Abstract
Genetic determinants of blood pressure are poorly defined. We undertook a large-scale gene-centric analysis to identify loci and pathways associated with ambulatory systolic and diastolic blood pressure.
We measured 24-hour ambulatory BP in 2020 individuals from 520 white European nuclear families (the GRAPHIC Study) and genotyped their DNA using the Illumina HumanCVD BeadChip array which contains approximately 50000 single nucleotide polymorphisms in >2000 cardiovascular candidate loci. We found a strong association between rs13306560 polymorphism in the promoter region of MTHFR and CLCN6 and mean 24-hour diastolic blood pressure - each minor allele copy of rs13306560 was associated with 2.6 mmHg lower mean 24-hour diastolic blood pressure (P=1.2×10−8). rs13306560 was also associated with clinic diastolic blood pressure in a combined analysis of 8129 subjects from the GRAPHIC Study, the CoLaus Study and the Silesian Cardiovascular Study (P=5.4×10−6). Additional analysis of associations between variants in Gene Ontology-defined pathways and mean 24-hour blood pressure in the GRAPHIC Study showed that cell survival control signalling cascades could play a role in blood pressure regulation. There was also a significant over-representation of rare variants (minor allele frequency <0.05) amongst polymorphisms showing at least nominal association with mean 24-hour blood pressure indicating that a considerable proportion of its heritability may be explained by uncommon alleles.
Through a large scale gene-centric analysis of ambulatory blood pressure, we identified an association of a novel variant at the MTHFR/CLNC6 locus with diastolic blood pressure and provided new insights into the genetic architecture of blood pressure.
Keywords: gene, genetics, blood pressure, single nucleotide polymorphism, association, heritability
Introduction
Raised blood pressure (BP) is the single most important risk factor for cardiovascular diseases worldwide (1). BP is a complex trait with significant heritability (2-4). However, a majority of the causative genes and related molecular mechanisms remains largely unknown. Recent candidate gene studies and the first genome-wide association scans (GWAS) have revealed that at least a fraction of BP-associated genes map to functionally and/or clinically important signalling cascades of cardiovascular regulation (5-9). This indicates that BP gene discovery may be greatly facilitated by large-scale systematic analysis of variants in pathways of cardiovascular regulation. The recently developed Illumina HumanCVD BeadChip, herewith called the 50K IBC array, permits simultaneous genotyping of approximately 50000 common and low frequency single nucleotide polymorphisms (SNPs) in >2000 candidate genes/loci with the highest functional relevance to cardiovascular system (10). The overall genetic coverage for many genes related to cardiovascular disease is denser on this array than on traditional genotyping platforms used in GWAS. Such a selection of genetic variants enables investigations of major cardiovascular pathways at an unprecedented scale without compromising the depth of coverage and allows important questions about the genetic architecture of BP to be examined.
Here we report the results of the first large-scale experiment using the new 50K IBC array in relation to mean 24-hour systolic and diastolic BP (SBP and DBP) in a cohort of subjects whose clinic BP values and the prevalence of hypertension are representative of the British general population (2,5,11).
Methods
Studies, subjects and phenotypes
The major stages of our research strategy are presented in Figure 1. The primary analysis using the 50K IBC array was performed in subjects recruited as part of the Genetic Regulation of Arterial Pressure of Humans in the Community (GRAPHIC) Study. Validation of the main findings from the GRAPHIC Study was carried out in subjects from two additional European Caucasian cohorts, the CoLaus Study and the Silesian Cardiovascular Study.
Figure 1.
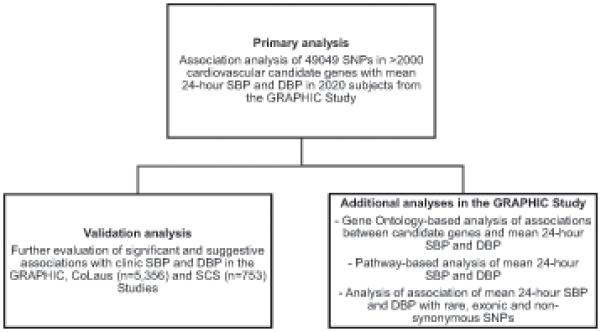
Major stages of the research strategy.
GRAPHIC Study
Details of recruitment and phenotyping of the GRAPHIC subjects are described elsewhere (5). Briefly, nuclear families (all of white European ancestry) with both parents (aged 40-60 years) and two adult (aged ≥18 years) offspring were identified through general practices in Leicestershire, UK (5). Participants had a detailed history taken and were examined by research nurses following standard protocol. Three clinic BP readings were made using an Omron HEM-705CP digital BP monitor. Clinic BP was defined as the mean of the second and third readings. Ambulatory BP was measured using a Spacelabs 90207 monitor (Spacelabs, Wokingham, UK) for 26 hours. The first two hours of each record was discarded to avoid an alerting response (5). Readings were taken at 30 minute intervals between 8 am and 10pm and hourly between 10pm and 8am. The ambulatory BP data were summarised weighing each time period proportional to its length (2). 2037 subjects from 520 families underwent successful ambulatory BP monitoring and were genotyped in this project. Because of low overall genotyping call rate (<90%) or incomplete phenotypic information, 17 individuals were excluded from the current association analysis.
CoLaus Study and Silesian Cardiovascular Study (SCS)
The recruitment strategy and phenotyping of these cohorts have been described elsewhere (12-13) and further details are given in online Data Supplement (please see http://hyper.ahajournals.org). Full genetic and phenotypic information was available for 5356 and 753 biologically unrelated subjects from CoLaus and SCS, respectively.
The studies were conducted according to the principles expressed in the Declaration of Helsinki, approved by their respective local bioethical committees and all subjects gave informed written consents for participation.
Genotyping and quality control filters
Genotyping platform
Detailed description of the strategy for the selection of genes on the Illumina HumanCVD BeadChip are provided elsewhere (10). A list of the loci on the array is available at http://bmic.upenn.edu/cvdsnp/. Further description of tagging of loci is provided in Online Data Supplement (http://hyper.ahajournals.org).
Primary genetic association analysis
200 ng of leukocyte DNA from each subject in GRAPHIC Study was hybridised to 50K IBC arrays (version 2) on an Illumina Bead Station 500 (Illumina, Inc.), using protocols specified by the manufacturer. Alleles were called using the Illumina BeadStudio (v3) Genotyping Module (based on GenCall Software algorithms for clustering, calling and scoring genotypes). The quality of cluster plots was visually inspected by 2 independent investigators.
DNA analysis in the CoLaus cohort and the SCS
Genotypes of SNPs showing significant and suggestive associations with mean 24-hour BP in GRAPHIC Study (n=17) were extracted in silico from GWAS conducted in the CoLaus cohort using the Affymetrix Genome-Wide Human SNP Array 5.0. Of these 5 SNPs were directly genotyped by the Affymetrix array and 10 were imputed. Information on 2 SNPs (rs4648310 and rs17037388) was not available. The SNP that showed consistent association with BP in GRAPHIC and CoLaus (rs13306560) was genotyped in the SCS using commercially available TaqMan assays on ABI PRISM 7900HT Sequence Detection System (Applied Biosystems).
Statistical analysis
Heritability of BP was estimated using an algorithm implemented in Sequential Oligogenic Linkage Analysis Routines (SOLAR v2.0) software (14). The χ2 test was used in the parental generation of the GRAPHIC Study to examine whether distributions of genotypes for each SNP on the 50K IBC array were concordant with Hardy-Weinberg equilibrium. SNPs which passed this and additional filters (see Online Data Supplement http://hyper.ahajournals.org) were examined for their associations with BP under an additive model of inheritance using generalized estimating equations (GEE) with exchangeable correlation structure to account for familial correlations and with adjustment for age, age2 and sex. Association analysis of top SNPs in the GRAPHIC Study with clinic BP in the CoLaus and SCS cohorts was based on age, age2 and sex adjusted linear regression models fitted in PLINK http://pngu.mgh.harvard.edu/purcell/plink/ (15) under additive model of inheritance. BP values of subjects on antihypertensive treatment in all cohorts were adjusted for BP-lowering effect of therapy using a semi-parametric algorithm in GRAPHIC and SCS (16-17) or adding a constant of 15 mmHg (to SBP) and 10 mmHg (to DBP) in CoLaus (9). Genetic effects are shown as β-coefficients (β) per each extra minor allele copy of a SNP with the respective standard error (SE).
Two sensitivity analyses [(i) no treatment correction, (ii) exclusion of subjects on antihypertensive treatment] were undertaken to examine if antihypertensive therapy had any impact on the observed associations.
To correct for multiple testing in the GRAPHIC Study for each association with the two principal phenotypes (mean 24-hour SBP and DBP) we calculated false positive discovery rate (q-values) based on the method proposed by Storey and Tibshirani (18) and available in the QVALUE software (http://genomine.org/qvalue/). The q-value thresholds of 0.05 and 0.25 were selected to identify the findings that represent significant and suggestive associations, respectively.
Fixed-effect meta-analysis of association between rs13306560 and clinic BP in the GRAPHIC, CoLaus and SCS studies was conducted under additive model of inheritance using inverse variance weighted averages of β-coefficients and SE in METAL (http://www.sph.umich.edu/csg/abecasis/metal/index.html). The between-study heterogeneity was evaluated using χ2 test.
Two-tail Fisher’s exact test was used to analyse the distributions of rare, exonic and non-synonymous variants across different strata of associations with mean 24-hour mean SBP and DBP in the GRAPHIC Study.
The power estimates of the GRAPHIC Study to detect both nominal (P<0.05) and Bonferroni adjusted (33577 tests) effects of various sizes for mean 24-hour SBP and DBP across a range of different minor allele frequencies were derived using the average SE observed at each allele frequency and constructed in STATA.
An extended description of the statistical analysis is available in Online Data Supplement (http://hyper.ahajournals.org).
Bioinformatic analysis
All SNPs on the 50K IBC array were annotated to specific loci using resources available in public domain including information on design of the array at http://bmic.upenn.edu/cvdsnp/, NCBI (Built 36.3) and SNP Function Portal - web-based application for analysis of human genetic variants (19). To examine in more detail associations between mean 24-hour BP and genes previously linked to SBP and DBP we identified such genes using the Gene Ontology Annotation Database (GOA) (http://www.ebi.ac.uk/GOA/downloads.html). The details of this GOA-based strategy are shown in Table S1. Identification of processes associated with mean 24-hour SBP and DBP was conducted using Gene set-based analysis of polymorphisms (GeSBAP) web interface (20). Gene ontology (GO) database was used as a reference repository of functionally annotated biological processes.
An extended description of the bioinformatic analysis is provided in Online Data Supplement (http://hyper.ahajournals.org).
Results
Subjects
The characteristics of the GRAPHIC subjects included in the analysis are shown in Table 1. The relevant characteristics of the CoLaus and SCS cohorts are listed in Table S2.
Table 1.
Characteristics of the GRAPHIC Study
| Variable | Fathers (n=511) | Mothers (n=512) | Sons (n=508) | Daughters (n=489) |
|---|---|---|---|---|
| Age (years) | 53.8 (4.3) | 51.8 (4.4) | 25.0 (5.1) | 25.9 (5.4) |
| Body mass index (kg/m2) | 27.8 (3.9) | 27.1 (4.5) | 24.9 (4.1) | 24.6 (5.0) |
| Clinic SBP (mmHg) | 137.8 (18.4) | 128.5 (18.3) | 127.7 (13.0) | 113.8 (11.9) |
| Mean 24-hour SBP (mmHg) | 124.3 (11.5) | 117.0 (11.4) | 120.8 (8.1) | 112.8 (7.2) |
| Clinic DBP (mmHg) | 86.0 (10.7) | 80.8 (10.4) | 75.9 (9.6) | 73.6 (8.6) |
| Mean 24-hour DBP (mmHg) | 77.7 (7.2) | 71.7 (7.6) | 69.2 (6.5) | 67.9 (5.2) |
| Antihypertensive treatment (%) | 77 (15.1) | 53 (10.4) | 3 (0.6) | 1 (0.2) |
Data are means and standard deviations or counts and percentages
Heritability of mean 24-hour and clinic BP in the GRAPHIC Study
The narrow sense heritability (h2) of mean 24-hour SBP was marginally higher than that of clinic SBP (h2=0.33, SE=0.05, p=8.4×10−14 vs. h2=0.31, SE=0.04, p=1.2×10−13, respectively). The mean 24-hour DBP showed more significant heritability than clinic DBP (h2=0.41, SE=0.05, p=3.7×10−20 vs. h2=0.32, SE=0.04, p=3.3×10−15, respectively).
Associations between SNPs on the 50K IBC array and mean 24-hour SBP and DBP in the GRAPHIC cohort
Of 49094 genotyped SNPs, 15517 SNPs were excluded from analysis for reasons given in Online Data Supplement (http://hyper.ahajournals.org). Of 33577 SNPs that passed all quality filters and were included in further analysis, 32939 were annotated to 3036 loci. The remainder (n=638) were mapped to hypothetical genes.
The distributions of nominal P-values for the associations of mean 24-hour SBP and DBP are shown in Figure S1. The power to detect associations with each principal phenotype is shown in Figure S2. The genomic control (λ) coefficients computed for mean 24-hour SBP and DBP showed no inflation of the association statistic driven by stratification (λ=0.972 and λ=0.973, respectively). 1782 and 1842 SNPs that passed quality filters showed at least nominal (p<0.05) association with mean 24-hour SBP and DBP, respectively. These nominal associations mapped to 738 (mean 24-hour SBP) and 762 (mean 24-hour DBP) loci.
After calculation of false positive discovery rate, only one SNP (rs2797221) in the Usher syndrome type IIa protein gene (USH2A) showed a significant association with mean 24-hour SBP - each extra minor allele copy of rs2797221 was associated with 3.9 mmHg lower adjusted mean 24-hour SBP (p=8.6×10−7, q=0.0285; Table 2). Four other SNPs (rs9892909 in SPHK1, rs2283210 in KCNQ1, rs2623410 in PDE1A and rs443095 in THBS4) showed suggestive associations with mean 24-hour SBP (Table S3). The associations of these 5 SNPs with mean 24-hour DBP are presented in Table 2 and Table S4.
Table 2.
SNPs showing significant associations with mean 24-hour BP in the GRAPHIC Study
| Ch | Locus | SNP | Minor (coded)/ major allele |
MAF | HWE | β (SE) | P-value | q-value | β (SE) | P-value | q-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| mean 24-hour SBP | mean 24-hour DBP | ||||||||||
| 1 | USH2A | rs2797221 | T/C | 0.0112 | 1 | −3.92 (0.79) |
8.6×10−7 | 0.0285 | −2.44 (0.98) |
0.0131 | 0.7796 |
| 1 | MTHFR/CLCN6 | rs13306560 | A/G | 0.0527 | 0.1963 | −3.27 (0.72) |
6.1×10−6 | 0.0815 | −2.63 (0.46) |
1.2×10−8 | 0.0008 |
Ch – chromosome; SNP – single nucleotide polymorphism; MAF – minor allele frequency; HWE – statistical significance of Hardy-Weinberg equilibrium test (in parents); β – estimated quantitative effect of each SNP minor allele copy on mean 24-hour blood pressure (adjusted for age, age2, sex and antihypertensive medication); a negative β indicates lower BP in carriers of the minor allele; SE – standard error; P-value – level of statistical significance; q-value – false positive discovery rate; USH2A – Usher syndrome 2A (autosomal recessive, mild); MTHFR – 5,10-methylenetetrahydrofolate reductase (NADPH); CLCN6 - chloride channel 6
Only one SNP [rs13306560 in the region between 5,10-methylenetetrahydrofolate reductase (NADPH) gene (MTHFR) and chloride channel 6 gene (CLCN6)] showed a significant association with mean 24-hour DBP after calculation of false positive discovery rate - each minor allele copy of rs13306560 was associated with 2.6 mmHg lower mean 24-hour DBP (p=1.2×10−8, q=0.0008, Table 2). Four additional MTHFR SNPs (rs17037388, rs17037390, rs17367504 and rs13306561) as well as 8 other polymorphisms (rs4648310 in PTGS2, rs11801879 in NPPB, rs925596 in NLRP1, rs6993502 in CPA6, rs2280232 in STAT1, rs1035798 in AGER, rs873985 in CISH, rs12603626 in NOG) were associated with mean 24-hour DBP at a suggestive level of statistical significance after calculation of false positive discovery rate (Table S5). The associations of these 13 SNPs with mean 24-hour SBP are presented in Table 2 and Table S4.
In sensitivity analyses, the significant and suggestive SNPs retained their associations with mean 24-hour SBP and DBP even without using antihypertensive treatment correction or after exclusion of subjects on antihypertensive treatment (Tables S6-S7).
Evaluation of significant and suggestive association signals identified in analysis of mean 24-hour BP using clinic BP in the GRAPHIC Study, CoLaus cohort and Silesian Cardiovascular Study
The majority of SNPs showing significant or suggestive associations with mean 24-hour SBP (n=5) and DBP (n=13) were also associated (in most instances at a lower level of significance) with clinic BP in the GRAPHIC Study (Table 3 and Table S4). However, only one of the associations identified in analysis of mean 24-hour BP in the GRAPHIC Study was confirmed in relation to clinic BP in the CoLaus cohort (rs13306560 and clinic DBP; p=0.0322, r2hat imputation coefficient – 0.88, Table 3). The other SNPs were not associated with clinic BP in the CoLaus Study (Table S8). Further analysis based on direct genotyping of rs13306560 in Silesian Cardiovascular Study confirmed the association of rs13306560 with clinic DBP (p=0.0094; Table 3). The direction of allelic association of rs13306560 and clinic DBP was consistent across all examined populations. In inverse variance weighted fixed effects meta-analysis of the three studies each minor allele copy of rs13306560 was associated with an approximately 1.9 mmHg lower clinic DBP (P=5.4×10−6) (Table 3).
Table 3.
rs13306560 and clinic BP in the GRAPHIC Study and validation cohorts
| Study | Informative subjects |
Minor (coded) allele |
clinic SBP | clinic DBP | ||
|---|---|---|---|---|---|---|
| β (SE) | P-value | β (SE) | P-value | |||
| GRAPHIC | 2,020 * | A | −3.07 (1.06) | 0.0039 | −2.60 (0.68) | 0.0001 |
| CoLaus † | 5,356 * | −1.20 (0.85) | 0.1569 | −1.16 (0.54) | 0.0322 | |
| SCS | 753 * | −6.17 (3.01) | 0.0407 | −4.53 (1.74) | 0.0094 | |
| Combined ‡ | 8,129 * | −2.12 (0.65) | 0.0011 | −1.87 (0.41) | 5.4×10−6 | |
CoLaus – Swiss population-based study; SCS – Silesian Cardiovascular Study; β - individual β-coefficient; SE – standard error; P – values were obtained from tests adjusted for age, age2, sex, antihypertensive treatment and familial correlations (where appropriate)
number of subjects with available genotype and phenotype information
rs13306560 was imputed in the CoLaus cohort (r2hat=0.88)
based on inverse variance-weighted fixed effects meta-analysis; P-values of heterogeneity test were 0.1512 and 0.0744 for SBP and DBP, respectively
rs13306560 and 24-hour BP in the GRAPHIC Study – additional analyses
Body mass index did not have an effect on the association of rs13306560 with mean 24-hour DBP (Table S9). We also examined the association of rs13306560 with mean daytime (10.00-22.00) and nocturnal (00.00-6.00) DBP in the GRAPHIC Study. The SNP was associated with DBP at both time-periods; the association with nocturnal DBP was stronger than that of daytime DBP (daytime: β=−2.30, SE=0.56, P=3.4×10−5 and night-time: β=−3.37, SE=0.51, P=2.9×10−11).
MTHFR/CLCN6/NPPB locus – linkage disequilibrium and functional analysis in silico
rs13306560 maps to the promoter region of MTHFR and CLCN6 (http://genome.ucsc.edu/cgi-bin/hgTracks). Four other MTHFR SNPs (rs17037388, rs17037390, rs17367504, rs13306561) were also associated with mean 24-hour DBP at q<0.25 (Table S5). One of these SNPs, rs17367504, was identified as the lead association signal at this locus in a recent GWAS meta-analysis of clinic BP (9). These four additional SNPs are in strong LD [as measured by r2 (r2>0.8)] with each other (Figure 2). However, they are only in a very weak LD with rs13306560 (r2=0.27-0.28). rs13306560 is also in weak LD (r2=0.3 in the GRAPHIC Study) with rs11801879 - natriuretic peptide precursor B (NPPB) polymorphism showing suggestive association with mean 24-hour DBP in our discovery cohort (Figure 2 and Table S5). Genetic variation within this locus was previously associated with BP in subjects of European ancestry (7). However, our conditional analysis (see Online Data Supplement http://hyper.ahajournals.org) indicated that the association signal at rs17367504 and rs11801879 were both driven by their linkage disequilibrium (LD) with rs13306560. HapMap-based LD examination in Utah residents with ancestry from northern and western Europe (CEU population) revealed only one polymorphism (rs13306567) as a statistically similar proxy of rs13306560 (r2=0.83) within its adjacent 500 KB (Figure S3). The proxy maps to intron 3 of MTHFR (5178 bps from rs13306560) and is not present on the 50K IBC array. All the other SNPs within the 500 KB distance of rs13306560 exhibit very weak LD with it (all r2<0.4) (Figure S3). rs13306560 is the only confirmed polymorphism within the intergenic junction of MTHFR and CLCN6 (Figure S4). This short segment shows a high GC content (64.4%) and is a part of the larger 1104-bp CpG island spanning the 5′ flanking regions of both genes (Figure S4). The region lies within 6 quantitative trait loci (QTLs) for BP identified in rats and 1 QTL for atherosclerosis in mouse (Figure S4). It exhibits significant conservation across placental mammals and scores high in evolutionary and sequence pattern extraction through reduced representations (ESPERR) programme (that discriminates enhancer elements from neutral DNA sites with >90% accuracy) (21). rs13306560 lies in the centre of the most conserved part of this region – its ancestral allele is conserved not only in primates and other mammals but also in higher vertebrates (opossum) .
Figure 2. MTHFR/CLCN6/NPPB locus - association with mean 24-hour DBP in the GRAPHIC Study.
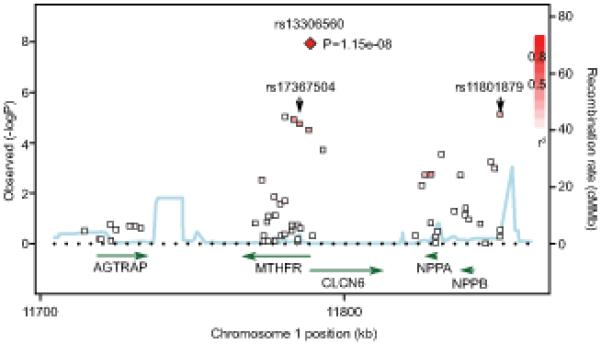
Associations of individual SNPs with mean 24-hour DBP in the GRAPHIC Study are plotted as −log10P against chromosomal base pair position. rs13306560 is shown as a red diamond and its LD relationship with the other markers (including rs17367504 and rs11801879) is indicated by colour shading (whereby: red - r2>0.8, white - r2<0.2). The locations of known genes in the region are shown in green and the recombination hotspots are represented as blue peaks. MTHFR - 5,10-methylenetetrahydrofolate reductase (NADPH) gene, CLCN6 – chloride channel 6 gene, NPPB - natriuretic peptide precursor B gene, NPPA - natriuretic peptide precursor A gene, AGTRAP - angiotensin II receptor-associated protein gene.
Associations between mean 24-hour SBP and DBP and BP-relevant candidate genes
To specifically investigate genes on the 50K IBC array with direct relevance to BP, we first identified gene ontology (GO) categories that include “blood pressure” terms using the GOA database (Table S1). This revealed 113 GO processes with 129 genes, of which 110 were present on the array. 105 of these genes were well-tagged (Tier 1 or 2) (Tables S10-S11). In total, 1982 SNPs were typed in these genes. Of 105 sufficiently tagged genes, only 3 (2.9%) (NPPB, KCNQ1 and PTGS2) had SNPs within the stratum of suggestive associations (q<0.25) with mean 24-hour SBP or DBP. Of the remaining 102 genes only one (NPPA) had SNPs associated with mean 24-hour BP below the threshold of significance above which associations could be attributed to pure chance only (q≥0.5) (Tables S10-S11).
We also examined the coverage on the 50K IBC array for 17 SNPs recently implicated in BP regulation through large scale meta-analyses of GWAS (8,9). Apart from the MTHFR variant discussed earlier, only 4 other SNPs were directly represented or had good proxies on the array. Of these, 2 SNPs (rs3184504 in SH2B3 locus and rs17696736 in the C12orf30 locus; both on chromosome 12) showed evidence of association. Both the direction of allelic association and effect size estimates of these 2 SNPs were consistent with the data from the meta-analyses of GWAS (Table S12).
Pathwayomic analysis of mean 24-hour SBP and DBP
GeSBAP analysis identified 5 biological processes as significantly associated with mean 24-hour BP after correction for multiple testing (Table S13). Of these, 2 processes (protein kinase cascade - GO:0007243 and regulation of cell proliferation - GO:0042127) were implicated in the regulation of both mean 24-hour SBP and DBP (Table S13).
Rare, exonic and non-synonymous variants and mean 24-hour SBP and DBP
There were no significant differences in distribution of exonic or non-synonymous variants among SNPs showing at least nominal (p<0.05) associations with mean 24-hour BP and those represented overall on the genotyping platform (Table 4). On the other hand, rare SNPs (those with MAF<0.05) were over-represented within the stratum of variants nominally associated with both mean 24-hour SBP and DBP (P=8.7×10−5 and P=0.0036, respectively) (Table 4). This difference became even more striking when applying a lower threshold for definition of uncommon polymorphisms (MAF<0.02); SNPs with MAF below this cut-off were almost twice more common amongst variants associated with mean 24-hour BP than expected by chance (P=3.0×10−6 and P=2.4×10−7 for mean 24-hour SBP and DBP, respectively) (Table 4). However, neither exonic nor exclusively non-synonymous SNPs were over-represented within the stratum of significant rare variants (Table 4), irrespective of the criterion of rare variant definition. Sensitivity analysis conducted using a lower level of statistical significance (p<0.01) confirmed these findings (Table S14).
Table 4.
Distribution of SNPs associated nominally (P<0.05) with mean 24-hour BP according to their characteristic and frequency
| Descriptor | Illumina gene- centric array |
SNPs associated with mean 24-hour SBP at p<0.05 |
P-value | SNPs associated with mean 24-hour DBP at p<0.05 |
P-value |
|---|---|---|---|---|---|
| All SNPs | 33,577 | 1,782 | - | 1,842 | - |
| Exonic SNPs | 2,298 (6.8%) | 126 (7.1%) | 0.7004 | 138 (7.5%) | 0.2770 |
| Non-synonymous SNPs | 1,634 (4.9%) | 84 (4.7%) | 0.8211 | 94 (5.1%) | 0.6175 |
| All SNPs with MAF<0.05 | 4,355 (13.0%) | 290 (16.3%) | 8.7×10−5 | 283 (15.4%) | 0.0036 |
| Exonic SNPs with MAF<0.05* | 516 (11.8%) | 32 (11.0%) | 0.7778 | 34 (12.0%) | 0.9244 |
| Non-synonymous SNPs with MAF<0.05* | 406 (9.3%) | 24 (8.3%) | 0.6021 | 24 (8.5%) | 0.7507 |
| All SNPs with MAF<0.02 | 1,188 (3.5%) | 104 (5.8%) | 3.0×10−6 | 112 (6.1%) | 2.0×10−7 |
| Exonic SNPs with MAF<0.02† | 162 (13.6%) | 14 (13.5%) | 1.0 | 20 (17.9%) | 0.2530 |
| Non-synonymous SNPs with MAF<0.02† | 128 (10.8%) | 10 (9.6%) | 0.8685 | 14 (12.5%) | 0.5292 |
SNP – single nucleotide polymorphism, MAF – minor allele frequency
percentage calculated in relation to total number of SNPs with MAF<0.05
percentage calculated in relation to total number of SNPs with MAF<0.02
Discussion
Our study has uncovered novel alleles, genes and pathways of BP regulation whilst showing that a majority of the previously examined candidates are not (or only weakly) associated with mean 24-hour BP. Mean 24-hour BP probably does not provide distinct information compared with clinic SBP and DBP. However, it shows higher reproducibility, closer correlations with indices of target organ damage and better predictive value of cardiovascular morbidity (22,23). The higher heritability of mean 24-hour BP than that of clinic measurements demonstrated particularly well for DBP in the GRAPHIC families suggests that 24-hour ambulatory BP monitoring (ABPM) may be particularly informative in gene discovery. However, because of unavailability of replication resource with 24-hour ABPM we validated our primary association findings by using clinic BP measurements in other independent cohorts.
Our findings need to be interpreted in the context of the power of our study. At a nominal level of significance, given the greater precision of our BP phenotypes and the population size, our study had a good power to detect effect sizes of as low as 1 mmHg for variants with MAF>10% (Figure S2). With Bonferroni correction, the power was significantly diminished because of the large number of SNPs analyzed (Figure S2). However, Bonferroni adjustment is probably over-conservative in context of SNPs on this array, given their significant LD-driven correlations and non-independence. Nonetheless, our top signal (rs13306560) from analysis of mean 24-hour DBP survived correction for multiple testing based on q-value calculations. The identified association would retain its statistical significance even after application of more conservative thresholds based on Bonferroni correction (1.5×10−6) or those used in previous GWAS (5×10−8). rs13306560 also gave consistent associations in relation to clinic BP in >8000 subjects from 3 European populations making it unlikely that it represents a false positive finding.
Indeed, our conditional analysis indicates that the association with rs13306560 may largely explain the association between BP and other SNPs at the MTHFR/CLCN6/NPPB locus reported in recent GWAS (8-9). Quantitatively, the magnitude of rs13306560 effect on BP is one of the largest genetic effects that have been reported to date for a relatively common variant. Furthermore, rs13306560 shows a significant functional potential itself in silico. Indeed, the alleles of rs13306560 are conserved in mammals and the SNP maps to CpG island in the MTHFR/CLCN6 promoter with evidence of operation of selective pressure (to maintain the sequence in this DNA fragment) throughout mammalian evolution. One possible mechanism by which rs13306560 could therefore mediate the association with BP is through differential methylation of the MTHFR/CLCN6 promoter in the two alleles. MTHFR has a strong biological potential to influence BP regulation. It encodes an enzyme that catalyses conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, a co-substrate for homocysteine remethylation to methionine (24). Circulating concentrations of homocysteine have been associated with risk of cardiovascular diseases including hypertension, possibly through promotion of free radical-mediated endothelial damage and dysfunction (25). One of the common non-synonymous MTHFR polymorphisms (rs1801133, −C677T or A222V) known to produce a thermolabile enzyme isoform with reduced activity in TT homozygotes (25) was over-represented in patients with hypertension (26). However, rs1801133 is not in LD with rs13306560 (r2=0.02) and showed only nominal association with mean 24-hour DBP in the GRAPHIC study. rs1801133 was also only weakly associated with clinic DBP in the recent GWAS meta-analysis of BP (9). Thus, it is unlikely that rs13306560 mediates its effect on BP through the thermolabile form of MTHFR.
CLCN6 belongs to a family of molecules that function as either transmembrane ion channels or electrogenic Cl−/H+ exchangers (27). CLCN6 protein is mainly expressed in the central and peripheral nervous system where it resides within vesicular endosomes (27). Experimental disruption of CLCN6 leads to a lysosomal storage disease; CLCN6 knockout mice show reduced pain sensitivity and mild behavioral abnormalities (27). CLCN6 mRNA expression was confirmed in the kidney but whether, like other members of the chloride transporters family (CLCNKB, CLCN5), it may contribute to renal ion handling is not known. Importantly, mutations in several chloride channels genes underlie monogenic forms of low blood pressure (CLCNKB) (5) and defects in tubular ion reabsorption leading to chronic kidney disease (CLCN5) (28). While the location of rs13306560 in the promoter of both MTHFR and CLNC6 makes them strong candidates, recent studies have shown that polymorphisms can act by influencing more distally located genes (29). Therefore, at this stage we cannot exclude MTHFR, CLCN6 or in fact other genes in this locus (i.e. NPPA and NPPB) as the mediator(s) of the identified association (Figure 2).
A notable finding in our study is that very few variants of the most frequently examined candidates for BP (such as genes of the renin-angiotensin and sympathetic nervous systems) which were present on 50K IBC array showed associations with mean 24-hour BP. These data are consistent with recent GWAS which also did not report any definite associations with such genes (9). The very good genetic coverage (exceeding that of the GWAS platforms) for a majority of these strong candidate genes makes it unlikely that any of their common variants (although not genotyped directly) were not captured in this analysis. This makes our investigation one of the most comprehensive and detailed studies of these systems for their genetic impact on BP. Collectively, these results clearly suggest that common genetic variants underlying familial predisposition to hypertension may reside outside classical systems of BP regulation (30). In respect to this, our pathwayomic analysis has uncovered that signalling cascades that control cell survival through promotion of cellular growth (i.e. mitogens activated through protein kinase cascades) and death (via apoptosis) may play a role in BP regulation. Future systematic analyses of these pathways may identify novel genetic determinants of human hypertension and illuminate novel molecular mechanisms of BP regulation.
Our data also show a consistent over-representation of rare alleles amongst the SNPs associated with mean 24-hour BP. Indeed, irrespective of the threshold of rare variant definition, stratum of statistical significance and the phenotype, rare SNPs were associated with BP more frequently than expected by chance. Most strikingly, rare SNPs (MAF<0.05) constitute also the majority of significant and suggestive associations with mean 24-hour SBP (Table 2 and Table S3). Interestingly, these over-represented rare variants do not lead to amino acid substitutions within the encoded proteins and map to extra-exonic regions. These results lend support to the hypothesis that rare alleles play a role in genetic predisposition to blood pressure elevation(31) and indicate that more subtle mechanisms than those leading to changes in amino acid structure of encoded proteins may underlie human hypertension.
Perspectives.
This large scale gene-centric analysis identified a novel association between a potentially functional variant at the MTHFR/CLNC6/NPPB locus and BP. We also show that both rare variants and genes that reside outside the classical physiological pathways of BP regulation may be important elements of the genetic architecture of BP. Future studies should focus on replication of these findings in larger cohorts and elucidation of the functional mechanisms that underlie the uncovered associations.
Supplementary Material
Acknowledgments
We thank the research nurses and other staff who undertook the recruitment and phenotyping in the studies.
Source of funding
Recruitment and genotyping of the GRAPHIC cohort was funded by the British Heart Foundation (BHF). The CoLaus study was supported by research grants from GlaxoSmithKline and from the Faculty of Biology and Medicine of Lausanne, Switzerland, and is currently supported by the Swiss National Science Foundation (33CSCO-122661). Silesian Cardiovascular Study was supported by NIH Fogarty International Research Collaboration Award (R03 TW007165 to M.T.). N.J.S. holds a BHF Chair of Cardiology and V.C is supported by the BHF. M.D.T. holds a Medical Research Council Clinician Scientist Fellowship (G0501942). This study is part of the research portfolio supported by the Leicester NIHR Biomedical Research Unit in Cardiovascular Disease.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest/Disclosures
Drs Dawn Waterworth, Kijoung Song and Vincent Mooser are employees of GSK.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guilbert JJ. The world health report 2002 - reducing risks, promoting healthy life. Educ Health (Abingdon) 2003;16:230. doi: 10.1080/1357628031000116808. [DOI] [PubMed] [Google Scholar]
- 2.Tobin MD, Raleigh SM, Newhouse S, Braund P, Bodycote C, Ogleby J, Cross D, Gracey J, Hayes S, Smith T, Ridge C, Caulfield M, Sheehan NA, Munroe PB, Burton PR, Samani NJ. Association of WNK1 gene polymorphisms and haplotypes with ambulatory blood pressure in the general population. Circulation. 2005;112:3423–3429. doi: 10.1161/CIRCULATIONAHA.105.555474. [DOI] [PubMed] [Google Scholar]
- 3.Luft FC. Twins in cardiovascular genetic research. Hypertension. 2001;37:350–356. doi: 10.1161/01.hyp.37.2.350. [DOI] [PubMed] [Google Scholar]
- 4.Charchar F, Zimmerli L, Tomaszewski M. The pressure of finding human hypertension genes: New tools, old dilemmas. J Hum Hypertens. 2008;22:821–828. doi: 10.1038/jhh.2008.67. [DOI] [PubMed] [Google Scholar]
- 5.Tobin MD, Tomaszewski M, Braund PS, Hajat C, Raleigh SM, Palmer TM, Caulfield M, Burton PR, Samani NJ. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension. 2008;51:1658–1664. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 6.Tomaszewski M, Charchar FJ, Lynch MD, Padmanabhan S, Wang WY, Miller WH, Grzeszczak W, Maric C, Zukowska-Szczechowska E, Dominiczak AF. Fibroblast growth factor 1 gene and hypertension: From the quantitative trait locus to positional analysis. Circulation. 2007;116:1915–1924. doi: 10.1161/CIRCULATIONAHA.107.710293. [DOI] [PubMed] [Google Scholar]
- 7.Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, Guiducci C, Kathiresan S, Benjamin EJ, Struck J, Morgenthaler NG, Bergmann A, Blankenberg S, Kee F, Nilsson P, Yin X, Peltonen L, Vartiainen E, Salomaa V, Hirschhorn JN, Melander O, Wang TJ. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41:348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Wellcome Trust Case Control Consortium. Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;44:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joint Health Surveys Unit . Health Survey for England 2003. Volume 2: Risk factors for cardiovascular disease. The Stationery Office; London: 2004. [Google Scholar]
- 12.Firmann M, Mayor V, Vidal PM, Bochud M, Pecoud A, Hayoz D, Paccaud F, Preisig M, Song KS, Yuan X, Danoff TM, Stirnadel HA, Waterworth D, Mooser V, Waeber G, Vollenweider P. The CoLaus study: A population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomaszewski M, Charchar FJ, Barnes T, Gawron-Kiszka M, Sedkowska A, Podolecka E, Kowalczyk J, Rathbone W, Kalarus Z, Grzeszczak W, Goodall AH, Samani NJ, Zukowska-Szczechowska E. A common variant in low-density lipoprotein receptor-related protein 6 gene (LRP6) is associated with LDL-cholesterol. Arterioscler Thromb Vasc Biol. 2009;29:1316–1321. doi: 10.1161/ATVBAHA.109.185355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almasy L, Blangero J. Multipoint quantitative trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH. Evidence for a gene influencing blood pressure on chromosome 17. genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the Framingham Heart Study. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 17.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: Antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–35. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 18.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Dai M, Xuan W, McEachin RC, Jackson AU, Scott LJ, Athey B, Watson SJ, Meng F. SNP function portal: A web database for exploring the function implication of SNP alleles. Bioinformatics. 2006;22:e523–529. doi: 10.1093/bioinformatics/btl241. [DOI] [PubMed] [Google Scholar]
- 20.Medina I, Montaner D, Bonifaci N, Pujana MA, Carbonell J, Tarraga J, Al-Shahrour F, Dopazo J. Gene set-based analysis of polymorphisms: Finding pathways or biological processes associated to traits in genome-wide association studies. Nucleic Acids Res. 2009;37:W340–344. doi: 10.1093/nar/gkp481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor J, Tyekucheva S, King DC, Hardison RC, Miller W, Chiaromonte F. ESPERR: Learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmas W, Moran A, Pickering T, Eimicke JP, Teresi J, Schwartz JE, Field L, Weinstock RS, Shea S. Ambulatory pulse pressure and progression of urinary albumin excretion in older patients with type 2 diabetes mellitus. Hypertension. 2006;48:301–308. doi: 10.1161/01.HYP.0000232644.98208.65. [DOI] [PubMed] [Google Scholar]
- 23.Khattar RS, Swales JD, Banfield A, Dore C, Senior R, Lahiri A. Prediction of coronary and cerebrovascular morbidity and mortality by direct continuous ambulatory blood pressure monitoring in essential hypertension. Circulation. 1999;100:1071–1076. doi: 10.1161/01.cir.100.10.1071. [DOI] [PubMed] [Google Scholar]
- 24.Verhoeff BJ, Trip MD, Prins MH, Kastelein JJ, Reitsma PH. The effect of a common methylenetetrahydrofolate reductase mutation on levels of homocysteine, folate, vitamin B12 and on the risk of premature atherosclerosis. Atherosclerosis. 1998;141:161–166. doi: 10.1016/s0021-9150(98)00156-7. [DOI] [PubMed] [Google Scholar]
- 25.Bellamy MF, McDowell IF. Putative mechanisms for vascular damage by homocysteine. J Inherit Metab Dis. 1997;20:307–315. doi: 10.1023/a:1005377310872. [DOI] [PubMed] [Google Scholar]
- 26.Qian X, Lu Z, Tan M, Liu H, Lu D. A meta-analysis of association between C677T polymorphism in the methylenetetrahydrofolate reductase gene and hypertension. Eur J Hum Genet. 2007;15:1239–1245. doi: 10.1038/sj.ejhg.5201914. [DOI] [PubMed] [Google Scholar]
- 27.Poet M, Kornak U, Schweizer M, Zdebik AA, Scheel O, Hoelter S, Wurst W, Schmitt A, Fuhrmann JC, Planells-Cases R, Mole SE, Hubner CA, Jentsch TJ. Lysosomal storage disease upon disruption of the neuronal chloride transport protein ClC-6. Proc Natl Acad Sci U S A. 2006;103:13854–13859. doi: 10.1073/pnas.0606137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milliner DS. Stones, bones, and heredity. Acta Paediatr Suppl. 2006;95:27–30. doi: 10.1111/j.1651-2227.2006.tb02411.x. [DOI] [PubMed] [Google Scholar]
- 29.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, Yao K, Kehoe SM, Lenz HJ, Haiman CA, Yan C, Henderson BE, Frenkel B, Barretina J, Bass A, Tabernero J, Baselga J, Regan MM, Manak JR, Shivdasani R, Coetzee GA, Freedman ML. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomaszewski M, Zimmerli L, Charchar FJ, Dominiczak AF. Genetic information in the diagnosis and treatment of hypertension. Curr Hypertens Rep. 2006;8:309–316. doi: 10.1007/s11906-006-0070-3. [DOI] [PubMed] [Google Scholar]
- 31.Ji W, Foo JN, O’Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


