Abstract
The DNA glycosylase MutY homolog (Myh1) excises adenines misincorporated opposite guanines or 7,8-dihydro-8-oxo-guanines on DNA by base excision repair thereby preventing G:C to T:A mutations. Schizosaccharomyces pombe (Sp) Hst4 is a NAD+-dependent histone/protein deacetylase involved in gene silencing and maintaining genomic integrity. Hst4 regulates deacetylation of histone 3 Lys 56 (H3K56) at the entry and exit points of the nucleosome core particle. Here we demonstrate that hst4 mutant is more sensitive to H2O2 than the wild-type cells. H2O2 treatment results in an SpMyh1 dependent decrease in SpHst4 protein level and hyper-acetylation of H3K56. Furthermore, SpHst4 interacts with SpMyh1 and with cell cycle checkpoint Rad9-Rad1-Hus1 (9-1-1) complex. SpHst4, SpMyh1, and SpHus1 are physically bound to telomeres. Following oxidative stress, there is an increase in the telomeric association of SpMyh1. Conversely, the telomeric association of spHst4 is decreased. Deletion of SpMyh1 strongly abrogated telomeric association of SpHst4 and SpHus1. However, telomeric association of SpMyh1 is enhanced in hst4Δ cells in the presence of chronic DNA damage. These results suggest that SpMyh1 repair regulates the functions of SpHst4 and the 9-1-1 complex in maintaining genomic stability.
Keywords: DNA glycosylase, MYH, Hst4, histone deacetylase, Hus1, checkpoint, DNA repair
Introduction
Reactive oxygen species (ROS) from endogenous and external sources produce thousands of both cytotoxic and mutagenic base lesions and DNA strand breaks per cell per day.1 The most frequently encountered lesion is 8-oxo-7,8-dihydroguanine (8-oxoG or GO), which can mispair with adenine during DNA replication and lead to a G:C to T:A transversion.2,3 In the absence of DNA repair, these events can lead to aging, cancer, and other diseases.4 Base excision repair (BER) is the major pathway utilized to repair these lesions.5,6 To initiate BER, DNA glycosylases detect and excise the damaged or incorrectly incorporated bases to generate apurinic/apyrimidinic (AP) sites.7,8 The BER mechanism employed in cellular defense against the 8-oxoG-induced mutagenesis is well conserved among organisms2,9.
In fission yeast Schizosaccharomyces pombe, the DNA glycosylase MutY homolog (SpMyh1) removes adenines that are misincorporated opposite to GO or G,3,10,11 thus reducing reduces G:C to T:A transversions. Correspondingly, SpMyh1 knockout cells (myh1Δ) have a higher mutation frequency than wild-type cells.12,13 This role of SpMyh1 in mutation avoidance12 is conserved in the human MutY homolog (hMYH or hMUTYH) [reviewed in 3]. The genomic instability associated with hMYH mutation can lead to MYH-associated polyposis (MAP), a hereditary form of colon cancer.14-18
The silent information regulator 2 (Sir2) is a class III NAD+-dependent histone/protein deacetylase (HDAC) and gene silencer that regulates transcription, recombination, genomic stability, and aging in multiple model organisms.19,20 Sir2 homologues (sirtuins) are conserved from yeast to mammals.21-23 The S. pombe Sir2 family consists of three members (Sir2, Hst2, and Hst4)21, whereas mammals encode seven members (SIRT1-SIRT7) [reviewed in 20,21]. S. pombe Hst4 (SpHst4) is required for deacetylation of the histone H3 core domain residue Lys56 (H3K56)24 and several other Lys residues in histone tails.25 Histone H3K56 acetylation plays important roles in preserving genomic integrity,26,27 and may disrupt histone-DNA interactions at the entry and exit points of the nucleosome core particle.28 Interestingly, Hst4 represses genes which are involved in amino-acid biosynthesis and oxidoreductase activity.25 SpHst4 defective cells have elongated cell morphology, chromosomal abbreviations, and a defect in silencing telomeres and centromeres.24,29 Moreover, hst4 mutants are more sensitive to many DNA damaging agents.24,25,29 These results clearly demonstrate the importance of SpHst4 in maintaining genomic stability.
DNA repair processes are coordinated by cell cycle checkpoint control30,31 and are controlled by chromatin structure.32 This coordinated regulation in response to DNA damage increases DNA repair, arrests the cell cycle to allow more time for DNA repair, or triggers apoptosis in cases of extreme DNA damage.33-36 Rad9, Rad1, and Hus1 are checkpoint sensors that form a heterotrimeric complex (the 9-1-1 complex).37,38 The sliding clamp structure of the 9-1-1 complex39-41 shares significant structural homology with the proliferating cell nuclear antigen (PCNA).42-44 Interestingly, the 9-1-1 complex regulates MYH repair in both S. pombe and human cells.45,46 The role of histone modifications in DNA repair and checkpoint signaling has been previously investigated.47,48 To study the role of SpHst4 in the repair of oxidative DNA damage and checkpoint signaling, we have investigated whether it functions in the SpMyh1 BER pathway. Here, we demonstrate that SpHst4 interacts with SpMyh1 and the 9-1-1 complex. H2O2 treatment results in an SpMYH1 dependent decrease in SpHst4 protein level and hyper-acetylation of H3K56. In addition, we show that the telomeric association of SpHst4 and SpMyh1 is dependent on oxidative stress. Significantly, deletion of SpMyh1 strongly abrogated telomeric association of SpHst4 and SpHus1 suggesting that SpMyh1 may act as an adaptor for these proteins. Our results provide new insights into the roles of DNA repair, histone acetylation, and checkpoint regulation in the maintenance of genomic stability.
Results
Hst4 defective cells are more sensitive to hydrogen peroxide
S. pombe Hst4 plays a critical role in preserving genomic integrity.24,25,29 hst4 mutants have been shown to be more sensitive to hydroxyurea (HU), phleomycin, ultraviolet light (UV), methyl methane sulphonate (MMS), and the microtubule destabilizing agent tiabendazole (TBZ) than wild-type cells.24,25,29 However, its sensitivity to oxidative stress has not been demonstrated. In Fig. 1a, we showed that hst4 mutant cells were more sensitive to H2O2 than wild-type cells. H2O2 sensitivity was observed for concentrations higher than 1 mM. We also tested two additional S. pombe mutants lacking the histone deacetylases Clr6 and Sir2 respectively. Clr6 (cryptic loci regulator) is a class I HDAC involved in epigenetic regulation 49 and Sir2 belongs to the same class III HDAC family as Hst4.29,50 As shown in Fig. 1b, clr6Δ was as sensitive to 2 mM H2O2 as the hst4Δ mutant, however, sir2Δ did not show an increase in H2O2 sensitivity.
Fig. 1.
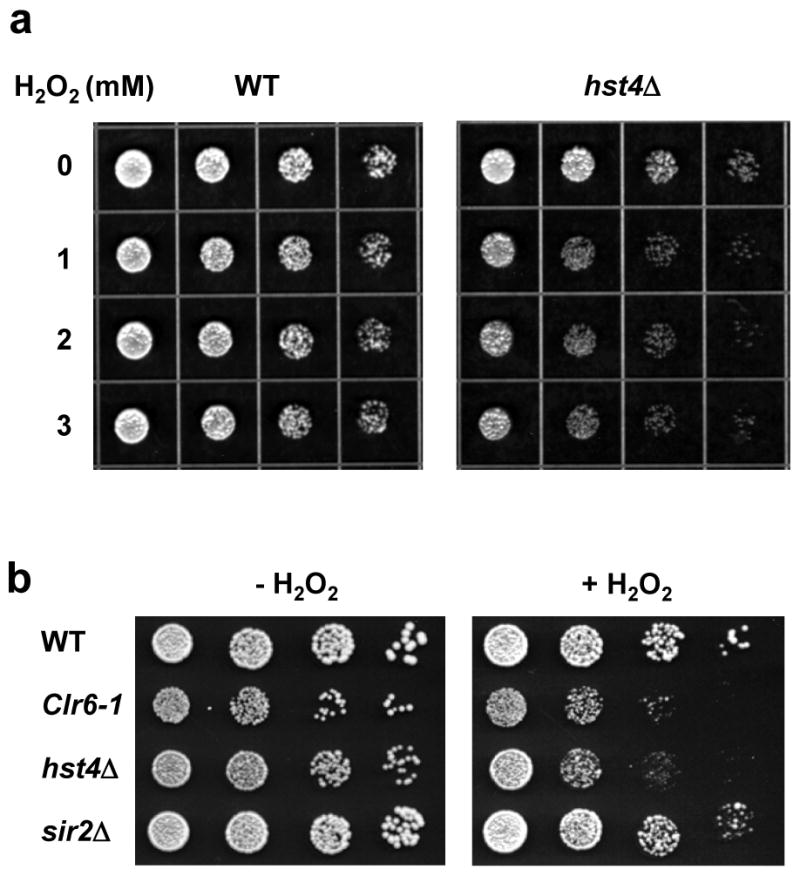
S. pombe hst4Δ cells are more sensitive to H2O2. (a) Wild-type (WT) and hst4Δ S. pombe cells in exponential growth were treated with 0, 1, 2, and 3 mM of H2O2 for 30 min, diluted for every 4-fold, and 4 μl are spotted onto YES media. (b) Wild-type (WT) cells and three HDAC mutants clr6Δ, hst4Δ, and sir2Δ were treated with or without 2 mM of H2O2 for 30 min, diluted for every 5-fold, and 4 μl are spotted onto YES media.
Oxidative damage alters SpHst4 expression and histone H3K56 acetylation
To determine whether the SpHst4 protein level is altered after oxidative stress, we prepared total cell extracts from a strain expressing Myc-tagged SpHst4 and monitored the SpHst4 protein by Western blotting with c-Myc antibody (Fig. 2). The SpHst4 protein levels were normalized to the amounts of histone H3. Upon treatment with 5 mM H2O2 for 30 min, the level of SpHst4 decreased by 4-folds (Fig. 2a, lane 2, upper panel). The level of SpHst4 continued to decrease by 7-folds after recovery in H2O2-free media for 1 h (Fig. 2a, lane 3, upper panel) but returned to a normal level after a 3 h recovery (Fig. 2a, lane 4, upper panel). Because SpHst4 controls the acetylation of histone H3K56 24, the decreased level of SpHst4 observed after treatment with H2O2 may contribute to the up-regulation of H3K56 acetylation. To test this, we monitored the acetylation level of H3K56. In wild-type cells, the acetylation of H3K56 increased immediately after H2O2 treatment (Fig. 3, lane 2, upper panel) then continued to increase during the first three hours of the recovery period (Fig. 3, lanes 3 and 4, upper panel). The level of H3K56 acetylation was very high in untreated hst4 cells (Fig. 3, lane 5, upper panel). This observation is consistent with the finding that Hst4 can deacetylate H3K56.24 However, the level of H3K56 acetylation in hst4 cells did not change significantly after H2O2 treatment (Fig. 3, lanes 5-8, upper panel).
Fig. 2.
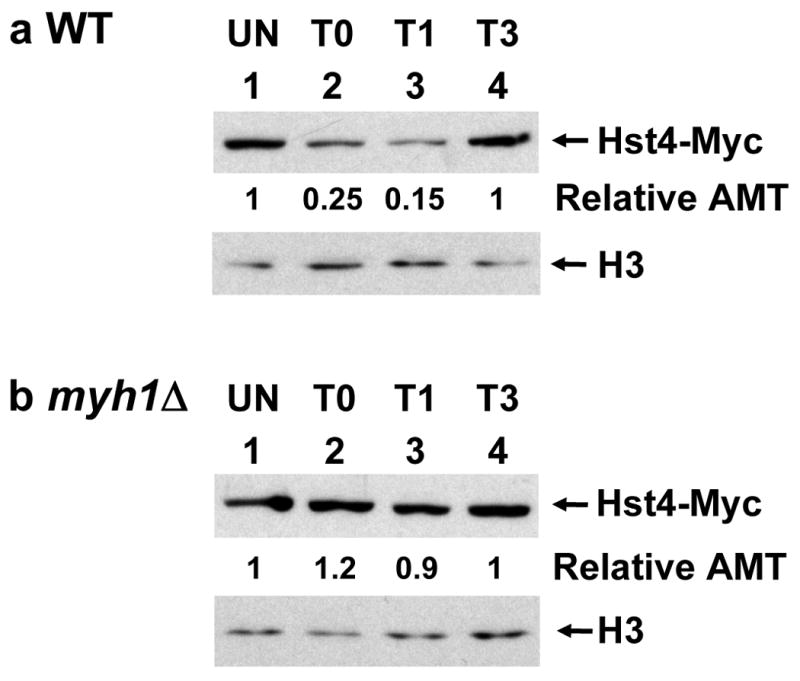
SpHst4 protein level decreases following H2O2 treatment in wild-type (WT) but not in myh1Δ cells. S. pombe Hu1500 (a) and Hst4-MYC myh1Δ (b) were treated with 5 mM of H2O2 for 30 min and then recovered for 0 (T0), 1 (T1), or 3 (T3) hours or remained untreated (UN). Total cell extracts were prepared, separated on 4-20% gradient SDS-polyacrylamide gels and subjected for Western blotting with a mixture of antibodies against c-Myc and histone H3. The amounts of SpHst4-Myc were quantitated relative to those of H3 (Relative AMT).
Fig. 3.
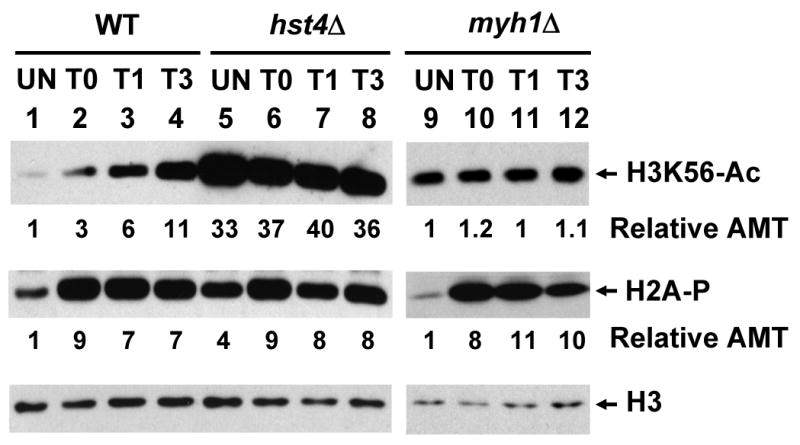
The levels of acetylated H3K56 (H3K56-Ac) and phosphorylated H2A (H2A-P) increase following H2O2 treatment in WT cells. S. pombe Hu303, Hu1481 (hst4Δ), and JSP303-Y4 (myh1Δ) were treated with 5 mM of H2O2 for 30 min and then recovered for 0 (T0), 1 (T1), or 3 (T3) hours or remained untreated (UN). Total cell extracts were prepared and separated on two sets of 15% SDS-polyacrylamide gels. Set 1 gel was subject for Western analysis with H3K56Ac antibody. The membrane was then stripped and probed with H2A-P antibody. Set 2 gel was subject for Western analysis with H3 antibody. Lanes 1-8 are in the same membrane while lanes 9-12 are on a separated membrane and Western blotting was performed separately. The amounts of H3K56Ac and H2A-P were quantitated relative to those of H3 (Relative AMT).
Because S. pombe Hst4 is important in DNA repair and genomic stability24,25,29 and hst4 mutants are more sensitive to H2O2, we asked whether the changes in SpHst4 protein levels was dependent on SpMyh1. As shown in Fig. 2b, the SpHst4 protein levels were not altered in myh1Δ cells following H2O2 treatment. Consistent with the finding that SpHst4 can regulate H3K56 acetylation, the H3K56 acetylation levels were not altered in myh1Δ cells following H2O2 treatment (Fig. 3, lanes 9-12, upper panel).
To determine whether the decreased SpHst4 protein level following H2O2 treatment is controlled at the level of transcription, we examined the hst4 mRNA level. As shown in Table 1, using act1 as a control and RT-qPCR analysis, the hst4 mRNA level of H2O2-treated wild-type (Hu1500) cells is 1.08-fold of that of the untreated cells while the hst4 mRNA level of H2O2-treated myh1Δ cells is 0.91-fold of that of the untreated cells. Thus, the level of hst4 mRNA was neither altered in wild-type cells nor in myh1Δ cells following 1 h recovery after treatment with H2O2. Therefore, the decreased SpHst4 protein level observed was not the result of hst4 mRNA transcriptional inhibition.
Table 1.
The mRNA level of hst4+ does not change following H2O2 treatment and is independent of SpMyh1.
| Ct | ΔCt | Folda | Foldb | |||
|---|---|---|---|---|---|---|
| hst4+ | act1+ | |||||
| HU1500 | UN | 20.01 | 15.07 | 5.04 | ||
| HU1500 | T1 | 20.22 | 15.28 | 4.94 | 1.07 | 1.08 ± 0.05 |
| myh1Δ | UN | 20.22 | 14.72 | 5.50 | ||
| myh1Δ | T1 | 20.09 | 14.68 | 5.41 | 1.06 | 0.91 ± 0.20 |
Cells at an OD600 of 0.8 were exposed to 5 mM H2O2 for 30 min and recovered for 1 h (T1) or left untreated (UN). RNA were prepared and RT-qPCR were performed with primers specific for hst4+ and act1+. The amount of hst4+ mRNA was quantitated relative to that of act1+. Fold changes of T1 relative to UN are presented.
Average of three separate experiments, with errors reported as the standard deviations.
Oxidative damage stimulates histone H2A phosphorylation
Mammalian ATR and ATM checkpoint kinases modulate chromatin structures near DNA breaks by phosphorylating histone H2AX.51 S. pombe H2A is similarly phosphorylated following ionizing radiation (IR) by the ATR/ATM-related kinases Rad3 and Tel1.52 The phosphorylated forms of human H2AX and S. pombe H2A are thought to be important for DNA repair and checkpoint maintenance. To check whether H2A is phosphorylated following oxidative stress, we monitored the H2A phosphorylation in wild-type, hst4Δ, and myh1Δ cells following H2O2 treatment. In wild-type and myh1Δ cells, the H2A phosphorylation increased approximately 8-fold immediately after H2O2 treatment and remained high during the next 3 h as the cells recovered (Fig. 3, lanes 1-4 and lanes 9-12, middle panel). Thus, H2A phosphorylation in response to oxidative damage is independent of SpMyh1. The result is consistent with the previous finding that H2A phosphorylation occurs at double strand breaks induced by oxidative stress.52,53 In untreated hst4Δ cells, the level of H2A phosphorylation was higher than that in the wild-type cells (Fig. 3, compare lanes 1 and 5, middle panel). This result suggests that the DNA damage checkpoint is activated in hst4Δ cells and is consistent with the previous reports that hst4Δ cells contain DNA damage24,29 and elevated levels of Chk1 phosphorylation.24 The level of H2A phosphorylation increased only 2-fold in hst4Δ cells after H2O2 treatment as compared to untreated cells (Fig. 3, lanes 5-8, middle panel) and was compatible to treated wild-type cells (Fig. 3, compare lanes 2-4 with lanes 6-8, middle panel).
SpHst4 interacts with SpMyh1 and the Rad9-Rad1-Hus1 (9-1-1) complex
Because the change in SpHst4 protein levels following oxidative stress is dependent on SpMyh1, we asked whether SpHst4 interacts with SpMyh1 DNA glycosylase. We demonstrated that SpHst4 physically interacted with SpMyh1 by glutatathine S-transferase (GST) pull-down (Fig. 4a) and co-immunoprecipitation (Fig. 4b). Treatment with H2O2 resulted in approximately 3-fold increased SpHst4-SpMYH1 interaction (Fig. 4b, compare lanes 2 and 4).
Fig. 4.
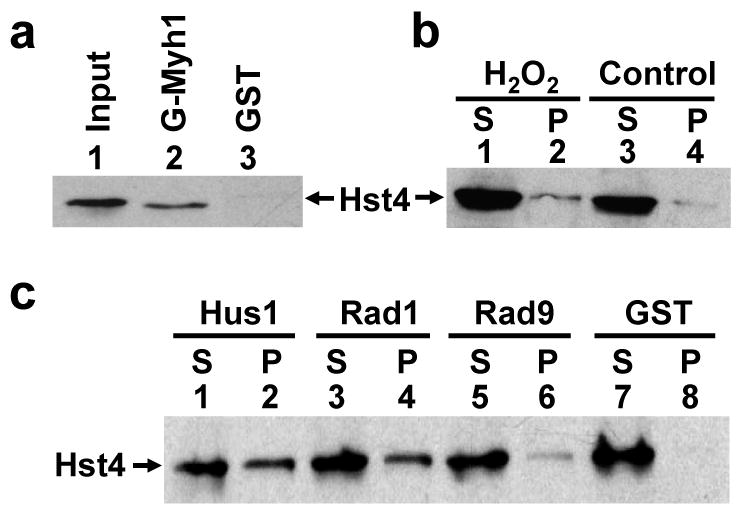
SpHst4 interacts with SpMyh1 and the Rad9/Rad1/Hus1 subunits. (a) Pull-down of SpHst4 by GST-SpMyh1. GST-SpMyh1 (lane 2), and GST alone (lane 3) were immobilized to glutathione-Sepharose and incubated with extracts from Hu1500 containing SpHst4-Myc. The pellets were fractionated by a 10% SDS-PAGE followed by Western blot analysis with the c-Myc antibody. Lane 1 contains 10% of the input yeast extracts. (b) The interaction of SpHst4 and SpMyh1 was enhanced following oxidative stress. Hu1500 cells were exposed to 5 mM H2O2 for 30 min and recovered for 1 hour or left untreated (control). Total soluble cell extracts were prepared and analyzed by immunoprecipitation with antibody against SpMyh1. Western blotting was performed to detect SpHst4. S represents the supernatant and P represents the pellet. (c) Pull-down of SpHst4 by GST-SpHus1, GST-SpRad1, and GST-SpRad9. GST-SpHus1 (lanes 1 and 2), GST-SpRad1 (lanes 3 and 4), GST-SpRad9 (lanes 5 and 6), and GST alone (lanes 7 and 8) were immobilized to glutathione-sepharose and incubated with purified SpHst4-Myc. The supernatants (S, 10%) and pellets were fractionated by a 10% SDS-PAGE followed by Western blot analysis with the c-Myc antibody.
It has been shown that SpHst4 functions in the DNA damage response pathway.24 Moreover, several checkpoint components are essential for the survival of the hst4Δ mutant.24 We have shown that MYH glycosylase is associated with the 9-1-1 complex in both S. pombe and human cells.45,46 Because SpHst4 interacts with SpMyh1, we asked whether SpHst4 interacts with the 9-1-1 complex. We demonstrated that purified SpHst4 interacted with all three subunits of the 9-1-1 complex (Fig. 4c). The interaction of SpHst4 with SpHus1 and SpRad1 was stronger than the SpHst4-SpRad9 interaction. These results suggest that SpHst4 and SpMyh1 form a complex with the 9-1-1 complex and that SpHst4 may have a direct role in BER and the DNA damage response.
SpHst4 and SpMyh1 are bound to telomeric DNA
There is increasing evidence that supports the importance of regulating DNA repair at telomeres.54 Oxidative damage to CG-rich telomeric DNA requires efficient BER to maintain its integrity.55,56 The S. pombe telomeres of chromosomes I and II are ∼300 base pairs in length with variable repeat unit of consensus T1-3AC0-1A0-2C0-1G1-8 and TTACAGG is the most common repeats. The telomeres of chromosome III of S. pombe consists of ∼150 rDNA repeats separated in two clusters at both ends. One rDNA repeat (∼10.5 kb) consists of a transcription unit for the 35S rRNA precursor and a non-transcribed sequence, which contains an autonomously replicating sequence (ars) element (ars3001) and a replication fork barrier (RFB).57 A recent report has shown that SpHst4 binds to over 300 open reading frames and heterochromatin regions including mat, rDNA, and telomeres.25
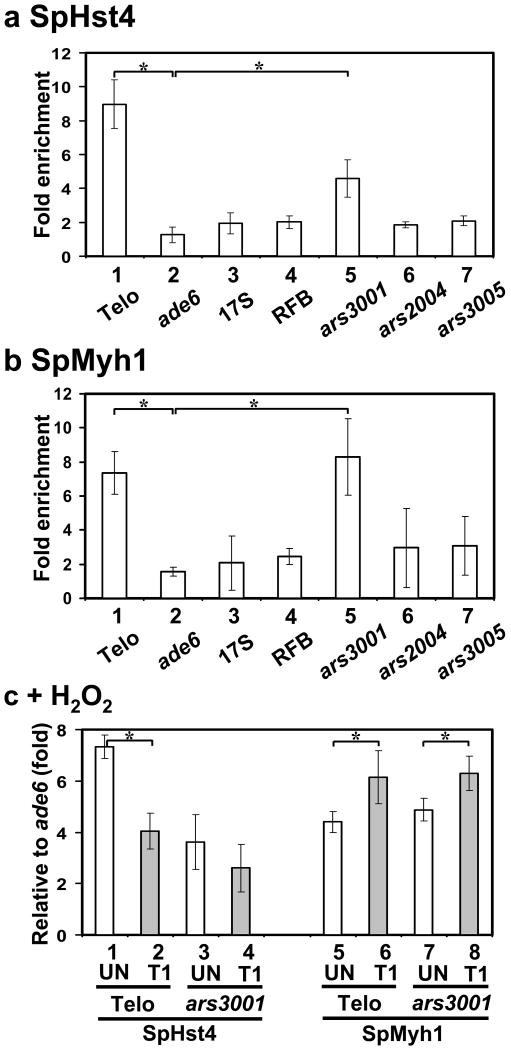
We have confirmed that SpHst4 is physically bound to telomeres by performing chromatin immunoprecipitation (ChIP) assay with S. pombe expressing Myc-tagged SpHst4 (Fig. 5a). Precipitated DNA was quantitated by quantitative PCR (qPCR) with primers for the telomere-associated sequences (Telo). Telomeric DNA was specifically enriched 8-fold from Hst4-Myc immunoprecipitant and the association of SpHst4 at telomeres was 7-fold over that at ade6 region (Fig. 5a, columns 1 and 2). Further ChIP analyses on rDNA regions (17S, RFB, and ars3001) and the non-telomere-adjacent regions (ars2004, and ars3005) indicated that SpHst4 was also enriched at ars3001 but not at other tested sites (Fig. 5a). The association of SpHst4 at ars3001 was 4-fold over that at ade6 region (Fig. 5a, columns 2 and 5).
Fig. 5.
The associations of SpHst4 and SpMyh1 at telomeres and ars3001 of the rDNA repeat are altered following oxidative stress. (a) S. pombe cells (Hu1500) were subjected to ChIP assay with c-Myc antibody to precipitate SpHst4-Myc associated DNA. Precipitated DNA (IP) samples were amplified with primers listed in Table S2 by quantitative PCR (qPCR). Enrichment (n-fold) was calculated according to the formula 2ΔCt-ΔCtControl, in which ΔCt is the difference between the number of cycles required to go above background in input and IP samples. The control is the sample with beads only. Statistical significances between telomeres (Telo) and ars3001 relative to ade6+ are indicated with stars. (b) ChIP was performed similarly to (a) except that SpMyh1 antibody was used to precipitate SpMyh1 associated DNA. (c) ChIP was performed similarly to (a) and (b) except that Hu1500 cells was treated with 5 mM H2O2 for 30 min and then recovered for one hour (T1) or left untreated (UN). c-Myc antibody (columns 1-4) and SpMyh1 antibody (columns 5-8) were used to precipitate SpHst4 and SpMyh1, respectively, associated DNA. Fold enrichment at Telo and ars3001 was calculated relative to ade6+. Statistical significance (P < 0.02) between wild-type and hst4Δ is indicated with a star.
The ChIP assay with SpMyh1 polyclonal antibody demonstrated that SpMyh1 was also enriched at telomeres of chromosomes I and II as well as at the ars3001 region (Fig. 5b, columns 1 and 5) but not at other tested sites (Fig. 5b). The associations of SpMyh1 at telomeres and ars3001 were about 4.5-fold over that at ade6 region. Interestingly, the pattern of SpMyh1 association with telomeres is similar to that of SpHst4.
The recruitment of SpMyh1 and SpHst4 to telomeres is altered by oxidative stress
Because both myh1 and hst4 defective cells are more sensitive to H2O2 [Fig. 1 and 12], we tested whether their association at the telomeres was affected by oxidative stress. Fig. 5c indicated that the association of SpHst4 at telomeres decreased by 45% when S. pombe cells were treated with H2O2 and then recovered for 1 hour (Fig. 5c, column 1 vs. column 2). This result is consistent with Fig. 2a which shows that the total SpHst4 protein level decreased by 85%. We have reported that SpMyh1 protein levels remain unchanged following H2O2 treatment45. However, the association of SpMyh1 at telomeres increased by 40% in oxidatively damaged S. pombe cells (Fig. 5c, column 5 vs. column 6). The changes in the association of SpHst4 and SpMyh1 at ars3001 were similar to their association at telomeres of chromosomes I and II but were less significant when S. pombe cells were treated with H2O2 (Fig. 5c, columns 3, 4, 7, and 8).
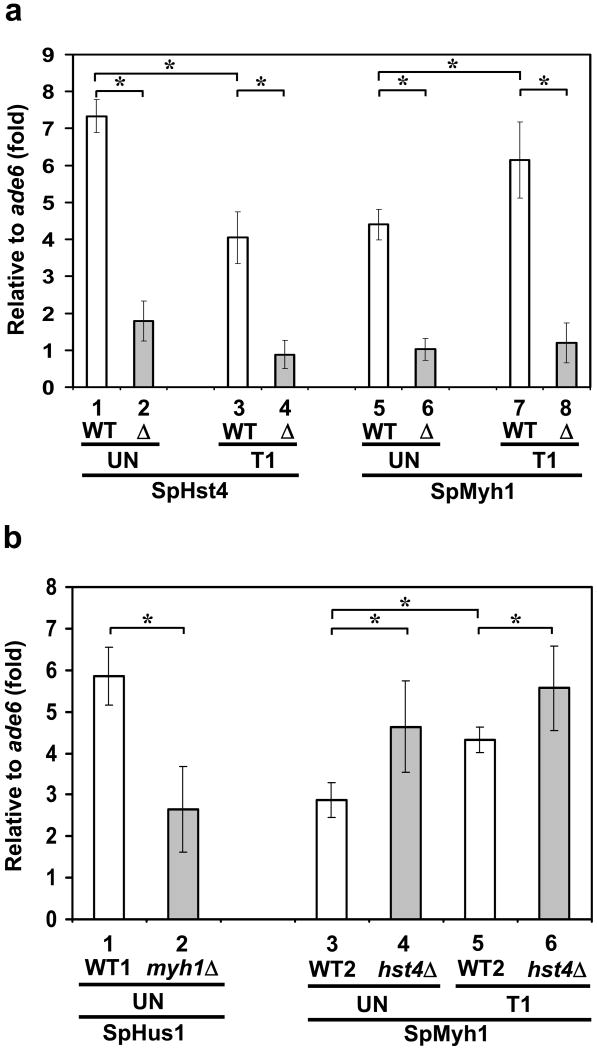
SpHst4 and SpHus1 association at telomeres is substantially dependent on SpMyh1
The correlated association of SpHst4 and SpMyh1 at telomeres and ars3001 (Figs. 5a and 5b) prompted us to investigate their mutual dependence. First, we examined whether the recruitment of SpHst4 to telomeres was dependent on SpMyh1. Using ChIP assay with c-Myc antibody, we showed that deletion of SpMyh1 abrogated SpHst4 association to telomeres (Fig. 6a, compare columns 1 and 2, and columns 3 and 4). In untreated cells, the association of SpHst4 at telomeres reduced from 7-fold in wild-type cells to 2-fold in myh1Δ cells. In H2O2-treated cells, the association of SpHst4 at telomeres reduced from 4-fold in wild-type cells to the basal level in myh1Δ cells. As a control, the enrichment of SpMyh1 to telomeres was lost in the myh1Δ mutant (Fig. 6a, compare columns 5 and 6 and columns 7 and 8). Thus, SpHst4 association at telomeres is dependent on SpMyh1.
Fig. 6.
The association of SpHus1 and SpHst4 to telomeres is dependent on SpMyh1 and the association of SpMyh1 to telomeres is enhanced in hst4Δ cells. (a) ChIP was performed similarly to Fig. 5 except that both Hu1500 (WT) and Hst4-MYC myh1Δ (Δ) cells were used. Cells were treated with 5 mM H2O2 for 30 min and then recovered for one hour (T1) or left untreated (UN). c-Myc antibody (columns 1-4) and SpMyh1 antibody (columns 5-8) were used to precipitate SpHst4 and SpMyh1 associated DNA, respectively. Fold enrichment at Telo was calculated relative to ade6+. Statistical significance between wild-type and myh1Δ is indicated with a star. (b) ChIP was performed similarly to (a) except that Hus1-MYC (WT1), Hus1-MYC myh1Δ (myh1Δ), Hu303 (WT2) and Hu1481 (hst4Δ) cells were used. c-Myc antibody (columns 1-2) and SpMyh1 antibody (columns 3-6) were used to precipitate SpHus1 and SpMyh1 associated DNA, respectively. Fold enrichment at Telo was calculated relative to ade6+. Statistical significance (P < 0.02) between wild-type and hst4Δ is indicated with a star.
The S. pombe 9-1-1 complex has been shown to associate with telomeres and is important in telomere maintenance.58,59 Since SpHst4 and SpMyh1 form a complex with the 9-1-1 complex, we examined whether the recruitment of SpHus1 to telomeres is dependent on SpMyh1. Myc-SpHus1 was particularly enriched at telomeres of chromosomes I and II (Fig. 6b, column 5) confirming the previous report59. Deletion of SpMyh1 strongly abrogated but did not completely prevent SpHus1 association to telomeres (Fig. 6b, compare columns 1 and 2), suggesting the possible involvement of other factors for SpHus1 association at telomeres.
SpMyh1 association at telomeres is enhanced in hst4Δ cells
Next, we examined whether the recruitment of SpMyh1 at telomeres was dependent on SpHst4. As shown in Fig. 6b (lanes 3-6), the association of SpMyh1 at telomeres was increased either in the untreated or H2O2-treated hst4Δ cells. This result is in agreement with the finding that the untreated hst4Δ cells contain DNA damage which is indicated by elevated levels of H2A phosphorylation (Fig. 3, lane 5, 2nd panel) and Chk1 phosphorylation.24 This increased DNA damage may lead to the enhanced association of SpMyh1 with telomeres.
Discussion
S. pombe Hst4, a member of the class III HDACs (sirtuins), plays an important role in maintaining genomic stability.24,25,29 Sirtuins are gene silencers/regulators that impact the aging process [reviewed in 20,21,60]. S. pombe hst4 mutants are more sensitive to many DNA damaging agents24,25,29 including H2O2 (Fig. 1). S. pombe hst4 mutants have constitutively elevated levels of H3K56 acetylation following treatment with both H2O2 (Fig. 3) and methylating agents.24 In this study, we show that SpHst4 interacts with SpMyh1 DNA glycosylase and the 9-1-1 checkpoint sensor. Moreover, Myh1 is required for SpHst4 association at the telomeres, decrease in SpHst4 protein level, and hyper-acetylation of histone H3K56 following oxidative stress. Thus, SpMyh1 repair is correlated with SpHst4-mediated histone modification. Mammalian sirtuins play a role in many biological processes, such as insulin secretion, fat mobilization, response to stress, and lifespan regulation.19,20 Particularly, sirt6 knockout mice display premature aging.61,62 Similar to S. pombe hst4 mutants, SIRT6-deficient cells are more sensitive to DNA damaging agents (methylating agents, H2O2, and IR) and have an increased frequency of genomic aberrations.62 Moreover, SIRT6 has a function in modulating telomeric chromatin.63 Furthermore, SIRT6 and SpHst4 share similar substrate specificity on H3K9 and H3K56.24,25,63-65 Most importantly, it has been shown that SIRT6 plays a role in BER because expression of DNA polymerase β in SIRT6-deficient cells restores resistance to DNA damaging agents.61,62 Based on the similar phenotypes of Hst4 defective S. pombe cells and sirt6 knockout mice62 and their roles in histone deacetylation and BER, we hypothesize that SpHst4 may be the functional homolog of mammalian SIRT6.
Telomeres contain DNA double-strand ends that do not trigger the DNA damage response, nevertheless they require DNA repair and checkpoint proteins for maintenance. The CG-rich telomere sequence is particularly susceptible to oxidative damage.66-68 Oxidative damage is repaired inefficiently in telomeric DNA in comparison to the rest of the chromosome.68 Single and multiple GO lesions in the human tandem telomeric sequence repeats disrupt recognition by the telomere repeat binding factors TRF1 and TRF2.69 TRF2 has been shown to interact with enzymes involved in BER (DNA Polymerase β and FEN-1).55 Furthermore, OGG1 DNA glycosylase is involved in repairing GO lesions in telomeres.56 We hypothesize that Myh1, Hst4, and the 9-1-1 complex are involved in the reduction of G:C to T:A transversions at the telomeres based on the following results. First, both myh1 and hst4 mutants are more sensitive to H2O2 [12 and Fig. 1]. Second, Hst4 interacts with Myh1 and the 9-1-1 complex [Fig. 4]; and Myh1 interacts with the 9-1-1 complex.46 Third, Myh1, Hst4, and the 9-1-1 complex are enriched at telomeres [25,58,59 and Figs. 5 and 6]. The association of SpMyh1 at telomeres and ars3001 (Fig. 5b) is a new and interesting finding and indicates that SpMyh1 is enriched at certain defined sites of the genome. The telomeric presence of SpMyh1 likely indicates that it plays a role in maintaining the CG-rich regions. When telomeres are damaged, Myh1 recognizes the A/GO mismatch and recruits SpHst4 and the 9-1-1 complex for efficient repair (Figs. 4 and 6). However, further investigation is necessary to determine the biological significance of SpMyh1 enrichment at ars3001. It appears that SpMYH1 association at ars3001 is not related to replication origin because SpMYH1 does not bind to all autonomously replicating sequences such as ars2004 and ars3005.
It has been shown that SpHst4 can deacetylate H3K56 and histone tails.24,25 Sufficient data support that a balanced level of H3K56 acetylation is critical for genome integrity. Unacetylatable histone H3 (H3K56R), complete loss of H3K56 acetylation (Rtt109 deletion), acetylation mimic H3K56Q mutant, and H3K56 hyperacetylation in the hst4 mutant (or hst3 hst4 double mutant in S. cerevisiae) all elicit similar phenotypes that exhibit sensitivity to DNA-damaging agents.24,70-74 Recent studies proposed a model that H3K56 acetylation plays a role in replication-coupled chromatin assembly and chromatin reassembly during double-strand break repair and checkpoint recovery.75,76 H3K56 acetylation is enriched on chromatin fractions undergoing DNA repair and has been proposed to open chromatin for DNA repair.77 Our results support this notion that DNA damage alters H3K56 acetylation. We have shown that when wild-type S. pombe cells are treated with H2O2 with a one-hour recovery, the H3K56 acetylation is up-regulated via down-regulation of SpHst4 (Figs. 2 and 3); and SpHst4 association at the telomeres is reduced (Fig. 5c). However, after 3 h of recovery, the Hst4 protein level is restored, but the H3K56 acetylation level is increased. It is possible that a histone acetyltransferase (such as Rtt109)74 may be activated, other histone deacetylases may be reduced, or Hst4 deacetylase activity may be inhibited. Because SpMyh1 repairs replication errors, its repair is coupled with DNA replication through interaction with PCNA.78 Interestingly, acetylation of H3K56 occurs during S-phase.24,70,74 Thus, SpMyh1 repair may occur before deposition of newly assembled nucleosomes.
It has been shown that loss of ScHst3-mediated regulation of H3K56 acetylation in S. cerevisiae results in a defect in the S phase DNA damage checkpoint and that the increased level of H3K56 acetylation is accomplished by SpHst4 down-regulation in response to the methylating agent MMS24,73. In S. cerevisiae, ScHst3 undergoes rapid ScMec1-dependent phosphorylation and is targeted for ubiquitin-mediated proteolysis following MMS treatment.24,73 We have shown that down-regulation of SpHst4 following H2O2 treatment is not accompanied by down-regulation of hst4 mRNA. Thus, the mechanism of regulating SpHst4 expression level following oxidative stress may follow a similar mechanism as ScHst3 by MMS treatment.24,73 Our results indicate that Myh1 is required for SpHst4 association at the telomeres, the decrease in SpHst4 protein level, and hyper-acetylation of histone H3K56 following oxidative stress. Therefore, we propose that by recruiting SpHst4 to mismatched DNA sites, SpMyh1 may expose Hst4 protein to targeted degradation by the activated DNA damage checkpoint. Targeted H3K56 acetylation could facilitate DNA repair by creating a more open chromatin or by recruiting DNA damage-signaling proteins.73 The direct physical interactions of SpHst4 with SpMyh1 and the 9-1-1 complex support this notion. Thus, the increased genomic instability in hst4Δ with global overacetylation of H3K56 or Rtt109Δ without H3K56 acetylation may be caused by the inability to mark H3K56 on the chromatin containing damaged or mismatched DNA.
Hst4 defective S. pombe cells have elevated chromosome abbreviation and DNA damage.24,25,29 Our data (Fig. 6b) is consistent with the idea that the association of SpMyh1 at telomeres is increased in order to repair DNA damage in hst4Δ cells. Moreover, the interaction of SpHst4 with the 9-1-1 complex links histone acetylation with checkpoint regulation. It has been shown that several checkpoint components are essential for the survival of hst4Δ mutant.24 In addition, Celic et al.79 have observed that the temperature sensitive phenotype of hst3 hst4 S. cerevisiae can be suppressed by inactivation of the 9-1-1 complex. In conclusion, our results suggest that SpMyh1 repair coupled with cell cycle checkpoint and histone acetylation is critical for genomic stability and/or telomere maintenance.
Materials and Methods
Yeast S. pombe strains and growth
The yeast strains used in this study are listed in Table S1 in appendices. Standard procedures and media were used for culture growth, transformation, and genetic analysis.80 Yeast cells were grown in YES medium (5 g of yeast extract, 30 g of glucose, and 100 mg each of adenine, histidine, leucine, lysine, and uracil per liter) for regular maintenance. The Hu1500 strain was constructed by epitope-tagging of the endogenous hst4+ gene using the procedure described.81 The protein was tagged at the C-terminus with 6xMyc (i.e. natural promoter and expression levels). The myh1Δ mutant containing hst4+-Myc was constructed by two genetic crosses. The marker his3D1 from FY526 was introduced to Hu1500, and then the resulting strain was crossed with JSP303-Y4 (myh1Δ∷his3+) to generate the Hst4-MYC myh1Δ strain. Both strains were verified by PCR for correct integration and by Western blotting for expression of Hst4-MYC.
H2O2 treatment of yeast cells
For H2O2 sensitivity, 1 ml of an overnight yeast culture grown in YES was added to 20 ml of YES medium. At an OD600 of 0.5, 2 ml of the culture were aliquoted into each 30 ml test tube and hydrogen peroxide was added to each culture at various concentrations. After incubating for 30 min, the cells were spun down and resuspended in peroxide-free fresh medium. Diluted cells (4 μl) were spotted onto YES plates. The plates were placed in a 30°C incubator for 2-3 days. For cell extract and mRNA preparations, cells at an OD600 of 0.6-0.8 were exposed to 5 mM H2O2 for 30 min and recovered for different time intervals or left untreated (control).
Preparation of cell extracts and Western blotting
Total cell extracts were prepared from 10 ml of S. pombe culture. The cells was washed with 20% TCA and frozen at -80°C. The cell pellet was thawed and resuspended in 0.25 ml of 20% TCA and lysed by glass beads. After precipitation with 5% TCA, the pellet was washed with 0.75 ml ethanol and resuspended in 40 μl of 1 M Tri-HCl, pH 8.0. After adding 80 μl of 2X SDS loading buffer, the solution was boiled for 10 min and then centrifuged at 14,000 rpm for 5 min. The supernatants were collected and portions were fractionated on a 15% SDS-polyacrylamide gel for histones or 4-20% gradient gel for SpHst4-Myc. The proteins were transferred onto a nitrocellulose membrane. Western blot analyses were performed with antibodies against c-Myc-tag (Santa Crutz Biotechnology), H3 (Abcam), K56 acetylated H3 (Active Motif), S129 phosphorylated H2A (Abcam), and tubulin (Abcam). Soluble cell extract were prepared as described by Chang et al.12 except without ammonium sulphate precipitation.
Preparation of RNA and RT-PCR
S. pombe cells were grown to log phase, treated with 5 mM H2O2 for 30 min and then recovered for one hour (T1) in fresh YES media or left untreated. Total RNA were isolated with acid phenol procedures82 and further cleaned up by RNeasy Mini Kit (Qiagen). The RNA (100 ng) was used as a template for reverse transcriptase quantitative PCR (RT-qPCR) reactions using primers for hst4 and act1 (see Table S2) and iScript one-step RT-PCR kit with SYBR Green SuperMix (Bio-Rad). The reactions were carried out with Roche LightCyler 480 with 50°C for 10 min and 95°C for 5 min for RT and then followed by PCR cycles (95°C for 30 sec, 55°C for 30 sec, and 70°C for 1 min). The mRNA level of hst4 was calculated relative to that of act1 as ΔCt which is the difference between the number of cycles required to go above background in hst4 and act1 samples. The fold difference of hst4 mRNA levels of treated cells over untreated cells (UN) is calculated according to the formula 2ΔCt(un)- ΔCt(T1). The reactions were carried out in duplicate and data are averaged from three independent experiments.
Cloning, expression, and purification of SpHst4
The cDNA of SpHst4 was amplified by PCR from an S. pombe cDNA library in pGADGH (kindly provided by D. Beach, Cold Spring Harbor Laboratory) using Pfu DNA polymerase (Stratagene) with the appropriate primers (listed in Table S2). The PCR products were digested with BamHI and SalI, cloned into pET21a (BamH1 and Xho1 digested), transformed into E. coli DH5α cells (Invitrogen), and selected via ampicillin resistance. All clones were confirmed through DNA sequencing.
To express the His-tagged SpHst4 protein, the plasmid was transformed into the Rosetta cells (Invitrogen). The cells were cultured in Luria-Bertani broth containing 100 μg/ml ampicillin and 35 μg/ml chloramphenicol at 37°C. Protein expression was induced at an A590 of 0.6 by the addition of isopropyl 1-thio-β-D-galactopyranoside to a final concentration of 0.4 mM. After 16 hours at 25°C, the cells were harvested by centrifugation at 10,000 × g for 20 min. The SpHst4–His protein was purified by Ni-NTA resin (Qiagen) under native conditions according to the manufacturer's protocol. The SpHst4–His protein was dialyzed twice with 1 L TEG buffer (50 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, 50 mM KCl, 10% glycerol, 0.5 mM dithiothreitol and 0.1 mM PMSF) and further purified by two connected 1 ml SP column (GE Health) equilibrated with TEG buffer. Upon washing with 12 ml of equilibration buffer, the column was eluted using a step gradient of 10 ml each with 0.4 M, 0.55 M, and 1 M step gradient of KCl in TEG buffer. The fractions that contain most of the SpHst4–His protein (confirmed by SDS-polyacrylamide gel analysis) were pooled and dialyzed with 1 L TEG buffer and loaded onto 1 ml DEAE column (GE Health) equilibrated with TEG buffer. Upon washing with 3 ml of equilibration buffer, the column was eluted with a 20 ml linear gradient of KCl (0.05–0.6 M) in TEG buffer. Most of SpHst4 was found in the flow through fraction (Fig. S1), which was divided into small aliquots and stored at -80°C. The SpHst4–His protein was approximately 98% pure (Fig. S1, lane 4) and its concentration was determined by the Bradford method.
GST pull-down assay
Expression, immobilization of GST fusion constructs, and GST-pull-down assay were similar to the procedures described previously.45 E. coli (BL21Star/DE3) cells (Stratagene) harboring the pGEX4T-SpMyh1 plasmid were cultured in Luria-Bertani broth containing 100 μg/ml ampicillin. Protein expression was induced as described above. The cell extracts from a 0.5-liter culture were immobilized onto glutathione-Sepharose 4B (GE Health). Immobilized GST-SpMyh1 and GST alone GST beads were incubated with 0.5 mg of cell extracts from HU1500 containing Hus1-Myc overnight at 4°C. After washing, the pellets were fractionated on a 10% SDS-polyacrylamide gel and transferred onto a nitrocellulose membrane. Western blot analyses were performed with antibody against c-Myc-tag (Santa Crutz Biotechnology).
Co-immunoprecipitation
HU1500 cell extracts (1 mg) were precleared by incubation with protein A Sepharose (50 μl) in phosphate buffered saline (PBS) with protease inhibitors (Sigma/Aldrich) for 4 hours at 4°C. After removal of the beads, the supernatant was mixed with polyclonal SpMyh1 antibody for 16 hours at 4°C. Then, protein A Sepharose (50 μl) was added to precipitate SpMyh1. After centrifugation at 1,000 × g, the supernatant was collected and the pellet was washed. Both the supernatant (10% of total volume) and pellet fractions were resolved on a 10% SDS-polyacrylamide gel. The Myc-tagged SpHst4 that co-precipitated with SpMyh1 was verified with Western blot analysis using antibodies against c-Myc (Santa Crutz Biotechnology).
Chromatin immunoprecipitation (ChIP) assay
The ChIP method was performed according to the published procedures.83 Briefly, S. pombe cells were grown to log phase, treated with 5 mM H2O2 for 30 min and then recovered for one hour in fresh YES media or left untreated. Cells were washed with PBS and treated with 1% formaldehyde for 15 min at room temperature to crosslink protein-DNA. Chromatin was purified and sonicated to shear the DNA to 0.5-1 kb. Immunoprecipitation was performed with antibodies against SpMyh1 (polyclonal) or c-Myc (Covance, monoclonal 9E10). After crosslink reversal at 65°C for over 3 hr, the DNA was recovered by Qiagen PCR purification kit and used as a template for quantitative PCR (qPCR) reactions. The qPCR reactions contained templates, primers (see Table S2), and PerfeCta SYBR Green SuperMix (Quanta) and were carried out with Roche LightCyler 480 with PCR cycles (95°C for 30 sec, 55°C for 30 sec, and 70°C for 1 min). The relative protein enrichment at each site was calculated according to the formula 2ΔCt-ΔCtControl, in which ΔCt is the difference between the number of cycles required to go above background in input and immunoprecipitant samples. The control is the sample with protein A Sepharose beads only. qPCR reactions were carried out in duplicate and ChIP data averaged over more than three independent experiments.
Supplementary Material
Acknowledgments
We thank Drs. Susan Forsburg (University of Southern California) and Charles Hoffman (Boston College) for kindly providing the S. pombe strains. We are grateful to Dr. David Beach, a Howard Hughes Medical Institute Investigator at Cold Spring Harbor Laboratory for providing a S. pombe cDNA library. We appreciate the critical reading from Dr. Amrita Madabushi and Mr. Randall Gunther. This work was supported by grants (GM35132 and CA78391) from National Institute of Health to AL. The Swedish Cancer Society, Swedish Research Council (VR) and the Göran Gustafssons Foundation for Research in Natural Sciences and Medicine support KE.
Abbreviations used
- 9-1-1
Rad9-Rad1-Hus1
- AP
apurinic/apyrimidinic
- ars
autonomously replicating sequence
- BER
base excision repair
- ChIP
chromatin immunoprecipitation
- Clr6
cryptic loci regulator
- GO
8-oxo-7,8-dihydroguanine
- GST
glutatathine S-transferase
- H3K56
histone H3 Lys56
- Hst
homolog of Sir2
- HU
hydroxyurea
- IR
ionizing radiation
- MAP
MYH-associated polyposis
- MMS
methyl methane sulphonate
- Myh1
MutY homolog
- PCNA
proliferating cell nuclear antigen
- qPCR
quantitative polymerase chain reaction
- RFB
replication fork barrier
- ROS
reactive oxygen species
- RT
reverse transcription
- Sir2
silencing information regulator 2
- Sc
Saccharomyces cerevisiae
- Sp
Schizosaccharomyces pombe
- TBZ
tiabendazole
- Telo
telomere-associated sequences
- UV
ultraviolet light
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci USA. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu AL, Li X, Gu Y, Wright PM, Chang DY. Repair of oxidative DNA damage. Cell Biochem Biophy. 2001;35:141–170. doi: 10.1385/CBB:35:2:141. [DOI] [PubMed] [Google Scholar]
- 3.Lu AL, Bai H, Shi G, Chang DY. MutY and MutY homologs (MYH) in genome maintenance. Front Biosci. 2006;11:3062–3080. doi: 10.2741/2033. [DOI] [PubMed] [Google Scholar]
- 4.Loeb LA, Christians FC. Multiple mutations in human cancers. Mutat Res. 1996;350:279–286. doi: 10.1016/0027-5107(95)00117-4. [DOI] [PubMed] [Google Scholar]
- 5.Barzilai A, Yamamoto K. DNA damage responses to oxidative stress. DNA Repair (Amst) 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 7.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Mol CD, Parikh SS, Putnam CD, Lo TP, Tainer JA. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu REv Biophys Biomol Struct. 1999;28:101–128. doi: 10.1146/annurev.biophys.28.1.101. [DOI] [PubMed] [Google Scholar]
- 9.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 10.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxo-guanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tchou J, Grollman AP. Repair of DNA containing the oxidatively-damaged base 8-hydroxyguanine. Mutat Res. 1993;299:277–287. doi: 10.1016/0165-1218(93)90104-l. [DOI] [PubMed] [Google Scholar]
- 12.Chang DY, Gu Y, Lu AL. Fission yeast (Schizosaccharomyces pombe) cells defective in the MutY-homologous glycosylase activity have a mutator phenotype and are sensitive to hydrogen peroxid. Mol Genet Genomics. 2001;266:336–342. doi: 10.1007/s004380100567. [DOI] [PubMed] [Google Scholar]
- 13.Lu AL, Fawcett WP. Characterization of the recombinant MutY homolog, an adenine DNA glycosylase, from Schizosacchromyces pombe. J Biol Chem. 1998;273:25098–25105. doi: 10.1074/jbc.273.39.25098. [DOI] [PubMed] [Google Scholar]
- 14.Al Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C to T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 15.Halford SE, Rowan AJ, Lipton L, Sieber OM, Pack K, Thomas HJ, Hodgson SV, Bodmer WF, Tomlinson IP. Germline mutations but not somatic changes at the MYH locus contribute to the pathogenesis of unselected colorectal cancers. Am J Pathol. 2003;162:1545–1548. doi: 10.1016/S0002-9440(10)64288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C→T:A mutations. Hum Mol Genet. 2002;11:2961–2967. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- 17.Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, Bisgaard ML, Orntoft TF, Aaltonen LA, Hodgson SV, Thomas HJ, Tomlinson IP. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 18.Sampson JR, Dolwani S, Jones S, Eccles D, Ellis A, Evans DG, Frayling I, Jordan S, Maher ER, Mak T, Maynard J, Pigatto F, Shaw J, Cheadle JP. Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet. 2003;362:39–41. doi: 10.1016/S0140-6736(03)13805-6. [DOI] [PubMed] [Google Scholar]
- 19.Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med. 2007;39:335–345. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 20.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 22.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 23.Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins--novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 24.Haldar D, Kamakaka RT. Fission yeast Hst4 functions in DNA damage response by regulating histone H3 K56 acetylation. Eukaryot Cell. 2008;7:800–813. doi: 10.1128/EC.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durand-Dubief M, Sinha I, Fagerstrom-Billai F, Bonilla C, Wright A, Grunstein M, Ekwall K. Specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation and retrotransposon silencing. EMBO J. 2007;26:2477–2488. doi: 10.1038/sj.emboj.7601690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozdemir A, Masumoto H, Fitzjohn P, Verreault A, Logie C. Histone H3 lysine 56 acetylation: a new twist in the chromosome cycle. Cell Cycle. 2006;5:2602–2608. doi: 10.4161/cc.5.22.3473. [DOI] [PubMed] [Google Scholar]
- 29.Freeman-Cook LL, Sherman JM, Brachmann CB, Allshire RC, Boeke JD, Pillus L. The Schizosaccharomyces pombe hst4(+) gene is a SIR2 homologue with silencing and centromeric functions. Mol BiolCell. 1999;10:3171–3186. doi: 10.1091/mbc.10.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 31.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 32.Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004;271:2335–2349. doi: 10.1111/j.1432-1033.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 33.Canman CE. Replication checkpoint: Preventing mitotic catastrophe. Curr Biol. 2001;11:R121–R124. doi: 10.1016/s0960-9822(01)00057-4. [DOI] [PubMed] [Google Scholar]
- 34.Longhese MP, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 36.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 37.Hang H, Lieberman HB. Physical interactions among human checkpoint control proteins HUS1p, RAD1p, and RAD9p, and implications for the regulation of cell cycle progression. Genomics. 2000;65:24–33. doi: 10.1006/geno.2000.6142. [DOI] [PubMed] [Google Scholar]
- 38.St Onge RP, Udell CM, Casselman R, Davey S. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol Biol Cell. 1999;10:1985–1995. doi: 10.1091/mbc.10.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the Rad9-Rad1-Hus1 DNA damage checkpoint complex--implications for clamp loading and regulation. Cell Mol. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 40.Sohn SY, Cho Y. Crystal structure of the human Rad9-Hus1-Rad1 clamp. J Mol Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 41.Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T. Structure and functional implications of the human Rad9-Hus1-Rad1 cell cycle checkpoint complex. J Biol Chem. 2009;284:20457–20461. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J Biol Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 43.Shiomi Y, Shinozaki A, Nakada D, Sugimoto K, Usukura J, Obuse C, Tsurimoto T. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-Rad1-Hus1 and Rad17-RFC. Genes to Cells. 2002;7:861–868. doi: 10.1046/j.1365-2443.2002.00566.x. [DOI] [PubMed] [Google Scholar]
- 44.Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucl Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang DY, Lu AL. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J Biol Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- 46.Shi G, Chang DY, Cheng CC, Guan X, Venclovas C, Lu AL. Physical and functional interactions between MutY homolog (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez-Capetillo O, Nussenzweig A. Linking histone deacetylation with the repair of DNA breaks. Proc Natl Acad Sci USA. 2004;101:1427–1428. doi: 10.1073/pnas.0307342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iizuka M, Smith MM. Functional consequences of histone modifications. Curr Opin Genet Dev. 2003;13:154–160. doi: 10.1016/s0959-437x(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 49.Bjerling P, Silverstein RA, Thon G, Caudy A, Grewal S, Ekwall K. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol Cell Biol. 2002;22:2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freeman-Cook LL, Gomez EB, Spedale EJ, Marlett J, Forsburg SL, Pillus L, Laurenson P. Conserved locus-specific silencing functions of Schizosaccharomyces pombe sir2+ Genetics. 2005;169:1243–1260. doi: 10.1534/genetics.104.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 52.Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol. 2004;24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 54.Wright WE, Shay JW. Telomere-binding factors and general DNA repair. Nat Genet. 2005;37:116–118. doi: 10.1038/ng0205-116. [DOI] [PubMed] [Google Scholar]
- 55.Muftuoglu M, Wong HK, Imam SZ, Wilson DM, III, Bohr VA, Opresko PL. Telomere repeat binding factor 2 interacts with base excision repair proteins and stimulates DNA synthesis by DNA polymerase beta. Cancer Res. 2006;66:113–124. doi: 10.1158/0008-5472.CAN-05-2742. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Rhee DB, Lu J, Bohr CT, Zhou F, Vallabhaneni H, Souza-Pinto NC, Liu Y. Characterization of oxidative Guanine damage and repair in mammalian telomeres. PLoS Genet. 2010;6:e1000951. doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez JA, Kim SM, Huberman JA. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp Cell Res. 1998;238:220–230. doi: 10.1006/excr.1997.3835. [DOI] [PubMed] [Google Scholar]
- 58.Khair L, Chang YT, Subramanian L, Russell P, Nakamura TM. Roles of the checkpoint sensor clamp Rad9-Rad1-Hus1 (911)-complex and the clamp loaders Rad17-RFC and Ctf18-RFC in Schizosaccharomyces pombe telomere maintenance. Cell Cycle. 2010;9 doi: 10.4161/cc.9.11.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakamura TM, Moser BA, Russell P. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics. 2002;161:1437–1452. doi: 10.1093/genetics/161.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 61.Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J Intern Med. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 63.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oikawa S, Kawanishi S. Site-specific DNA damage at GGG sequence by oxidative stress may accelerate telomere shortening. FEBS Lett. 1999;453:365–368. doi: 10.1016/s0014-5793(99)00748-6. [DOI] [PubMed] [Google Scholar]
- 67.Rubio MA, Davalos AR, Campisi J. Telomere length mediates the effects of telomerase on the cellular response to genotoxic stress. Exp Cell Res. 2004;298:17–27. doi: 10.1016/j.yexcr.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 68.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 69.Opresko PL, Fan J, Danzy S, Wilson DM, III, Bohr VA. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucl Acids Res. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 71.Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 72.Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 73.Thaminy S, Newcomb B, Kim J, Gatbonton T, Foss E, Simon J, Bedalov A. Hst3 is regulated by Mec1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J Biol Chem. 2007;282:37805–37814. doi: 10.1074/jbc.M706384200. [DOI] [PubMed] [Google Scholar]
- 74.Xhemalce B, Miller KM, Driscoll R, Masumoto H, Jackson SP, Kouzarides T, Verreault A, Arcangioli B. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J Biol Chem. 2007;282:15040–15047. doi: 10.1074/jbc.M701197200. [DOI] [PubMed] [Google Scholar]
- 75.Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 78.Chang DY, Lu AL. Functional interaction of MutY homolog (MYH) with proliferating cell nuclear antigen (PCNA) in fission yeast, Schizosaccharomyces pombe. J Biol Chem. 2002;277:11853–11858. doi: 10.1074/jbc.M111739200. [DOI] [PubMed] [Google Scholar]
- 79.Celic I, Verreault A, Boeke JD. Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage. Genetics. 2008;179:1769–1784. doi: 10.1534/genetics.108.088914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 81.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 82.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons; New York, NY: 1994. [Google Scholar]
- 83.Ampatzidou E, Irmisch A, O'Connell MJ, Murray JM. Smc5/6 is required for repair at collapsed replication forks. Mol Cell Biol. 2006;26:9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.