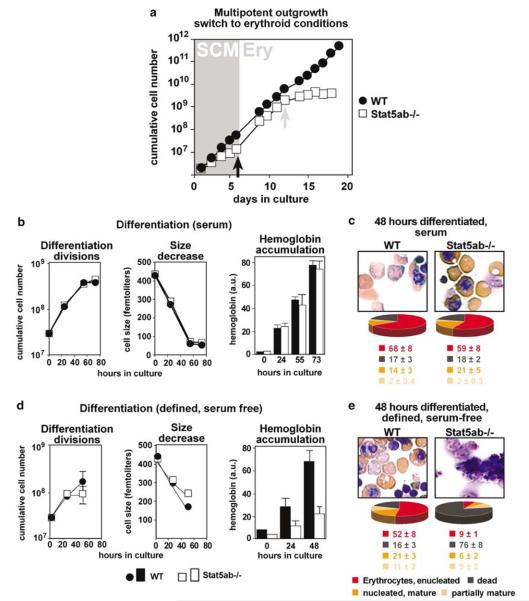
Activated Stat5 is a major downstream signal transducer of EpoR. We therefore addressed its potential role during erythroid progenitor renewal and differentiation, using fetal liver-derived erythroblasts of Stat5ab−/− mice (Teglund et al., 1998). As these cells also showed a marked proliferation defect in ‘erythroid proliferation medium’, they were pre-expanded as multipotent progenitors. The cultures proliferated slower than respective wild-type cells (Figure 3a; SCM), similar to avian myeloblasts expressing a dominant-negative Stat5ΔC mutant (Kieslinger et al., 2000). When switched to erythroid conditions (Figure 3a, black arrow, day 6), Stat5ab−/− cultures ceased to proliferate after about 8 days (=‘day 14’ in Figure 3a), containing fewer immature cells whereas numbers of partially mature/mature cells were massively enhanced.
Figure 3.
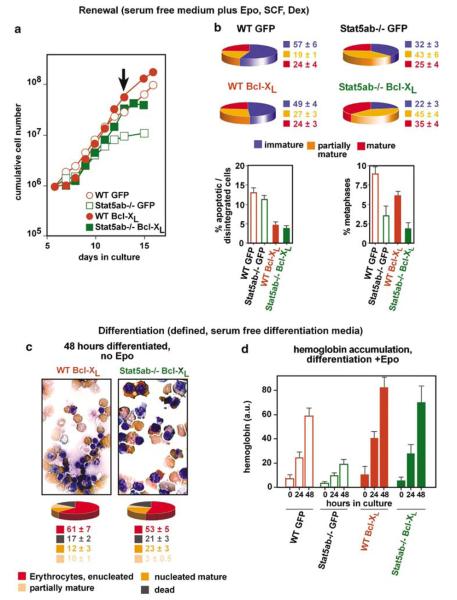
Erythroblasts from Stat5ab−/− mice: defects in renewal and terminal differentiation. (a) Fetal liver cells from Stat5ab−/− (white symbols) and wild-type mice (black) were expanded in SCM (gray area, ‘SCM’), switched to erythroid proliferation medium (white area, ‘Ery’), cultivated for another 6 days, and cumulative cell numbers were determined. (b, d) Stat5ab−/− and wild-type erythroid progenitors (day 12, gray arrow) analysed for differentiation parameters in both serum-containing (b) and defined, serum-free differentiation medium (d) as described in Figures 1 and 2 (error bars: s.d.s, n = 3). (c, e) Top panels: Cytospins from (b) and (d) subjected to neutral benzidine/histological staining after 48 h. Bottom: quantitation with respect to partially mature/mature/enucleated erythrocytes (percentages ±s.d.s, n = 3).
When seeded into standard, serum-containing differentiation medium 6 days after switching to erythroid conditions (Figure 3a, gray arrow, day 12), both Stat5ab−/− and wild-type cells exhibited similar differentiation kinetics (Figure 3b), with >75% of enucleated and nucleus-containing erythrocytes and o20% of dead/disintegrated cells in cytospins (Figure 3c). Unlike wild-type cells (Figure 3d and e, left panels) or GRdim/dim and EpoRH cells, Stat5ab−/− erythroblasts in defined, serum-free differentiation medium died within 48 h (Figure 3d and e). They showed proliferation arrest after 24 h, aberrant size decrease, massively reduced hemoglobin accumulation (Figure 3d) and >75% of dead cells in cytospins (Figures 3e, right panels). These results suggest that functional Stat5 is required for both erythroblast renewal and differentiation under defined, serum-free conditions.
STAT5-dependent regulation of bcl-XL transcription: Lineage-specific differences during differentiation and renewal
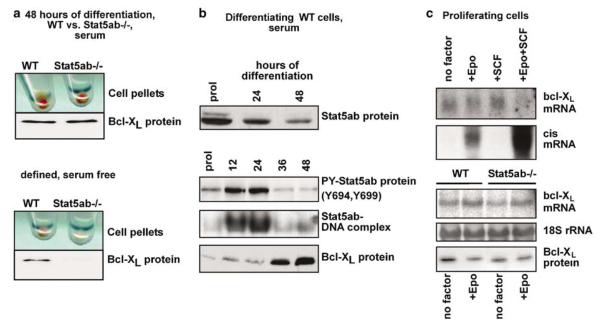
Earlier work had demonstrated an essential role of the antiapoptotic protein Bcl-XL in protecting differentiating erythroid and myeloid progenitors from apoptosis (Kieslinger et al., 2000; Dolznig et al., 2002). In Epo-responsive Friend erythroleukemia cells (HCD57) and primary myeloid cells dependent on IL-3/GM-CSF, apoptosis protection correlated with direct, STAT5 mediated transcriptional activation of bcl-XL (Socolovsky et al., 1999; Kieslinger et al., 2000). We therefore tested whether bcl-XL was a direct target gene of Stat5 in proliferating or differentiating erythroblasts and whether ablation of Stat5ab would prevent upregulation of Bcl-XL and survival in differentiating Stat5ab−/− cells. Surprisingly, Stat5ab−/− cells differentiating in serum-containing medium accumulated hemoglobin and upregulated Bcl-XL like wild-type cells (Figure 4a, top), but failed to do so in fully defined, serum-free medium (Figure 4a bottom, see also Figure 3c and e). These unexpected findings prompted us to analyse the relationship between Stat5 and Bcl-XL in more detail. In wild-type erythroblasts, total Stat5ab protein levels declined steadily during maturation (Figure 4b, top panel). Tyrosine phosphorylation and DNA-binding activity were transiently elevated during the first 12–24 h but declined to low levels after 36 h. In contrast, Bcl-XL protein was not upregulated until 36 h after differentiation induction, when Stat5 activity had already declined (Figure 4b, bottom panel), suggesting that Stat5 would not directly activate transcription of bcl-XL. To address this, proliferating wild-type erythroblasts were factor-deprived and restimulated with Epo, SCF or both. None of these cytokines induced bcl-XL mRNA, whereas Epo and Epo + SCF clearly induced cis mRNA, a well-defined Stat5 target gene (Verdier et al., 1998). Furthermore, proliferating, immature wild type as well as Stat5ab−/− erythroblasts expressed similar levels of bcl-XL mRNA after factor withdrawal and Epo restimulation (Figure 4c). Since bcl-XL was directly induced by Stat5 in IL-3-stimulated wild type but not Stat5ab−/− myeloid cells (Kieslinger et al., 2000), bcl-XL transcription in erythroid progenitors is probably coregulated by Stat5 plus other serum factors.
Figure 4.
Bcl-XL upregulation in Stat5ab−/− erythroblasts is dependent on serum and not subject to direct transcriptional activation by Epo-driven Stat5 activation. (a) Primary Stat5ab−/− and wild-type erythroblasts were induced to differentiate in serum-containing (upper panels) or defined, serum-free differentiation medium (lower). Respective cell pellets (viable cells only) show hemoglobinization (red color, top of panels) and upregulation of Ter119 expression (data not shown, (Dolznig et al., 2001)). Lysates were analyzed for Bcl-XL protein (bottom) (b) Primary wild-type erythroblasts were induced to differentiate in serum-containing differentiation medium and analyzed for total Stat5ab protein, tyrosine-phosphorylated Stat5ab, and Bcl-XL at the times indicated. Stat5ab-DNA-binding activity was determined in EMSAs using extracts from the same preparations. (c) Upper panels: proliferating erythroblasts kept for 4 h without cytokines (no factor) and restimulated for 1 h with Epo (+ Epo), SCF (+ SCF) or both (+ Epo + SCF) were analysed by Northern blotting for bcl-XL mRNA and – as positive control – cis mRNA expression. Lower panels: primary mouse erythroblasts from wild-type (WT) or Stat5ab−/− fetal livers factor-depleted for 4 h, restimulated with Epo for 3 h and analysed for bcl-XL-mRNA (loading control: 18S ribosomal RNA) and protein expression.
Exogenous Bcl-XL rescues the differentiation but not the proliferation defect of Stat5ab−/− erythroblasts
Exogenous overexpression of Bcl-XL enables terminal differentiation of primaryerythroblasts in the absence of Epo (Dolznig et al., 2002). To test whether Bcl-XL would also rescue the differentiation defect of Stat5ab−/− erythroblasts, cells were infected with a GFP-Bcl-XL retroviral construct, expanded as multipotent progenitors and switched to the erythroid lineage. More than 90% of the infected cells stably expressed Bcl-XL as shown by flow cytometry for GFP expression (data not shown). To analyse renewal, cells were cultivated in serum-free medium plus renewal factors (Epo/SCF/Dex), using empty vector-expressing cells as controls. Bcl-XL-transduced and control Stat5ab−/− erythroblasts both showed enhanced differentiation at the expense of renewal (Figure 5b, top), leading to premature proliferation arrest, as revealed by reduced or absent increase in cell numbers (Figure 5a) and a strong reduction of metaphases in cytospins (Figure 5b, bottom right). Exogenous Bcl-XL only slightly delayed growth arrest of renewing WT and Stat5 deficient cells, most likely due to enhanced survival of maturing cells (Figure 5b, bottom left). In contrast, wild-type cells proliferated exponentially (Figure 5a) as immature cells (Figure 5b), regardless of the presence or absence of exogenous Bcl-XL.
Figure 5.
Retrovirally expressed Bcl-XL rescues the differentiation but not the renewal defect of Stat5−/− erythroblasts. Freshly isolated wild type or Stat5ab−/− fetal liver cells were infected with MSCV-IRES-GFP or MSCV-Bcl-XL-IRES-GFP retroviral vectors (a) Cumulative cell numbers determined for Bcl-XL expressing wild-type or Stat5ab−/− erythroblasts expanded in ‘erythroid proliferation medium’. (b) Aliquots of proliferating cultures in (a) were subjected to cytospin analysis (arrow in a) for mature, partially mature, and immature cells as described for Figure 1 (top panels, viable cells only). The same cytospins were analysed for proportions of apoptotic/disintegrated cells and cells in mitosis, respectively (bottom) (c) The cell types shown in (a) and (b) were differentiated for 48 h in defined, serum-free medium lacking Epo. Cytospins (top) were quantified as in Figures 1 and 3 (percentages ±s.d.s, n = 3). (d) Aliquots from wild-type or Stat5ab−/− erythroblasts expressing empty vector (WT GFP, Stat5ab−/−) or exogenous Bcl-X (WT Bcl-XL, Stat5ab−/− Bcl-XL) were differentiated in fully defined medium plus Epo and analysed for hemoglobin content at the times indicated (error bars: s.d.s, n = 3).
We then analysed whether exogenous Bcl-XL could rescue differentiation of Stat5ab−/− cells in defined, serum-free media. Even in the absence of Epo, Bcl-XL-transduced Stat5ab−/−, and wild-type cells effectively underwent terminal differentiation in serum-free medium, as judged by histology and quantitative evaluation of cytospins (Figure 5c). Bcl-XL also rescued the defective hemoglobin accumulation of Stat5ab−/− cells in medium plus Epo/Insulin (Figure 5d). Together with the results described above, this demonstrated that the inability of Stat5ab−/− cells to upregulate bcl-XL during differentiation can be overcome by Stat5ab-independent pathways, induced by serum factors in cooperation with Epo.
Stat5ab−/− cells upregulate activated Stat3 and Stat1: compensation of Stat5ab−/− cell defects?
Asynchrony of Stat5ab activation versus Bcl-XL expression, failure of Epo to increase basal levels of Bcl-XL transcription in proliferating cells and serum-factor dependence of bcl-XL upregulation in differentiating Stat5ab−/− cells all pointed to an involvement of Stat5ab-independent pathways. Furthermore, the relatively mild renewal defect in proliferating Stat5ab−/− cells was difficult to reconcile with the expected, complete defect in erythroblast outgrowth from EpoR−/− or Jak2−/− fetal livers (FG et al, unpublished).
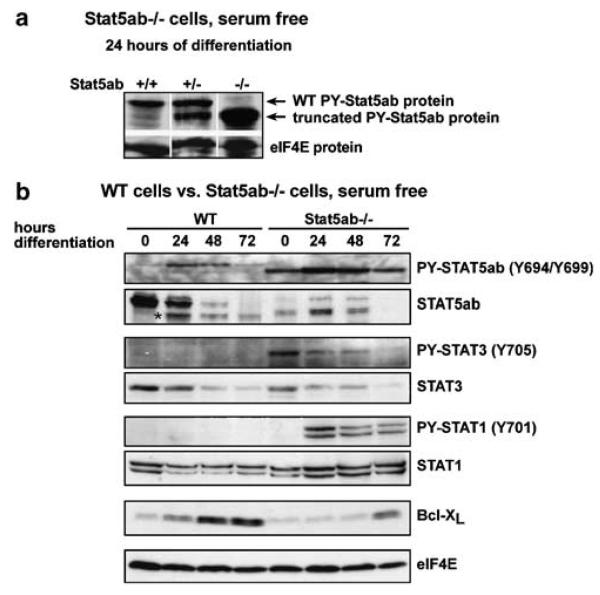
We therefore tested whether Stat5ab−/− cells might have residual Stat5ab activity and/or show enhanced expression/activation of other Stat family members. Phospho-Stat5-specific antibodies indeed detected high levels of a smaller, phosphorylated Stat5 protein in Stat5ab−/− cells 24 h after differentiation induction. Heterozygous Stat5ab+/− cells expressed the same small Stat5 together with apparently full-length wild-type Stat5, which in Stat5ab+/+ cells was the only form detected (Figure 6a). Tyrosine phosphorylation of truncated Stat5 in Stat5ab−/− cells occurred with similar kinetics than that in wild-type cells (maximum after 24 h of differentiation) but was stronger than that in wild-type cells at all time points tested (Figure 6a), while total expression levels of truncated Stat5 were lower than those of Stat5 in wild-type cells and downregulation of the truncated protein during erythroid differentiation was delayed when compared to the kinetics of wild-type Stat5 expression. In line with this, we observed a several-fold increase in DNA-binding activity of the truncated Stat5 protein from Stat5−/− cells during erythroid differentiation in EMSA assays when compared to wild-type cells (Supplementary Figure S1). Thus, unlike the recently reported Stat5ab-knockout animals (Cui et al., 2004), the Stat5ab−/− cells used here express an N-terminally truncated but phosphorylated Stat5 in erythroblasts, which might retain some biological functions of wild-type protein (Supplementary Figure S1 and (Moriggl et al., 2005)).
Figure 6.
Selective activation of Stat3 and Stat1 in proliferating and differentiating Stat5ab−/− erythroblasts. (a) Primary fetal liver cells from wild-type, Stat5ab+/−, and Stat5ab−/− mice were induced to differentiate in serum-free medium for 24 h and subjected to Western blot analysis for tyrosine-phosphorylated Stat5ab. (b) Primary wild-type- and Stat5ab−/− erythroblasts were induced to differentiate in defined serum-free medium for the times indicated and lysates were subjected to Western blot analysis for tyrosine phosphorylated Stat5, Stat3 and Stat1, as well as for total Stat5, Stat3, Stat1, and Bcl-XL protein. Loading control, eIF4E levels, *, nonspecific band.
Stat5ab−/− and wild-type cells showed identical levels of Stat3 protein, which gradually decreased during differentiation. Interestingly, Stat3 was strongly tyrosine-phosphorylated in renewing Stat5ab−/− erythroblasts but not in wild-type cells (Figure 6b). This suggested that absence of Stat5 might promote compensatory Stat3 activation. We also observed strong upregulation of phosphorylated Stat1 in differentiating but not in proliferating Stat5ab−/− cells, which, however, was obviously insufficient to restore Bcl-XL expression in Stat5ab−/− cells (Figure 6b). These results raise the possibility that Stat3-signaling might play a role in partially rescuing the renewal defect of Stat5ab−/− erythroblasts, perhaps also improving survival during differentiation.
Discussion
It is increasingly recognized that abnormal self-renewal in human leukemia can be caused by cooperation of (mutated) transcriptional regulators – including nuclear receptors – with mutated/amplified receptor tyrosine kinases and/or constitutively active Stat5 or Stat3 (Stirewalt and Radich, 2003; Tenen, 2003; Yu and Jove, 2004; Ren, 2005). Here, we provide genetic evidence that members of the same protein families – the transcriptional regulator GR cooperating with EpoR and its key signal transducer Stat5 – plus the RTK c-Kit – are essential for sustained renewal of primary erythroid progenitors. Initially, this was not supported by analysis of respective mutant mice (GR−/−, GRdim/dim, EpoRH,and Stat5ab−/−), none of which showed major defects in steady-state erythropoiesis in vivo (Cole et al., 1995; Reichardt et al., 1998; Teglund et al., 1998; Zang et al., 2001). Using novel in vitro approaches to expand and differentiate primary mutant erythroblasts, we could show that all of these mutant cell types displayed enhanced differentiation at the expense of immature progenitor proliferation, gradually arresting expansion of the cultures. GR-function and EpoR cytoplasmic domain signaling were, however, dispensable for erythroid differentiation. In contrast, Stat5ab−/− erythroblasts failed to differentiate under fully defined, serum-free conditions, instead undergoing apoptosis due to defective up-regulation of bcl-XL. This defect of Stat5ab−/− erythroblasts could be rescued by exogenous bcl-XL or serum factors. Interestingly, Stat5ab−/− erythroblasts also exhibited increased activation of Stat3 and Stat1. These findings raise the possibility that other Stat family members might compensate for renewal- and survival defects of Stat5−/− erythroid progenitors.
Similar players and mechanisms in normal and oncogene-driven renewal?
The avian leukemia oncogenes v-ErbB and v-Sea – causing acute erythroleukemia in chicks (Beug et al., 1996; Bauer et al., 2001) – activate both Stat5- and PI3K-signaling pathways (von Lindern et al., 2001). Thus, these oncogenes can substitute for c-Kit-dependent PI3K activation and EpoR-dependent Stat5 activation in normal progenitors. Furthermore, v-Sea induces renewal in wild type, but not in Stat5−/− erythroblasts (HB, unpublished data). Apparently, these leukemia oncogenes utilize the same signaling pathways to induce renewal as those driving stress erythropoiesis in normal progenitors.
Both Stat5 and Stat3 are frequently upregulated and constitutively activated in human leukemia (Yu and Jove, 2004). A constitutively active Stat5 mutant causes multilineage leukemia after expression in Stat5−/− mouse bone marrow cells (Moriggl et al., 2005). Conversely, the human leukemia oncogene Tel-Jak2 (Lacronique et al., 1997) needs Stat5 to induce a myeloproliferative disease in transgenic mouse models (Schwaller et al., 2000). In our cell model, lack of Stat5 caused an (probably compensatory) activation of Stat3. This might explain the puzzling finding that EpoR−/− and Jak2−/− erythroblasts completely lack self-renewal ability (the respective fetal liver cells cannot be expanded in vitro), while Stat5−/− cells showed only a mild defect in renewal. In line with this, erythroblasts from mice completely lacking Stat5ab (Cui et al., 2004) showed even stronger compensatory upregulation of Stat3 during renewal, and exhibited no renewal defect at all (MK et al., unpublished data). These mice do not display an early embryonic lethal phenotype similar to EpoR- or Jak2-deficient animals, but die perinatally, probably due to other defects besides a failure of erythropoiesis. This further supports the idea of unknown compensatory mechanisms enabling functional erythropoiesis in the absence of Stat5ab in vivo (FG and MK, unpublished data). In trials to interfere with such compensatory mechanisms, cells expressing a dominant negative version of Stat5 (Stat5ΔC, lacking the transactivation domain and thus also interfering with Stat5 functions in complexes with other transcriptional regulators) were employed. These cells failed to undergo functional erythroid differentiation and upregulation of Bcl-XL even in serum-containing medium, where no defect was observed in Stat5−/− cells (see Supplementary Material, Figure S2 and respective experimental description).
Erythroid differentiation: mechanisms distinct from those controlling renewal?
Apparently, several molecular players essential for erythroblast renewal also function in terminal erythroid differentiation but act via different mechanisms in the two processes. For instance, terminal erythroid differentiation did not require signaling via the EpoR cytoplasmic domain in vitro or in vivo, which was, however, essential for renewal. Conversely, cytoplasmic truncation of the EpoR was reported to attenuate Epo-dependent proliferation and apoptosis in another study (Li et al., 2003). Similarly, GR function is essential for renewal, and even inhibitory for erythroid differentiation (Reichardt et al., 1998; von Lindern et al., 2001).
Distinct signaling pathways controlling erythroblast renewal versus differentiation were also evident for Stat5. The renewal defect of Stat5ab−/− cells was clearly due to increased terminal differentiation at the expense of renewal, rather than enhanced apoptosis, and could not be rescued by Bcl-XL. In contrast, differentiating Stat5ab−/− erythroblasts failed to induce Bcl-XL and did not survive in the absence of serum factors, which rescued both normal differentiation and Bcl-XL induction in serum-containing media. One possibility to explain these puzzling results is that Stat5ab−/−, but not wild-type fetal liver erythroblasts, showed significant Stat3 tyrosine phosphorylation before and after differentiation induction. Therefore, Stat3 might contribute to survival and late Bcl-XL upregulation during differentiation, either alone or in cooperation with truncated Stat5 protein still expressed in the Stat5ab−/− erythroid cells used.
Regulation of Bcl-XL and apoptosis by Stat5: lineage-specific differences?
The complex regulation of bcl-XL transcription by Stats plus serum factors indicates that Bcl-XL induction in terminally differentiating, primary-erythroid cells is not directly induced by cytokines but appears to be already ‘on’ due to Stat 5-independent pathways. In contrast, primary myeloid progenitors (Kieslinger et al., 2000), myeloid and lymphoid cell lines (Packham et al., 1998; Dumon et al., 1999; Silva et al., 1999) and erythroleukemic cell lines (Socolovsky et al., 1999), exhibit ‘simple’, direct transcriptional induction of bcl-XL mRNA by activated Stat5. Apparently, control by Epo is not sufficient for erythropoiesis (requiring rapid modulation in response to stress or disease and generating >1011 erythrocytes per day in man) but is modulated by multiple factors that activate Stat5, Stat3, or even Stat1. This might explain why human erythroleukemias are very rare or why Tel-Jak2, although perturbing both myeloid and erythroid differentiation (Onnebo et al., 2005), causes myelo- and lympho-proliferative disease but no erythroleukemia in mice (Schwaller et al., 2000).
Materials and methods
Cultivation of murine erythroid progenitors
Erythroid progenitors from fetal livers of E12.5 mouse embryos were cultivated as described (Dolznig et al., 2005), using serum-free ‘erythroid proliferation medium’ (StemPro-34™; Invitrogen), plus 2 U/ml human recombinant Epo (‘Erypo’, Cilag AG), 100 ng/ml murine recombinant SCF (R&D Systems), 10−6 m dexamethasone (Dex; Sigma) and 40 ng/ml human recombinant insulin-like-growth-factor-1 (IGF-1; Promega). Mice used were: wild-type, GRdim/dim (Reichardt et al., 1998), GR−/− (Cole et al., 1995), EpoRH, EpoRHM (Zang et al., 2001), and Stat5ab−/− (Teglund et al., 1998). Cell numbers and size distributions were determined using an electronic cell counter (CASY-1; Schärfe-System). Immortal mouse erythroblasts (clone I/11; p53-deficient) were cultivated as described (Dolznig et al., 2001).
Generation of erythroid progenitors from immature (multipotent) progenitors
Fetal livers of E12.5 mouse embryos from GRdim/dim, GR−/−, EpoRH, EpoRHM, and Stat5ab−/− mice were seeded at 4 × 106 cells/ml in ‘SCM’ medium, consisting of StemPro-34™ supplemented with SCF (100 ng/ml), IL-3 (2 ng/ml), IL-6 (0.5 ng/ml), Flk2/Flt3 ligand (10 ng/ml), GM-CSF (3 ng/ml), and IGF-1 (40 ng/ml; all from R&D), and 10−6 m Dex. Cultures were maintained between 4–7 × 106 cells/ml by daily partial medium changes. Cells were purified by centrifugation through Ficoll (lymphocyte separation medium, 1.078 g/ml; Eurobio) when containing >20% of dead and/or differentiated cells.
To induce erythroid commitment, cells were transferred to ‘erythroid proliferation medium’ (2 × 106 cells/ml). After a lag phase of 1–2 days, exponential growth was re-established. Differentiating/dead cells were removed by Ficoll purification at day 4. After 6–7 days, cultures were subjected to cytofluorometry and cytospin analysis. Cell populations showing continuous proliferation and a clear erythroblast phenotype (>90% CD117high, CD71high, Ter119low; Mac-1-, GR-1-, CD34-, and Sca-1-negative; see Figure 1d) were selected for further experiments.
Differentiation induction
Terminal differentiation in serum-containing or serum-free media supplemented with 10 U/ml Epo, insulin (10 ng/ml, Actrapid-HM), and the glucocorticoid-antagonist ZK112993 (3 μM) (Mikulits et al., 1995) was performed as described (von Lindern et al., 2001; Dolznig et al., 2005). Where indicated, differentiation was carried out in fully defined medium (Dolznig et al., 2002). Cell numbers, cell sizes, hemoglobin contents, and cytospin analyses were analysed as described (von Lindern et al., 2001). Quantitation of cytospins for immature, partially mature, mature and apoptotic erythroid cells was carried out as described (Kolbus et al., 2002).
Flow cytometry
Cells were stained with fluorescence-labeled antibodies (all PharMingen), against murine Ter119 (PE-labeled), Sca-1 (PE-labeled), GR-1 (PE-labeled), CD71 (PE-labeled), CD45R/B220 (APC-conjugated), CD135/Flk-2/Flt-3 (PE-labeled), CD11b/Mac-1 (PE-labeled), CD117/c-Kit (FITC-conjugated), and CD34 (FITC-conjugated). Fluorescence-labeled antibodies against murine IgG were used as negative control. Surface marker expression was quantified by cytofluorometry (Facs-Scan, Becton Dickinson).
Retroviral infections
For infection of p53−/− (I/11) and primary erythroblasts from wild-type and Stat5−/− mice, a bicistronic retroviral vector (pMCSV) expressing human Bcl-XL (Dumon et al., 1999) or murine Stat5ΔC (Moriggl et al., 1996), coupled to GFP via an IRES sequence, was used (Kieslinger et al., 2000). Infection efficiencies were determined by flow cytometry three days after infection. Stat5ΔC-transduced cells were subjected to FACS sorting (FACS-Vantage, Becton Dickinson).
Cytokine stimulation of erythroblasts
Immortal (I/11) as well as primary erythroblasts from wild type and mutant mice were incubated at 20 × 106 cells/ml in plain DMEM for 4 h at 37°C. Cells were then restimulated in StemPro34™ medium containing 10-fold-concentrated cytokines (1 μg/ml SCF, 20 U/ml Epo, or SCF + Epo) for 1 (mRNA-) or 3 h (protein analysis) and frozen in liquid nitrogen until further analysis.
Western blot analysis
Frozen cell pellets were lysed and subjected to SDS PAGE and Western Blot analysis, using antibodies against murine Bcl-XL (Becton Dickinson), Stat5ab (Santa Cruz), tyrosine-phosphorylated Stat5ab (Upstate Biotechnology), Stat3 (Becton Dickinson), tyrosine-phosphorylated Stat3 (Cell Signaling Technologies), Stat1 (Santa Cruz), tyrosine-phosphorylated Stat1 (Cell Signaling), Erk1/2 (Sigma) and eIF4E (Cell Signaling).
RNA extraction and Northern analysis
Total RNA was prepared from 1–4 × 107 cytokine-stimulated or nonstimulated cells using Trizol (Sigma) and processed for Northern blot analysis using 10 μg of RNA per sample and labeled probes for murine bcl-XL- or murine cis cDNA. Equal loading was confirmed by hybridizing with murine GAPDH cDNA (not shown).
Stat5/DNA bandshift analysis
Nuclear extracts from differentiating I/11 erythroblasts were incubated with –labeled double-stranded DNA-oligonucleotides representing the Stat5 binding-site in the bcl-XL-promoter (Socolovsky et al., 1999) or the Stat5 binding site in the beta-casein promoter (Moriggl et al., 1996) and subjected to electrophoretic mobility-shift assays (EMSAs) (Kieslinger et al., 2000).
Supplementary Material
Acknowledgements
We thank H Reichardt and G Schütz for access to GRdim/dim mice, JN Ihle for Stat5ab−/−, EpoRH, and EpoRHM mice, and G Stengl and G Litos for expert technical assistance. This work was supported by the ‘Fonds zur Förderung der wissenschaftlichen Forschung, FWF’, Austria (to HB and EWM) and the Herzfelder Family Foundation (to EWM).
Footnotes
Supplementary Information accompanies the paper on Oncogene website (http://www.nature.com/onc)
References
- Bauer A, Gandrillon O, Samarut J, Beug H. In: Hematopoiesis: a developmental approach. Zon L, editor. Oxford University Press; Oxford: 2001. pp. 368–390. [Google Scholar]
- Bauer A, Tronche F, Wessely O, Kellendonk C, Reichardt HM, Steinlein P, et al. Genes Dev. 1999;13:2996–3002. doi: 10.1101/gad.13.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beug H, Bauer A, Dolznig H, von Lindern M, Lobmayer L, Mellitzer G, et al. Biochim Biophys Acta. 1996;1288:M35–M47. doi: 10.1016/s0304-419x(96)00032-7. [DOI] [PubMed] [Google Scholar]
- Beug H, Dahl R, Steinlein P, Meyer S, Deiner E, Hayman MH. Oncogene. 1995;11:59–72. [PubMed] [Google Scholar]
- Blau HM, Brazelton TR, Weimann JM. Cell. 2001;105:829–841. doi: 10.1016/s0092-8674(01)00409-3. [DOI] [PubMed] [Google Scholar]
- Broudy VC. Blood. 1997;90:1345–1364. [PubMed] [Google Scholar]
- Carotta S, Pilat S, Mairhofer A, Schmidt U, Dolznig H, Steinlein P, et al. Blood. 2004;104:1873–1880. doi: 10.1182/blood-2004-02-0570. [DOI] [PubMed] [Google Scholar]
- Cole PJ, Blendy JA, Monaghan AP, Kriegelstein K, Schmid W, Aguzzi A, et al. Genes Dev. 1995;9:1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, et al. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolznig H, Bartunek P, Nasmyth K, Müllner E, Beug H. Cell Growth Diff. 1995;6:1341–1352. [PubMed] [Google Scholar]
- Dolznig H, Boulme F, Stangl K, Deiner EM, Mikulits W, Beug H, et al. FASEB J. 2001;15:1442–1444. doi: 10.1096/fj.00-0705fje. [DOI] [PubMed] [Google Scholar]
- Dolznig H, Habermann B, Stangl K, Deiner EM, Moriggl R, Beug H, et al. Curr Biol. 2002;12:1076–1085. doi: 10.1016/s0960-9822(02)00930-2. [DOI] [PubMed] [Google Scholar]
- Dolznig H, Kolbus A, Leberbauer C, Schmidt U, Deiner EM, Mullner EW, et al. Methods Mol Med. 2005;105:323–344. doi: 10.1385/1-59259-826-9:323. [DOI] [PubMed] [Google Scholar]
- Doppler W, Geymayer S, Weirich HG. Adv Exp Med Biol. 2000;480:139–146. doi: 10.1007/0-306-46832-8_17. [DOI] [PubMed] [Google Scholar]
- Dumon S, Santos SC, Debierre-Grockiego F, Gouilleux-Gruart V, Cocault L, Boucheron C, et al. Oncogene. 1999;18:4191–4199. doi: 10.1038/sj.onc.1202796. [DOI] [PubMed] [Google Scholar]
- He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, et al. Nature Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- Kieslinger M, Woldman I, Moriggl R, Hofmann J, Marine JC, Ihle JN, et al. Genes Dev. 2000;14:232–244. [PMC free article] [PubMed] [Google Scholar]
- Kolbus A, Pilat S, Husak Z, Deiner EM, Stengl G, Beug H, et al. J Exp Med. 2002;196:1347–1353. doi: 10.1084/jem.20020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffe M, et al. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- Li K, Menon MP, Karur VG, Hegde S, Wojchowski DM. Blood. 2003;102:3147–3153. doi: 10.1182/blood-2003-01-0078. [DOI] [PubMed] [Google Scholar]
- Mikulits W, Chen D, Mullner EW. Nucleic Acids Res. 1995;23:2342–2343. doi: 10.1093/nar/23.12.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriggl R, Gouilleux-Gruart V, Jahne R, Berchtold S, Gartmann C, Liu X, et al. Mol Cell Biol. 1996;16:5691–5700. doi: 10.1128/mcb.16.10.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriggl R, Sexl V, Kenner L, Duntsch C, Stangl K, Gingras S, et al. Cancer Cell. 2005;7:87–99. doi: 10.1016/j.ccr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Motoyama N, Kimura T, Takahashi T, Watanabe T, Nakano T. J Exp Med. 1999;189:1691–1698. doi: 10.1084/jem.189.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Onnebo SM, Condron MM, McPhee DO, Lieschke GJ, Ward AC. Exp Hematol. 2005;33:182–188. doi: 10.1016/j.exphem.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Packham G, White EL, Eischen CM, Yang H, Parganas E, Ihle JN, et al. Genes Dev. 1998;12:2475–2487. doi: 10.1101/gad.12.16.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, Morrison SJ. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, et al. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, et al. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Ren R. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- Sawada K, Krantz SB, Dai CH, Koury ST, Horn ST, Glick AD, et al. J Cell Physiol. 1990;142:219–230. doi: 10.1002/jcp.1041420202. [DOI] [PubMed] [Google Scholar]
- Schulte CE, von Lindern M, Steinlein P, Beug H, Wiedemann LM. EMBO J. 2002;21:4297–4306. doi: 10.1093/emboj/cdf429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR, et al. Mol Cell. 2000;6:693–704. doi: 10.1016/s1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Shet AS, Jahagirdar BN, Verfaillie CM. Leukemia. 2002;16:1402–1411. doi: 10.1038/sj.leu.2402577. [DOI] [PubMed] [Google Scholar]
- Silva M, Benito A, Sanz C, Prosper F, Ekhterae D, Nunez G, et al. J Biol Chem. 1999;274:22165–22169. doi: 10.1074/jbc.274.32.22165. [DOI] [PubMed] [Google Scholar]
- Silva M, Grillot D, Benito A, Richard C, Nunez G, Fernandez-Luna JL. Blood. 1996;88:1576–1582. [PubMed] [Google Scholar]
- Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- Stirewalt DL, Radich JP. Nat Rev Cancer. 2003;3:650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, et al. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- Tenen DG. Nature Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Verdier F, Rabionet R, Gouilleux F, Beisenherz-Huss C, Varlet P, Muller O, et al. Mol Cell Biol. 1998;18:5852–5860. doi: 10.1128/mcb.18.10.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lindern M, Deiner EM, Dolznig H, Hayman MJ, Muellner EM, Beug H. Oncogene. 2001;20:3651–3664. doi: 10.1038/sj.onc.1204494. [DOI] [PubMed] [Google Scholar]
- Von Lindern M, Schmidt U, Beug H. Cell Cycle. 2004;3:876–879. [PubMed] [Google Scholar]
- von Lindern M, Zauner W, Steinlein P, Mellitzer G, Fritsch G, Huber K, et al. Blood. 1999;94:550–559. [PubMed] [Google Scholar]
- Wu H, Klingmuller U, Besmer P, Lodish HF. Nature. 1995;377:242–246. doi: 10.1038/377242a0. [DOI] [PubMed] [Google Scholar]
- Yu H, Jove R. Nat Rev Cancer. 2004;4:655–665. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Zang H, Sato K, Nakajima H, McKay C, Ney PA, Ihle JN. EMBO J. 2001;20:3156–3166. doi: 10.1093/emboj/20.12.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.