Abstract
Caspase-3 (CASP3) cleaves many proteins including protein kinases (PKs). Understanding the relationship(s) between CASP3 and its PK substrates is necessary to delineate the apoptosis signaling cascades that are controlled by CASP3 activity. We report herein the characterization of a CASP3-substrate kinome using a simple cell-free system to synthesize a library that contained 304 PKs tagged at their N- and C-termini (NCtagged PKs) and a luminescence assay to report CASP3 cleavage events. Forty-three PKs, including 30 newly identified PKs, were found to be CASP3 substrates, and 28 cleavage sites in 23 PKs were determined. Interestingly, 16 out of the 23 PKs have cleavage sites within 60 residues of their N- or C-termini. Furthermore, 29 of the PKs were cleaved in apoptotic cells, including five that were cleaved near their termini in vitro. In total, approximately 14% of the PKs tested were CASP3 substrates, suggesting that CASP3 cleavage of PKs may be a signature event in apoptotic-signaling cascades. This proteolytic assay method would identify other protease substrates.
Keywords: caspase, protein kinases, apoptosis, cell-free protein synthesis, protein library
On the basis of the corresponding genetic sequences, >500 human and mouse proteolytic enzymes have been predicted.1 This number is comparable with that found for protein kinases (PKs), which are the main signal-transduction enzymes.2, 3 Proteases are involved in the maturation, localization, stabilization, and complex formation of proteins, and in many biological processes, for example, normal development,4, 5 cancer,6, 7 infectious diseases,8 and cell death.9 Therefore, it is important to be able to identify protease substrates using simple assays.
Apoptosis requires the action of many different proteins that participate in apoptotic cell-signaling pathways.10 Caspases and PKs are critical components of growth and apoptosis signaling pathways.2, 10 Large-scale analyses of the biological networks involving PKs and caspases are vital for the elucidation of apoptosis signaling pathways. Recent whole-cell proteomic studies that used mass spectrometry attempted to identify substrates of caspases that are involved in apoptosis and have shown that the percentage of PKs found as caspase substrates during apoptosis is 3–6% of ∼300.11, 12 However, cellular protein expression levels may have biased the results.13 Furthermore, it is difficult to identify specific pairs of proteases and substrates because numerous cleavage events occur simultaneously in cells. Therefore, an in vitro approach that could identify specific proteases and their corresponding substrates would complement cell-based approaches. A diagram, derived from a comprehensive in vitro study, that illustrates the relationships between caspases and their PK substrates would help clarify the signal-transduction events that occur during apoptosis.
A collection of recombinant proteins, that is, a protein library, is needed to screen a large number of protein substrates. In addition, to screen a protein library comprehensively two in vitro high-throughput methods – one for protein synthesis and one for the detection of the targeted biochemical reaction – are required. Recently, we developed an automated protein synthesis system that uses a wheat cell-free system.14, 15, 16 Using this system, we were able to synthesize many human and Arabidopsis PKs.17, 18 Recent work by others suggested that the wheat cell-free system could produce 13 364 human proteins, which, because of the large number of proteins involved, represents an in vitro-expressed proteome.19 We also recently developed a method to label monobiotin proteins that had been synthesized in the wheat cell-free system.20 These monobiotin-labeled proteins were then used directly – without purification – to detect protein ubiquitination21 and an autoantibody in the serum.22 As the procedures used with many commercially available detection kits depend on biotin–streptavidin interactions, our purification-free, synthesis/biotin-labeling method provides a simple and highly specific system that can be used for biochemical analyses.
Caspase-3 (CASP3) cleaves many different proteins,23, 24 and its action in vivo irreversibly induces apoptosis. For the study reported herein, we delineated a CASP3-substrate kinome using a simple luminescent-based detection method to screen an N- and C-terminally tagged (NCtagged) PK library produced in the wheat cell-free system. This comprehensive characterization of a CASP3-substrate kinome is a resource that can be used to understand the roles of PKs in apoptosis.
Results
Generation of an NCtagged PK library used to identify CASP3 PK substrates
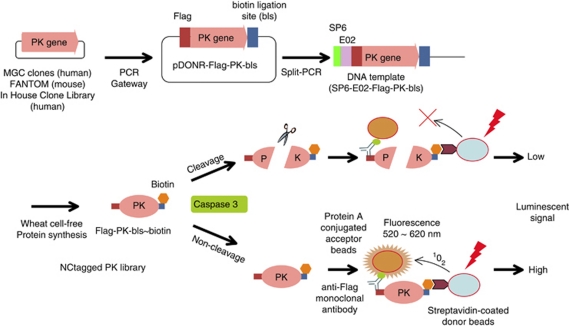
To identify PKs that are substrates of CASP3, we first made a library consisting of 248 human and 56 mouse PKs (Supplementary Table S1). The nucleotide sequences for the Flag-tag and the biotin ligation site (bls) were added upstream and downstream, respectively, of the PK open-reading frame by PCR incorporation of Gateway recombination tags. Each PCR product (attB1-Flag-PK-bls-attB2) was inserted into a pDONR221 vector using the Gateway BP Clonase II system (upper panel, Figure 1). The Flag-PK-bls nucleotide sequences from the Escherichia coli cultures were used without purification to construct, by split-primer PCR, the DNA templates for protein synthesis.14 The NCtagged PK library (304 PKs) was produced using an automated protein synthesizer (GenDecoder 1000; CellFree Sciences Co., Ltd., Matsuyama, Japan), with biotin and biotin ligase added into the synthesis mixtures for monobiotin labeling at the bls.20, 21 That the members of the protein library were NCtagged was confirmed by immunoblotting with anti-Flag antibodies and Alexa488-labeled streptavidin. To assess the suitability of the designed PKs to act as CASP3 substrates, we used NCtagged p21-activated kinase 2 (PAK2), which is a known CASP3 substrate,25 as the test case. The biotinylated NCtagged-PAK2 (Flag-PAK2-bls∼biotin) was treated with CASP3 and cleavage of PAK2 was confirmed by immunoblotting with Alexa488-conjugated streptavidin (Figure 2a). In addition, the cleavage site (319DELD↓S323), determined by amino-acid sequencing, was found to be the same as that reported previously.25 (The arrow indicates the hydrolytic bond.)
Figure 1.
Schematics of the DNA template construction and the CASP3-substrate-screening assay. Protein kinase (PK) genes were obtained from the human MGC and mouse FANTOM libraries, and from a library of PK genes that we had cloned. The PK genes were PCR amplified with the Flag and the biotin ligation site (bls) tags added to the upstream and downstream ends, respectively. The modified genes were each inserted into a Gateway pDONR221 vector (pDONR-Flag-PK-bls) and DNA templates (SP6-E02-Flag-PK-bls) were constructed by split-primer PCR and then expressed in the wheat cell-free protein synthesis system that included biotin ligase and -biotin to give Flag-PK-bls∼biotin constructs. The Flag and biotin tags were bound to protein A-conjugated acceptor beads via an anti-Flag antibody and streptavidin-conjugated donor beads, respectively. An intact complex luminesced strongly, whereas after CASP3 cleavage and dissociation of the protein fragments, the luminescence was abolished or reduced
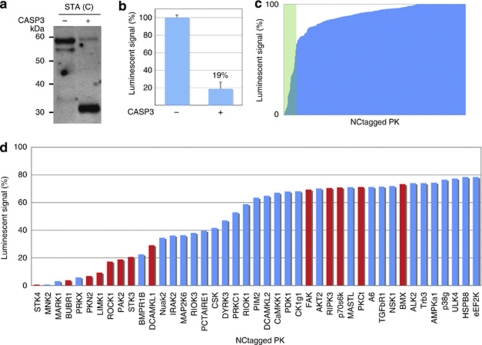
Figure 2.
Screening of CASP3-cleaved PK substrates from the NCtagged PK library. (a) Immunoblot of NCtagged PAK2 that had been incubated in the presence (+) or absence (−) of CASP3. Alexa488-labeled streptavidin (STA(C)) was used for detection. (b) Detection of CASP3-cleaved NCtagged PAK2 using the AlphaScreen system. The luminescence for the control (no CASP3) was set to 100%. The value of 19% indicated that most of the NCtagged PAK2 was cleaved by CASP3. Each value is the mean of three independent experiments, and the uncertainty is reported as the standard deviation. (c) Luminescent signals remaining after in vitro CASP3 treatment of NCtagged PKs that had been synthesized in the wheat cell-free system. The x axis lists the NCtagged PKs in ascending order of their luminescent signals after CASP3 treatment. (d) NCtagged PKs that returned luminescent signals of <78% of the control values. The plot contains the data of (c) within the green rectangle. Red bars are for PKs that were known to be substrates of CASP3 before this report
A luminescent assay to detect PK substrates of CASP3
A schematic of the assay used to monitor cleavage of the NCtagged PKs by CASP3 is shown in Figure 1. The PK construct is first incubated with CASP3. If the construct contains a sequence that can be cleaved by CASP3, cleavage occurs. Acceptor and donor beads are then added. The Flag-tag binds a protein A-conjugated acceptor bead via an anti-Flag antibody, and the biotin bound to the C-terminus of the PK construct binds a streptavidin-conjugated donor bead. If an acceptor bead is in close contact with the donor bead, as is the case when the construct is not a CASP3 substrate and both beads are therefore bound intramolecularly, the system luminesces. However, if CASP3 had cleaved the NCtagged PK, luminescence is suppressed because the beads are no longer in close contact. As a proof-of-concept experiment, cleavage of the test PK, NCtagged PAK2, was assessed using this system. CASP3 treatment decreased the luminescent signal to 19±7% that of the control (no CASP3; Figure 2b). Therefore, the system could detect CASP3 cleavage and can replace conventional immunoblotting procedures.
Screening of the CASP3-substrate kinome
Using the luminescent system, 304 NCtagged PKs were screened. The level of luminescence after CASP3 treatment is reported as the percentage of the corresponding control (no CASP3; Figure 2c and d). Thirteen of the NCtagged PKs for which luminescence was low after CASP3 treatment are known CASP3 substrates.23, 24, 26 The smallest and largest luminescent values were for STK4 (1%) and BMX (73%), respectively; we therefore examined the physical states of the PKs that had been treated with CASP3 and had associated luminescence values of ∼80% by immunoblotting with anti-Flag antibodies and Alexa488-streptavidin to detect the N- and C-termini of the NCtagged PKs, respectively. This ‘terminal detection' (TD) immunoblot assay identified 43 NCtagged PKs that had been cleaved (Supplementary Table S1). In addition to the 13 PKs that were known to be CASP3 substrates, 30 previously unidentified PK that were substrates of CASP3 were found (Figure 3 and Table 1). In addition, because the apparent molecular weights of the N- and C-terminal fragments could be estimated from their positions in the TD immunoblot, the CASP3 cleavage sites could be predicted (red arrowheads, Figure 3). For MASTL, the signal on the immunoblot with Alexa488-conjugated streptavidin was not detectable, probably indicating that the efficiency of biotinylation in MASTL proteins might be too low to detect for the immunoblot. Luminescent signal of this clone was also very low (see Supplementary Table S1).
Figure 3.
In vitro cleavage of NCtagged PKs by CASP3. The NCtagged PKs that had been incubated in the presence (+) or absence (−) of CASP3 and their cleavage products were detected using anti-Flag antibodies (FLAG(N)) and Alexa488-conjugated streptavidin (STA(C)), which bound to the N- and C-termini of the PK constructs, respectively. The cartoons of the proteins that are under the lanes show the locations of the conserved domains (colored boxes) and the predicted cleavage sites (red arrowheads). The conserved domains that are found in the Conserved Domains Database (http://www.ncbi.nlm.nih.gov/cdd) are: ACD, alpha-crystallin domain; activin, conserved domain for activin members; ADF, actin depolymerization factor/cofilin-like domain; α-kinase, conserved kinase domain for the α-kinase family; C1, phorbol esters/diacylglycerol binding domain; DCX, doublecortin domain; death, death domain; GS, GS motif; KA1, kinase-associated domain; kinase, catalytic domain of protein kinase; PB1, Phox and Bem1p domain; PH, pleckstrin homology domain; RIO, catalytic domain of eukaryotic RIO kinase family; SH2, src homology 2 domain; SH3, src homology 3 domain; UBL, ubiquitin-like domain
Table 1. Characteristics of the newly identified CASP3 PK substrates.
| Symbols | Kinome names | Groups | Clone origin | AAa | Cleavage sequence | Cleavage sites | Methodsb | Conservationc | Smallest frag.d | In vivo cleavagese |
|---|---|---|---|---|---|---|---|---|---|---|
| ACVR1 | ALK2 | TKL | Hs | 509 | IASD↓M | 269 | NT | Yes | C240 | Yes |
| AKT2 | AKT2 | AGC | Mm | 481 | DAMD↓Y | 121 | NT | Yes | N121 | Yes |
| BMPR1B | ALK6 | TKL | Hs | 502 | CSTD↓G | 50 | NT | Yes | N50 | Yes |
| DFVD↓G | 120 | NT | Yes | |||||||
| CaMKK1 | CaMKK1 | Other | Hs | 520 | EEAD↓G | 32 | NT | Yes | N32 | Yes |
| CSK | CSK | TK | Mm | 450 | DAPD↓G | 409 | MS | Yes | C41 | Yes |
| CSNK1G1 | CK1g1 | CK1 | Mm | 459 | VHVD↓S | 343 | MU | Yes | C116 | Yes |
| eEF2K | eEF2K | Atypical | Hs | 725 | EGVD↓G | 14 | MU | Yes | N14 | Yes |
| DHLD↓N | 430 | MU | Yes | |||||||
| HSPB8 | H11 | Atypical | Hs | 196 | MAD↓G | 3 | NT | Yes | N3 | Yes |
| MAP2K6 | MAP2K6 | STE | Mm | 334 | DFVD↓F | 289 | MU | Yes | C45 | Yes |
| MAPK12 | p38g | CMGC | Hs | 367 | SAVD↓G | 46 | NT | Yes | N46 | Yes |
| MARK1 | MARK1 | CAMK | Hs | 795 | SATD↓E | 52 | MU | Yes | N52 | Yes |
| MKNK2 | MNK2 | CAMK | Hs | 414 | DQPD↓H | 32 | MU | No | N32 | Yes |
| DIPD↓A | 58 | NT | Yes | |||||||
| PDPK1 | PDK1 | AGC | Hs | 556 | SHPD↓A | 552 | NT | Yes | C4 | Yes |
| PIM2 | PIM2 | CAMK | Hs | 334 | TDFD↓G | 198 | MU | Yes | C113 | Yes |
| PRKAA1 | AMPKa1 | CAMK | Hs | 550 | TSLD↓S | 520 | MS | Yes | C30 | Yes |
| PRKCI | aPKCi | PKC | Hs | 596 | TQRD↓S | 6 | NT | Yes | N6 | Yes |
| PRKX | PRKX | AGC | Hs | 358 | ETPD↓G | 25 | NT | No | N25 | Yes |
| RIOK1 | RIOK1 | Atypical | Mm | 568 | EKDD↓I | 37 | NT | Yes | N37 | Yes |
| RIOK3 | RIOK3 | Atypical | Hs | 516 | DTRD↓D | 139 | NT | Yes | N139 | Yes |
| RPS6KA5 | MSK1 | AGC | Hs | 549 | DGGD↓G | 20 | NT | Yes | N20 | Yes |
| DELD↓V | 344 | NT | Yes | |||||||
| TEMD↓P | 356 | NT | Yes | |||||||
| SNARK | NuaK2 | CAMK | Hs | 628 | VSED↓S | 546 | MU | Yes | C82 | Yes |
| TRIB3 | TRB3 | CAMK | Hs | 358 | VVPD↓G | 338 | NT | Yes | C20 | Yes |
| ULK4 | ULK4 | Other | Hs | 580 | SQID↓S | 473 | MU | Yes | C107 | Yes |
| DCAMKL2 | DCAMKL2 | CAMK | Hs | 695 | — | — | — | — | — | Yes |
| DYRK3 | DYRK3 | CMGC | Hs | 568 | — | — | — | — | — | ND |
| IRAK2 | IRAK2 | TKL | Mm | 622 | — | — | — | — | — | Yes |
| MASTL | MASTL | AGC | Hs | 879 | — | — | — | — | — | Yes |
| PCTK1 | PCTAIRE1 | CMGC | Hs | 496 | — | — | — | — | — | Yes |
| PTK9 | A6 | Atypical | Hs | 384 | — | — | — | — | — | Yes |
| TGFBR1 | TGFBR1 | TKL | Hs | 426 | — | — | — | — | — | Yes |
Abbreviations: Hs, human clone; Mm, mouse clone; MS, mass spectroscopy; MU, mutation; ND, not determined; NT, N-terminal sequencing.
Length of amino acids.
Methods for determination of cleavage site.
Very similar site conserving the Asp (D) of the hydrolytic bond was found between human and mouse PKs (Yes), whereas no similar sites was done (No).
The smallest N (N) or C (C) fragment in the cleaved PKs. Number is the length of amino acids of the fragment.
Data from Figure 4.
A comparison of the luminescent and immunoblot data correlated a luminescent signal of <78% with a positive immunoblot result. Forty-eight PK constructs with luminescent signals >78% were tested and returned negative immunoblot results (Supplementary Table S1). Therefore, a luminescent signal of ∼78% is the apparent divisor between PKs that can be cleaved by CASP3 and those that cannot be cleaved.
In vivo identification of the PKs that were identified as CASP3 substrates by the luminescent assay
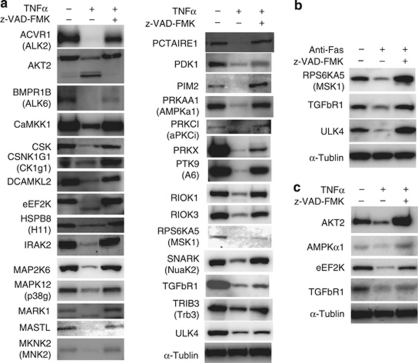
We investigated whether the newly identified PKs that were substrates of CASP3 were cleaved in HeLa cells that had been induced to undergo apoptosis by TNFα plus cycloheximide (TNFα)27 or anti-Fas antibody (anti-Fas).28 The genes encoding these PKs were each inserted into the transfection vector, pDEST26, using the Gateway system and expressed as (His)6-PK-Flag constructs. We were able to detect all expressed PK constructs, except DYRK3, by immunoblotting with anti-(His)6 or anti-Flag antibodies. Notably, they were detected as cleavage products and/or were found in smaller amounts when the cells had been induced to undergo apoptosis than when apoptosis had been inhibited by z-VAD-FMK (Figure 4a and b). Furthermore, apoptosis-induced cleavage of four endogenous PKs was found by immunoblotting with commercially available antibodies against the endogenous PKs (Figure 4c). These in vivo experiments validated the underlying concept of our in vitro cell-free system as the in vivo system found all of the PKs identified by the in vitro system.
Figure 4.
In vivo caspase cleavage of the newly identified PK substrates and of endogenous PK substrates. (a) In vivo cleavage of the (His)6-PK-Flag constructs expressed in apoptotic HeLa cells. The cells were treated with DMSO (control) or with TNFα and cycloheximide (TNFα) in the presence and absence of z-VAD-FMK (a CASP3 inhibitor) for 6 h and then lysed. The cell extracts were immunoblotted and the PK constructs were detected with anti-Flag antibodies, except for HSPB8 and MAP2K6. Anti-His tag antibody was used for the two PKs. (b) The cells were transfected with a plasmid of (His)6-PK-Flag constructs, and treated with DMSO (control), or with anti-Fas antibody (anti-Fas) in the presence and absence of z-VAD-FMK for 6 h and then lysed. Immunoblotting was carried out as (a). (c) HeLa cells were treated as in (a), but were not transfected with a (His)6-PK-Flag gene. Each endogenous PK was detected using an antibody specific for it. α-Tubulin was used as an internal marker
Characterization of the CASP3 cleavage sites in the newly identified PK substrates
We characterized the CASP3 cleavage sites in the newly identified PK substrates. As the positions of the cleaved PK fragments in the TD immunoblot could be used to estimate the size of the cleaved fragments and because the antibodies could be used to identify whether the fragments were derived from the N- or C-terminal regions of the PKs, we could predict the approximate positions of the CASP3 cleavage sites (red arrowheads, Figure 3). Each NCtagged PK that was a substrate for CASP3 was synthesized in the cell-free system and purified using Streptavidin Magnesphere Paramagnetic beads. Their C-terminal fragments that bound to the beads were recovered after CASP3 cleavage and their sequences were determined. Using this approach, the cleavage sites of ACVR1, AKT2, BMPR1B, CaMKK1, HSPB8, MAPK12, MKNK2(D58), PDPK1, PRKCI, PRKX, RIOK1, RIOK3, and RPS6KA5 were determined. We then attempted to determine the cleavage sites of the remaining PKs by other methods.
The NCtagged PKs that had low biotin-labeling efficiencies and were cleaved near their C-termini were genetically modified by the addition of a glutathione-S-transferase (GST) fragment at their C-termini to facilitate recovery with glutathione Sepharose 4B beads after CASP3 cleavage. CASP3 cleavage of the PK-GSTs produced the same size N-terminal fragments as those of the corresponding CASP3-cleaved NCtagged PKs, indicating that the GST tags did not alter the positions of the cleavage sites. In addition, the sequences of the cleaved c-src tyrosine kinase (CSK) and AMP-activated kinase-a1 (AMPKa1) fragments were determined using MALDI/TOF-MS. Other PK constructs that were synthesized in small amounts were subjected to D → A mutagenesis to determine their cleavage sites. In total, 28 cleavage sites in 23 PKs were identified (Table 1). Identical or similar cleavage sites were found in the corresponding human and mouse PKs, except for those of PRKX (Supplementary Table S2). (Sequence analysis showed that mouse PRKX does not have the N-terminal region that is found in human PRKX.) Therefore, the CASP3-substrate kinome may be highly conserved in mammals.
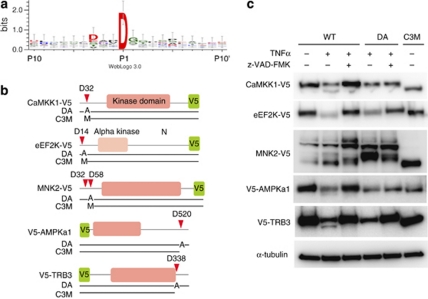
We also analyzed the common sequence attributes among the 28 cleavage sites and found that CASP3 prefers the sequence, DXXD↓G (Figure 5a). The consensus PK cleavage site for CASP3 in the MEROPS database is DXXD↓X. In the NCtagged PK library, 208 of the 304 PKs contain a DXXDX sequence. However, only 33 PKs were cleaved by CASP3; therefore, to be cleaved by CASP3, the DXXDX sequence and a structural element –probably accessibility – are required.
Figure 5.
The cleavage site logo and in vivo cleavage of five PKs that are cleaved by CASP3 near their N- or C-termini. (a) The 20 residues surrounding the D of the hydrolytic bond in 28 PKs were analyzed using WebLogo, version 3.0.36 (b) Cartoons of five PK sequences that have cleavage sites near their N- or C-termini. The corresponding PKs were used for the experiment shown in (c). The positions of the alanines in the D → A mutants (DA) are shown, as are the long fragments (C3M) produced by CASP3 cleavage. V5 tags were fused at the ends farther away from the cleavage sites. The first M in C3M of CaMKK1, eEF2K, and MNK2 indicates a methionine as a start amino acid. (c) Immunoblots of PK-V5s and V5-PKs that had been expressed in apoptotic HeLa cells. The cells were treated with DMSO (control) or with TNFα and cycloheximide in the presence or absence of z-VAD-FMK for 6 h and then lysed. The proteins were blotted and then detected with anti-V5 antibodies. WT indicates a wild-type protein
Characterization of the newly identified PKs that were cleaved near their N- or C-termini
Interestingly, 16 out of the 23 PKs, for which cleavage sites were characterized, have cleavage sites within 60 residues of their N- or C-termini. We investigated whether these sites were also cleaved in vivo when apoptosis was induced by TNFα. For these experiments, CaMKK1, eEF2K, MNK2, AMPKa1, and TRIB3, which were cleaved in vitro at D32, D14, D32/D58, D520 (30 residues away from the C-terminus), and D338 (20 residues away from the C-terminus), respectively, were used (Figure 5b and Table 1). Their genes (wild type, WT) were each reconstructed with a V5 tag added at the end opposite the cleavage site. The genes for their D → A mutants (DA), and for the sequences of their longer CASP3-cleaved fragments (C3M), were also constructed and all were expressed in control and in apoptotic cells (Figure 5c). Cleavage of the WT PKs produced long fragments corresponding to C3M in apoptotic cells, whereas z-VAD-FMK blocked cleavage. These cleavages near the N- and C-termini of the PKs suggest that CASP3 cleavage may regulate the activity level and/or cellular localization of the PKs, rather than simply inactivate the kinases.
Discussion
In 1995, PITSLRE,29 PKCδ,30 and DNA-PKcs31 were reported as the first PK-type substrates of CASP3. During the next 15 years, 36 additional PKs that can be cleaved by CASP3 were found.23, 24, 26 Notably, these authors showed that CASP3-cleaved PKs abrogate survival signals and accelerate apoptosis. In this study, we identified an additional 30 PKs that can be cleaved by CASP3. In addition, many of the cleavage sites were found in regulatory elements or in the regions near the N- and C-termini, rather than the kinase domain itself. Some of the newly identified CASP3-substrate PKs may be involved in apoptotic signal cascades. Sixteen PKs were shown to be cleaved in vitro near their N- or C-termini and at least five of them were also cleaved near their N- or C-termini in apoptotic cells (Figure 5). Using standard immunoblotting, proteins that are cleaved into a large and a small fragment may be overlooked because the mobilities of the large fragment and the intact protein will be nearly identical. Most of the PKs that had been previously reported to be cleaved by CASP3 were identified because the cleaved fragments had very different molecular weights than did the intact PK and were therefore easily detected by SDS-PAGE. Consequently, cleavages near the termini may have been overlooked. Taken together, our results suggest that CASP3 cleavage of some of the members of the CASP3-substrate kinome alters the function of the PKs and thereby signals apoptosis.
For the study reported herein, 304 out of 518 known PKs, synthesized as NCtagged PKs, were subjected to the in vitro cleavage assay (Supplementary Table S1). The relative number of PKs that were cleaved was ∼14%. A total of 69 PKs that are CASP3 substrates are now known, which suggests that at least ∼13% of the PKs in the human kinome are targets of CASP3. As ∼200 PKs have yet be tested as CASP3 substrates, an additional 26 PKs (13% of the 200) may be CASP3 substrates. The human genome contains 518 annotated PKs, which have been divided into 10 groups on the basis of their sequence homologies.3 Interestingly, the groups differ in terms of their susceptibilities to CASP3 cleavage (Table 2). Approximately 30% of the PKs in the AGC group are known CASP3 substrates, for example, AKT2, S6K, MSK, PKC, and PDK1. Many of the AGC-type PKs are commonly found in mammalian tissues,32 and their cleavage sites are located in their regulatory domains (Figure 4 and Table 1). Therefore, these abundant PKs may be activated when CASP3 cleaves them and then act as intracellular apoptosis signals. Conversely, CASP3 cleaved only a relatively small number (6∼8%) of the PKs in the CMGC group, which includes the kinases of the CDK and CDKL families, and the tyrosine kinase groups. Therefore, most members of these groups may only act indirectly as apoptosis signals after CASP3 activation.
Table 2. Characteristics of the protein kinases used in this study.
| Groups | Totala | Tested clones | Cleaved clones (newb) | Cleaved/test clones (%) |
|---|---|---|---|---|
| AGC | 63 | 33 | 10 (6) | 30 |
| CAMK | 74 | 52 | 8 (7) | 15 |
| CK1 | 12 | 8 | 1 (1) | 13 |
| CMGC | 61 | 39 | 3 (3) | 8 |
| Other | 83 | 46 | 3 (2) | 7 |
| STE | 47 | 25 | 4 (1) | 16 |
| TK | 90 | 51 | 3 (1) | 6 |
| TKL | 43 | 27 | 6 (4) | 22 |
| RGC | 5 | 2 | 0 | 0 |
| Atypicals | 40 | 21 | 5 (5) | 24 |
| Total | 518 | 304 | 43 (30) | 14 |
Each number is corresponding to human kinome.
Newly PK numbers found in this study.
Such ROCK1 and MST1, certain caspase cleavage products, work as apoptosis signaling.23, 24 In this study, we found at least six new CASP3 cleavage products, derived from AKT2, CaMKK1, eEF2 K, MARK1, MNK2, and TRB3, after 6 h from apoptosis induction (Figures 4 and 5). These cleavage products retain kinase domain, as in the case of ROCK1 and MST1. On the other hand, we could not detect any cleavage products from the other kinases in vivo. The reasons are not yet understood. However, recent proteomics approach has shown that the cleaved proteins displayed transient fragments in the apoptotic cells.12 Further analysis at multiple time points during the apoptotic cascade would be required for detection of the cleavage products from the remaining PKs.
For TRIB3, full-length TRIB3 (D338A) mutant was decreased in apoptotic condition (compared TNFα lane with TNFα plus z-VAD-FMK lane in Figure 5c). However, the mutant could not produce the CASP3 cleavage product found as the shorter form in TNFα lane of WT, indicating that the mutant was not cleaved by CASP3. The mutant was also not cleaved by CASP3 in vitro (data not shown). As TRIB3 has been known to receive proteasomal degradation,33 this unexpected reduction of the mutant TRIB3 in the apoptotic cells may be the effects of cycloheximide and/or caspase-inhibitor treatment on TRIB3 degradation.
Proteases often modify the activities of their targeted protein substrates. Identification of the specific substrate that is cleaved by a protease is necessary if the functions of both the protease and its substrate are to be understood. Proteomic studies have used mass spectrometry to exhaustively identify cellular proteins that have been cleaved by proteases.11, 12 However, it has been difficult to correlate specific proteases with their substrates because many proteases act at the same time in vivo.
Many full-length cDNAs derived from the genes of higher eukaryotes are available from many different sources. These cDNAs are potentially a great DNA template resource for in vitro syntheses of proteins. As a protein production system and for the functional analysis of proteins, the wheat cell-free system has many advantages: It can effectively use PCR-generated DNA templates.14 It is easily adapted to an automated system.15 It can be used to incorporate a single label into target proteins.20 Its synthesized proteins do not require purification before being assayed, and it has no detectable proteasome activity.21 In addition, the screening cost is very low (∼US$1/assay), which for our study translated to 10 cents to produce each NCtagged protein and 20 cents for the beads, CASP3, and disposable hardware used in one assay.
In summary, we showed that an NCtagged PK library synthesized in a cell-free system could be used to characterize a CASP3-substrate kinome. Analysis of the CASP3-cleavage sites indicated that CASP3 cleavage of PKs depends on both primary and tertiary structure. Almost all of the PK substrates that we identified in vitro were also identified in vivo. Systems similar to that used herein could be used to screen other protease substrates.
Materials and Methods
General
The following procedures have been described:14, 15, 16, 20, 21, 22, 34 wheat cell-free protein production; split-primer PCR synthesis of the DNA templates; parallel syntheses of mRNAs and their translated proteins; and measurements of the amounts of protein synthesized using densitometer scans of Coomassie brilliant blue-stained proteins or of radiolabeled proteins.
Construction of DNA templates for the expression of a PK protein library
The cloned genes encoding the PKs used in this study are listed in Supplementary Table S1. Their open-reading frames (without stop codons) were modified in two steps using PCR and the primers S1 (5′-CCACCCACCACCACCAatg(n)16-3′) and T1 (5′-TCCAGCACTAGCTCCAGA(n)19-3′) (lowercase letters indicate nucleotides of the gene) for the first step, and the primers attB1-Flag-S1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGACTACAAGGATGACGATGACAAGCT CCACCCACCACCACCAATG-3′) and T1-bls-STOP-attB2-anti (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTATTCGTGCCACTCGATCTTCTGGGCCTC GAAGATGTCGTTCAGGCCGCTTCCAGCACTAGCTCCAGA-3′) for the second step. The PCR-modified genes were each inserted into a pDONR221 vector using the Gateway BP Clonase II enzyme mix (Invitrogen, Carlsbad, CA, USA) to give pDONR-Flag-PK-bls vectors. Escherichia coli cells were transformed with these vectors and then cultured in wells of a 96-well plate that contained GYT medium (10% (v/v) glycerol, 0.125% (w/v) yeast extract, and 0.25% (w/v) tryptone) for 48 h without shaking. DNA templates for mRNA and protein expression were constructed using split-primer PCR14 in two steps. For the first step, the pDONR221-Flag-PK-bls plasmids that had not been isolated from the E. coli cells, and the primers pDONR221-1st_4080 (5′-ATCTTTTCTACGGGGTCTGA-3′) and deSP6E02-Flag-S1 (5′-GGTGACACTATAGAACTCACCTATCTCTCTACACAAAACATTTCCCTACATACAACTTTCAA CTTCCTATTATGGACTACAAGGATGACGATGACAAGCTCCACCCACCACCACCAATG-3′) were used, and for the second step, the amplified sequences of the first step and the primers SPu (5′-GGGTAGCATTTAGGTGACACT-3′) and pDONR221-2nd_4035 (5′-ACGTTAAGGGATTTTGGTCA-3′) were used to give SP6-E02-Flag-PK-bls DNA templates. (The E02 sequence is a translational enhancer,35 and the SP6 sequence is an SP6 RNA polymerase promoter.)
Cell-free protein synthesis
Cell-free protein synthesis used the reagents of an ENDEXT Wheat Germ Expression S Kit according to the manufacturer's instructions (CellFree Sciences Co., Ltd.), the bilayer translation method,15, 16, 34 and a robotic synthesizer (GenDecorder 1000; CellFree Sciences). Each DNA template was transcribed by SP6 RNA polymerase, then precipitated with ethanol, and collected by centrifugation (15 000 r.p.m. for 5 min., R10H rotor; Hitachi). Each mRNA (∼30–35 μg) was washed with 75% ethanol, added into a translation mixture, and translated in the bilayer mode31 with the following modifications. The translation mixture (25 μl) (bottom layer) contained 60 A260/ml of ENDEXT wheat germ extract, 1 × SUB-AMIX (24 mM Hepes-KOH, pH 7.8, 1.2 mM ATP, 0.25 mM GTP, 16 mM creatine phosphate, 2.5 mM DTT, 0.4 mM spermidine, 0.3 mM each of the 20 amino acids, 2.8 mM magnesium acetate, 100 mM potassium acetate), 2 μg creatine kinase (Roche Applied Science, Indianapolis, IN, USA), 500 nM -biotin (Nacalai Tesque, Kyoto, Japan), and 1 μl of the wheat cell-free translational mixture that expressed BirA biotin ligase (∼50 ng/μl, BirA: GenBank Accession No. NP_0312927). A 1 × SUB-AMIX solution (125 μl) was placed over the translation mixture. The bilayer was incubated at 26°C for 17 h to allow for protein synthesis. All steps including construction of the DNA templates were performed in the wells of a 96-well plate.
Cleavage assay
The cell-free-synthesized PKs that had luminescent signals >500 units (in the absence of CASP3) were studied. For each PK, 10 μl of the CASP3 cleavage buffer (20 mM Tris-HCl, pH 7.5, 0.2 mM DTT, 5 mM MgCl2, 3 mM ATP, 1 mg/ml BSA, 1 mU CASP3 (Sigma-Aldrich, St. Louis, MO, USA)) was mixed with 1 μl of the translation mixture that contained a Flag-PK-bls∼biotin construct, and the mixture was incubated at 30°C for 2 h in a well of a 384-well Optiplate (Perkin Elmer, Foster City, CA, USA). Using the reagents of an AlphaScreen IgG (protein A) detection kit (Perkin Elmer) according to the manufacturer's instructions, 15 μl of 20 mM Tris-HCl, pH 7.5, 0.2 mM DTT, 5 mM MgCl2, 5 μg/ml anti-FLAG M2 antibody (Sigma-Aldrich), 1 mg/ml BSA, 0.1 μl of streptavidin-coated donor beads and 0.1 μl of anti-IgG acceptor beads were added to the well. The solution was incubated at 23°C for 1 h. Luminescence was analyzed using the AlphaScreen detection program (Perkin Elmer). All repetitive mechanical procedures were performed by a Biomek FX robotic workstation (Beckman Coulter, Fullerton, CA, USA). The value of a luminescent signal is reported as the mean of three independent measurements.
TD immunoblotting
A mixture of each Flag-PK-bls∼biotin construct (3 μl of a translation mixture) and 7 μl of the CASP3 cleavage solution was incubated at 30°C for 1 h in a well of a 384-well Optiplate (Perkin Elmer). Then, the proteins were separated in SDS-PAGE gels and transferred to PVDF membranes (Millipore Bedford, MA, USA). The blotted proteins were prepared for detection using the reagents of an ECL-Plus Western Blotting Detection System kit (GE Healthcare, Piscataway, NJ, USA), anti-Flag M2 antibodies (Sigma-Aldrich) for N-TD, and Alexa488-streptavidin (Invitrogen) for C-TD. The labeled proteins were visualized using a Typhoon Imager (GE Healthcare) with a 532-nm laser and a 526-nm emission filter or an ImageQuant LAS-4000 mini CCD camera system (Fujifilm).
Sequencing and other purification procedures
When possible, long biotinylated C-terminal fragments produced by CASP3 cleavage were recovered attached to streptavidin beads, and then sequenced directly. When a PK construct had a low biotin-labeling efficiency and was cleaved near its C-terminus, a new construct was made by fusing the GST nucleotide sequence encoded in the pEU-E01-Gateway-GST vector to the C-terminal codon of the corresponding PK open-reading frame using the Gateway system and the pEU-E01-Gateway-GST vector. For purification, synthesized PKs (1.2 ml) were purified using Streptavidin Magnesphere Paramagnetic beads (Promega Corp., Madison, WI, USA) for the Flag-PK-bls∼biotin constructs or glutathione Sepharose 4B (GE Healthcare) for the PK-GST constructs. After washing the beads with PBS, the bound PKs were incubated with CASP3 (15 μl of total volume) as described above. The samples were boiled and the proteins separated by SDS-PAGE. After blotting and visualization (ProBlott, Applied Biosystems, Foster City, CA, USA), the membrane areas that contained the cleaved fragments were cut out and the fragments were sequenced (Applied Biosystems ABI 473A). CSK kinase (Carna Biosciences Inc., Kobe, Japan) and AMPKa1 (Cell Signaling Technology, Beverly, MA, USA) were cleaved with CASP3 (10 μl of total volume), and the cleavage products subjected to MALDI/TOF-MS (Shimazu Techno-Research Inc., Kyoto, Japan) for sequencing. D → A mutagenesis was carried out using the reagents of a PrimeSTAR Mutagenesis Basal kit (TakaraBio, Otsu, Japan) according to the manufacturer's instructions. The mutated genes were sequenced using an ABI PRISM 310 DNA sequencer (Applied Biosystems).
Construction of PK expression plasmids for the cell-based assay
Expression plasmids were produced using the Gateway method. To obtain the attB1-PK-Flag-(stop codon)-attB2 for Gateway BP Clonase II recombination, the open-reading frame products of the 30 newly identified PK substrates of CASP3 that had been produced by PCR using the S1 and T1 primers as described above were PCR amplified using the primers, attB1-S1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCACCCACCACCACCA-3′) and T1-Flag-stop-attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTACTTGTCATCGTCATCCTTGTAGTCGCTT CCAGCACTAGCTCCAGA-3′). These PCR products were each inserted into a pDEST26 vector (Invitrogen) using the Gateway system for construction of the His-PK-Flag nucleotide sequences. All sequences were confirmed by DNA sequencing as described above.
Cell-based assay
HeLa cells were cultured in Dulbecco's modified Eagle's medium, 10% fetal bovine serum, penicillin (100 mg/ml), and streptomycin (50 μg/ml). Transient transfections were carried out using Lipofectamine 2000 Transfection Reagent (Invitrogen) according to the manufacturer's instructions. At 24 h after transfection, cells were harvested after apoptosis was induced. Control cells were treated with DMSO and, for apoptosis induction or inhibition, with 20 ng/ml TNFα (Calbiochem, La Jolla, CA, USA) and 100 μM cycloheximide (Chemicon, Temecula, CA, USA) or 125 ng/ml anti-Fas antibody (Medical & Biological Laboratories Co., Ltd., Nagoya, Japan) in the presence (inhibition) or absence (induction) of 100 μM Z-VAD-FMK (Peptide Institute Inc., Osaka, Japan) for 6 h. Cells were washed with PBS and then lysed directly by adding one volume of 2 × SDS-PAGE sample buffer (125 mM Tris-HC1, pH 6.8, 20% glycerol, 4% SDS, 10% 2-mercaptoethanol, 0.001% bromphenol blue) before subjecting the cell extracts to SDS-PAGE and immunoblotting, which used anti-His antibodies (GE Healthcare) or anti-Flag M2 antibodies (Sigma-Aldrich). The following antibodies were employed to detect endogenous proteins: anti-α-tubulin (Sigma-Aldrich); anti-AKT2, anti-eEF2K, anti-AMPKa1, and anti-TGFbR1 (Cell Signaling Technology). Chemiluminescent signals, generated by ECL-Plus reagents (GE Healthcare), or Immobilon Western HRP substrate Luminol Reagent (Millipore), were detected using an LAS-4000 mini biomolecular imager (GE Healthcare).
Acknowledgments
We thank Professor Akihide Ryo (Yokohama City University, Japan) for his useful comments and suggestions concerning the cell analysis, Mr. Tatsuya Akagi for technical assistance, and Dr. Morishita (CellFree Sciences) for assistance with the robotic operations. This work was partially supported by the Special Coordination Funds for Promoting Science and Technology by the Ministry of Education, Culture, Sports, Science, and Technology, Japan (Nos. 19657041 and 22310127 to TS).
Glossary
- CASP3
caspase 3
- PK
protein kinase
- NCtagged
N- and C-terminally tagged
- TD
terminal detection
Dr. Endo is a founder of CellFree Sciences Co., Ltd. and a member of its scientific advisory board. Other authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Supplementary Material
References
- Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- Hunter T. Signaling – 2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Matrisian LM, Hogan BL. Growth factor-regulated proteases and extracellular matrix remodeling during mammalian development. Curr Top Dev Biol. 1990;24:219–259. doi: 10.1016/s0070-2153(08)60089-7. [DOI] [PubMed] [Google Scholar]
- Turgeon VL, Houenou LJ. The role of thrombin-like (serine) proteases in the development, plasticity and pathology of the nervous system. Brain Res Brain Res Rev. 1997;25:85–95. doi: 10.1016/s0165-0173(97)00015-5. [DOI] [PubMed] [Google Scholar]
- van Kempen LC, de Visser KE, Coussens LM. Inflammation, proteases and cancer. Eur J Cancer. 2006;42:728–734. doi: 10.1016/j.ejca.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Tube travel: the role of proteases in individual and collective cancer cell invasion. Cancer Res. 2008;68:7247–7249. doi: 10.1158/0008-5472.CAN-08-0784. [DOI] [PubMed] [Google Scholar]
- Abdel-Rahman HM, Kimura T, Hidaka K, Kiso A, Nezami A, Freire E, et al. Design of inhibitors against HIV, HTLV-I, and Plasmodium falciparum aspartic proteases. Biol Chem. 2004;385:1035–1039. doi: 10.1515/BC.2004.134. [DOI] [PubMed] [Google Scholar]
- Alnemri ES. Mammalian cell death proteases: a family of highly conserved aspartate specific cysteine proteases. J Cell Biochem. 1997;64:33–42. doi: 10.1002/(sici)1097-4644(199701)64:1<33::aid-jcb6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righetti PG, Boschetti E. Sherlock Holmes and the proteome – a detective story. FEBS J. 2007;274:897–905. doi: 10.1111/j.1742-4658.2007.05648.x. [DOI] [PubMed] [Google Scholar]
- Sawasaki T, Ogasawara T, Morishita R, Endo Y. A cell-free protein synthesis system for high-throughput proteomics. Proc Natl Acad Sci USA. 2002;99:14652–14657. doi: 10.1073/pnas.232580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawasaki T, Morishita R, Gouda MD, Endo Y. Methods for high-throughput materialization of genetic information based on wheat germ cell-free expression system. Methods Mol Biol. 2007;375:95–106. doi: 10.1007/978-1-59745-388-2_5. [DOI] [PubMed] [Google Scholar]
- Takai K, Sawasaki T, Endo Y. Practical cell-free protein synthesis system using purified wheat embryos. Nat Protoc. 2010;5:227–238. doi: 10.1038/nprot.2009.207. [DOI] [PubMed] [Google Scholar]
- Endo Y, Sawasaki T. Cell-free expression systems for eukaryotic protein production. Curr Opin Biotechnol. 2006;17:373–380. doi: 10.1016/j.copbio.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Sawasaki T, Hasegawa Y, Morishita R, Seki M, Shinozaki K, Endo Y. Genome-scale, biochemical annotation method based on the wheat germ cell-free protein synthesis system. Phytochemistry. 2004;65:1549–1555. doi: 10.1016/j.phytochem.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Goshima N, Kawamura Y, Fukumoto A, Miura A, Honma R, Satoh R, et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat Methods. 2008;5:1011–1017. doi: 10.1038/nmeth.1273. [DOI] [PubMed] [Google Scholar]
- Sawasaki T, Kamura N, Matsunaga S, Saeki M, Tsuchimochi M, Morishita R, et al. Arabidopsis HY5 protein functions as a DNA-binding tag for purification and functional immobilization of proteins on agarose/DNA microplate. FEBS Lett. 2008;582:221–228. doi: 10.1016/j.febslet.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Nozawa A, Seki M, Shinozaki K, Endo Y, Sawasaki T. A simple and high-sensitivity method for analysis of ubiquitination and polyubiquitination based on wheat cell-free protein synthesis. BMC Plant Biol. 2009;9:39. doi: 10.1186/1471-2229-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Komori H, Nose M, Endo Y, Sawasaki T. Simple screening method for autoantigen proteins using the N-terminal biotinylated protein library produced by wheat cell-free synthesis. J Proteome Res. 2010;9:4264–4273. doi: 10.1021/pr9010553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138:838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Murakawa M, Nishida E, Tsubuki S, Kawashima S, Sakamaki K, et al. Proteolytic activation of MST/Krs, STE20-related protein kinase, by caspase during apoptosis. Oncogene. 1998;16:3029–3037. doi: 10.1038/sj.onc.1201840. [DOI] [PubMed] [Google Scholar]
- Tomiyoshi G, Horita Y, Nishita M, Ohashi K, Mizuno K. Caspase-mediated cleavage and activation of LIM-kinase 1 and its role in apoptotic membrane blebbing. Genes Cells. 2004;9:591–600. doi: 10.1111/j.1356-9597.2004.00745.x. [DOI] [PubMed] [Google Scholar]
- Tatsuta T, Shiraishi A, Mountz JD. The prodomain of caspase-1 enhances Fas-mediated apoptosis through facilitation of caspase-8 activation. J Biol Chem. 2000;275:14248–14254. doi: 10.1074/jbc.275.19.14248. [DOI] [PubMed] [Google Scholar]
- Packard BZ, Komoriya A, Brotz TM, Henkart PA. Caspase 8 activity in membrane blebs after anti-Fas ligation. J Immunol. 2001;167:5061–5066. doi: 10.4049/jimmunol.167.9.5061. [DOI] [PubMed] [Google Scholar]
- Lahti JM, Xiang J, Heath LS, Campana D, Kidd VJ. PITSLRE protein kinase activity is associated with apoptosis. Mol Cell Biol. 1995;15:1–11. doi: 10.1128/mcb.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto Y, Manome Y, Meinhardt G, Kisaki H, Kharbanda S, Robertson M, et al. Proteolytic activation of protein kinase C delta by an ICE-like protease in apoptotic cells. EMBO J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casciola-Rosen LA, Anhalt GJ, Rosen A. DNA-dependent protein kinase is one of a subset of autoantigens specifically cleaved early during apoptosis. J Exp Med. 1995;182:1625–1634. doi: 10.1084/jem.182.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- Ohoka N, Sakai S, Onozaki K, Nakanishi M, Hayashi H. Anaphase-promoting complex/cyclosome-cdh1 mediates the ubiquitination and degradation of TRB3. Biochem Biophys Res Commun. 2010;392:289–294. doi: 10.1016/j.bbrc.2009.12.175. [DOI] [PubMed] [Google Scholar]
- Sawasaki T, Hasegawa Y, Tsuchimochi M, Kamura N, Ogasawara T, Kuroita T, et al. A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 2002;514:102–105. doi: 10.1016/s0014-5793(02)02329-3. [DOI] [PubMed] [Google Scholar]
- Kamura N, Sawasaki T, Kasahara Y, Takai K, Endo Y. Selection of 5′-untranslated sequences that enhance initiation of translation in a cell-free protein synthesis system from wheat embryos. Bioorg Med Chem Lett. 2005;15:5402–5406. doi: 10.1016/j.bmcl.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.