Abstract
Due to the importance of microbes as model organisms, biotechnology tools, and contributors to mammalian and ecosystem metabolism, there has been longstanding interest in measuring their metabolite levels. Current metabolomic methods, involving mass spectrometry-based measurement of cell extracts, enable routine quantitation of most central metabolites. Metabolomics alone, however, is inadequate to understand cellular metabolic activity: Flux measurement and proteomic, genetic, and biochemical approaches with a metabolomics bent are all needed. Here we highlight examples where these integrated methods have contributed to discovery of metabolic pathways, regulatory interactions, and homeostasis mechanisms. We also indicate enduring challenges concerning unstable and low abundance compounds, subcellular compartmentalization, and quantitative amalgamation of different data types.
Systems biology aims to explain physiological processes in terms of the concerted actions of numerous biochemicals. Microbial metabolism would seem to provide a promising arena for achieving this aim. Balanced growth in diverse environments a hallmark of microbial physiology is a metabolic capability. Microbes are relatively simple. Model microbes are also easy to grow and genetically tractable. The connections between metabolites, catalyzed by enzymatic transformations, are well mapped.
Consistent with this promise, microbial metabolism has been among the fastest areas of systems biology to develop. A key driver in this regard has been the constraints-based computational approach of flux balance analysis (FBA). A remarkable finding has been that E. coli (although not most other microbes) often maximizes growth per unit of carbon source consumed: Fluxes are optimally efficient [1]. This raises key questions: What regulatory mechanisms lead to flux optimality? How are these mechanisms different in organisms that have different metabolic objectives? How do these mechanisms coordinate appropriate responses to changing nutrient availability?
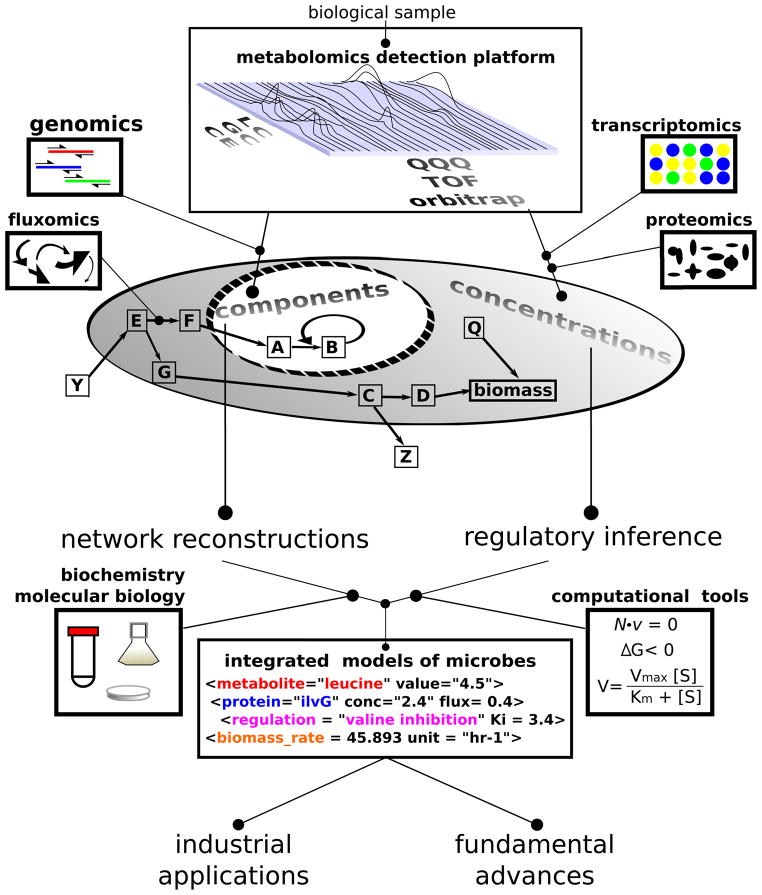
Metabolomics, defined broadly as the systems biology of metabolism, aims to address these questions. Metabolomics, defined more narrowly as comprehensive metabolite measurement, provides a critical tool for doing so. Figure 1 provides a schematic overview of microbial metabolomics and its integration with other experimental approaches. Metabolomics and genomics provide complementary tools for identifying metabolites and their reactions and thus reconstructing metabolic networks. Metabolomics, proteomics, and transcriptomics each provide data on the concentrations of cellular biochemicals. Fluxomics measures the cellular rates of enzyme-catalyzed reactions. This concentration and flux data, when overlaid on the metabolic network architecture, can be used to build and test quantitative, integrative models of microbes. The modeling process itself often leads to fundamental advances, e.g., when new reactions or regulation are required for the model to match experimental results. Moreover, the resulting predictive models should have value in the development and optimization of industrial processes, e.g., for producing desired metabolic products.
Figure 1. Overview of metabolomics in systems microbiology.
Metabolomic and genomics provide complementary information for identifying an organism’s metabolic capabilities. Concentration data for metabolites, proteins, and transcripts can be used for regulatory inference. Computational integration of such data aims to enable the development of mechanistically accurate, predictive metabolic models.
Here we briefly review the current state of metabolomic methods for microbes. We then examine case studies where comprehensive metabolite measurement has been successfully used in concert with other experimental methods and quantitative modeling to enable pathway discovery, regulatory inference, and understanding of homeostatic capabilities. Throughout, we point out areas where technological advances are particularly needed.
Metabolomic methods
Prototrophic microbes have a core metabolome consisting of approximately 700 compounds involved in central metabolism and essential biosynthetic pathways [2]. Including secondary metabolism and lipids, the actual number of small molecule species found in a typical microbe is substantially greater. Many core metabolites are biosynthetic intermediates that are present in low levels, often more than 10,000-fold less than the most abundant metabolites [3]. Such trace metabolites remain challenging to measure by any analytical technique. Instrumentation with improved sensitivity, as well as methods for their concentration (e.g., by solid phase extraction), are needed. More abundant core metabolites, including central carbon intermediates, amino acids, nucleotides, and cofactors, can be measured en masse by current LC-MS methods [4]. GC-MS and NMR provide complementary information by enabling quantitation of uncharged metabolites that are hard to measure by LC-MS [5,6]. In skilled hands, they, as well as CE-MS [7], also can largely substitute for LC-MS.
Metabolome measurement is a multiple-step process. Typical steps include cell growth, metabolism quenching, metabolome extraction, sample concentration, detection by LC-MS or another advanced instrument, and data analysis. As metabolite levels can change within seconds in microbes, special care must be taken to avoid artifacts due to cell handling steps prior to metabolome quenching [8]. For absolute metabolite quantitation, it is important to add isotopic internal standards at the time of metabolome quenching, so that the standards experience equal chances for degradative and absorptive losses as the endogenous compounds [9]. Many of the known approaches for non-disruptive, rapid metabolism quenching also initiate the extraction process [10]. For extraction of core metabolites from microbes, a 40:40:20 mixture of acetonitrile:methanol:water at cold temperature generally works well [11]. This mixture, especially with addition of formic acid, effectively preserves adenylate energy charge. Methods that similarly preserve NADH and NADPH are still needed. Once an extract is obtained, it can be concentrated using a nitrogen gas stream or freeze drier. Although risking metabolome alterations, the concentration step can both enhance signals and remove solvents that may interfere with subsequent analysis. For example, organic solvent in the sample can impair reversed phase LC separation.
In terms of actual analysis, many methods will give adequate results, and a combination of methods is often best [12]. A particularly effective single LC-MS method involves reversed phase LC separation with tributylamine ion pairing agent to enhance retention of anionic compounds, coupled to MS by negative mode electrospray ionization (ESI) [13–15]. This method covers, in one shot, most abundant core metabolites. A disadvantage is that tributylamine will interfere with positive mode ESI and is hard to wash out of an LC. Thus an LC dedicated to negative mode analysis is required.
In terms of mass spectrometers, triple quadrupole instruments are robust and good for measuring targeted metabolites. High resolution full scan detectors (time-of-flight or orbitrap) provide similar quantitation of known metabolites while also providing data on unexpected compounds. Accordingly, they are emerging as leading metabolomic tools. A continuing push in the mass spectrometry field is for more sensitive instruments with better mass resolution. Improved sensitivity will increase the range of metabolites that can be measured; better mass resolution will filter out interferences and reduce the risk of compound misidentification.
Pathway mapping
Despite complete sequencing of numerous microbial genomes, the functions of many genes remain unknown and many catalyzed transformations are performed by unknown gene products. This represents a major barrier to building accurate genome-scale models of metabolism. Accordingly, a major and long-standing goal of metabolomics is gene function assignment [16,17], recently reviewed in [18]. A simple approach involves examining metabolic changes in knockout strains [19,20]. For enzyme knockouts, substrate accumulation and/or product depletion is expected. For example, the compound changing most in a yeast deletion strain for YKL215C was oxoproline, leading to subsequent biochemical confirmation of YKL215C as an oxoprolinase [14]. Enhanced metabolomic throughput may soon enable such analyses at the genome-scale [21].
For known pathways, the mere observation of pathway metabolites can be a powerful aid in demonstrating the pathway’s activity. For example, in the promising bioenergy organism Clostridium acetobutylicum, the presence of citrate provided immediate evidence for existence of a functional TCA cycle, despite the paucity of annotated TCA enzymes. A complete, bifurcated cycle was then revealed using isotope tracers [22].
It is also possible for well known pathways to operate in a new manner. For example, feeding of 13C-labeled glutamine to the malaria parasite Plasmodium falciparum revealed a variant of the TCA cycle that bifurcates at α-ketoglutarate. In addition to carbon flowing clockwise from α-ketoglutarate through succinate to malate, it also flows counterclockwise through citrate to acetyl-CoA and malate [23]. The resulting bifurcated cycle is redox neutral and produces two carbon units. Enzymes involved uniquely in Plasmodium metabolism, such as the yet-to-be-defined citrate-cleaving enzyme, could become important drug targets.
For novel substrates and pathways, the hard work remains compound structure identification and pathway assembly. Better ability to predict structures from MS/MS data computationally would have great value. While some such methods exist [24], current work typically relies substantially on experimentation, as exemplified in recent mapping of a new pathway of pyrimidine catabolism in E. coli. Building from the observation that knockout of the rut operon blocks use of uracil as a sole nitrogen source, Loh et al. used a combination of 14C tracing and GC-MS to demonstrate that enzymes of the rut operon degrade uracil to 3-hydroxypropionic acid with concomitant release of two molecules of ammonia [25]. Follow-up studies have begun to elucidate the underlying enzymatic mechanisms [26].
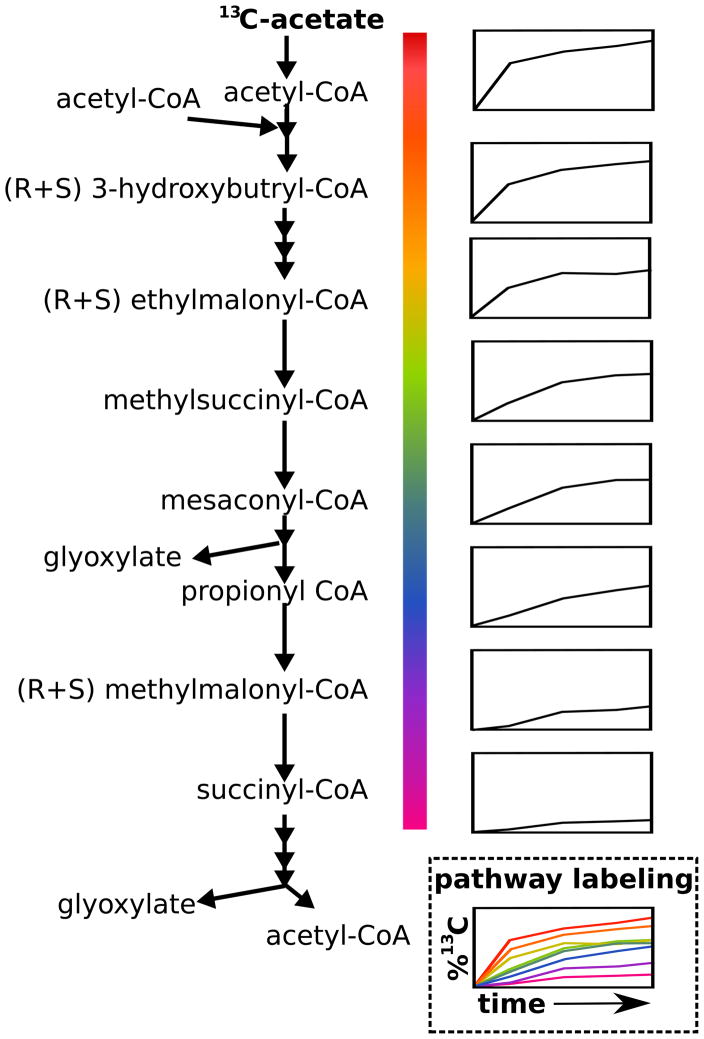
A set of organisms known as methylotrophs are of industrial interest for their ability produce complex molecules from methane or methanol. Methanol is assimilated by its reaction with glycine to yield serine. In the organism Methylobacterium extorquens AM1, there was a long-standing question regarding the pathway used to produce glyoxlyate, the carbon skeleton for the glycine. One hypothesis involved a series of nine steps known as the ethylmalonyl-CoA pathway, which had not been demonstrated in any organism. Recently, Peyraud et al. were able to show the presence of every intermediate of this putative pathway in M. extorquens using high resolution LC-MS [27••]. Furthermore, they showed that these compounds label from supplemented 13C-acetate in the temporal order of the proposed pathway (Figure 2). This was quite a technical tour-de-force given that the intermediates are liable CoA thioesters, and that labeling occurred very quickly reaching completion within 10 s. But, with the use of acid to stabilize the thioesters and a careful manual method for direct quenching and harvesting of metabolites, definitive support for the pathway was obtained. An important aspect of the pathway is its overall stoichiometry, which is capable of building two trioses from three methanol and three CO2 molecules (Figure 2). This costs less methanol and assimilates more CO2 than the serine cycle used in some other methylotrophs. Thus, metabolomics-driven identification of the ethylmalonyl-CoA pathway has elevated overall interest in M. extorquens as an industrial organism. In sum, these investigations demonstrate the power of combining metabolomics with other techniques for elucidating novel pathway architectures.
Figure 2. Elucidation of the methanol assimilation pathway in Methylobacterium extorquens AM1.
Metabolomics identified the intermediates of the novel ethylmalonyl-CoA pathway for methanol assimilation. Isotopic tracing from 13C-acetate confirmed the reaction sequence. Small graphs on the right side of the figure show percent 13C-labeling versus time.
Regulatory inference
More than simply resolving the connections of species, metabolomics provides a quantitative tool to investigate their dynamics. These can then provide insight into metabolic regulation. The resulting knowledge of regulatory mechanisms can then help guide bioengineering efforts to control metabolic flux rationally and expand our comprehension of microbial physiology, which is often involves a series of evanescent or even oscillating states [28].
A simple proof-of-concept study of metabolic dynamics involves the nitrogen assimilation network of E. coli. While there are only three central nitrogen metabolites (glutamine, glutamate, and α-ketoglutarate) and three enzymes, these are tied to all of central metabolism through the TCA cycle and energy cofactors (ATP and NADPH).
One of the enzymes, glutamine synthetase, is controlled by a cascade of protein covalent modification that has been studied extensively [29,30]. This cascade is allosterically regulated by glutamine and α-ketoglutarate. In examining the metabolomic response to increased ammonia availability, Yuan et al. observed the strongest metabolic changes in these two compounds [31•]. The changes percolated into other amino acids and TCA cycle compounds but not more broadly. Cofactor and glycolytic intermediate concentrations were nearly constant. Kinetic modeling of the concentrations of the central nitrogen compounds revealed that even this simple pathway contains unappreciated regulatory interactions. Transcription, protein covalent modification, and allostery were all known from genetics and biochemistry to play a role. However, the rate of glutamate biosynthesis was instead controlled primarily by competition between different amino acids (glutamine, glutamate, and aspartate) for the active site of glutamate synthase. Given the crowded nature of the cytosol, such active site competition may be a common contributor to in vivo metabolic regulation. Identifying cases of such competition may help in bioengineering efforts. For example, when such competition occurs, increasing the concentration of an enzyme’s substrate far above the in vitro Km value may be required to achieve near-maximal metabolic flux.
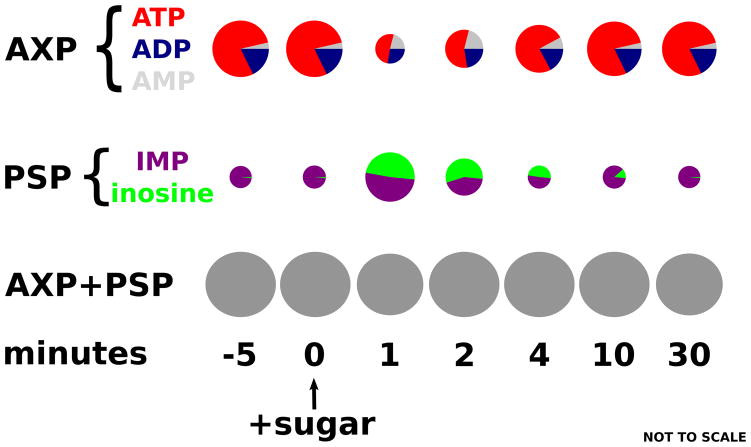
Akin to nitrogen assimilation, the respiro-fermentative transition in Saccharomyces cerevisiae has been studied extensively [32,33]. A mystery arising from these investigations was the rapid depletion of adenine nucleotides upon addition to glucose to respiring yeast. Abundant glucose would be expected to lead to rapid conversion of ATP to ADP as the hexokinase reaction rushes forward. However, depletion of ATP occurred without a corresponding increase in ADP or AMP. To investigate, Walther et al. [34••] used LC-MS. They determined that the “missing” ATP could precisely be accounted for by increases in the purine salvage pathway intermediates inosine and inosine monophosphate (Figure 3). ADP made by hexokinase is converted by adenylate kinase to AMP, which is then deaminated by Amd1. This pathway for degradation of AMP is selectively active in the presence of a hexose sugar. While wasteful of energy, it has the benefit of transiently helping to maintain adenylate energy charge. Since the presence of hexose sugar guarantees adequate long-term energy, it is important for cells to avoid being fooled by a temporary build-up of AMP into acting as if they are energy limited. In addition, AMP degradation helps ensure that ATP wins the competition for enzyme active sites. This case study highlights the importance that microbes place on properly matching internal metabolite levels with environmental conditions in this case, low AMP with an energy rich sugar-containing environment.
Figure 3. Metabolome dynamics reveal a role for purine salvage in the yeast respiro-fermentative transition.
Using isotopic standards and LC-MS to quantitate purine salvage pathway intermediates, Walther et al. solved the long-standing mystery of counter-intuitive total adenosine phosphate (AXP) depletion following glucose addition to respiring yeast. AXP is shuttled through IMP and inosine to prevent AMP accumulation and associated impairment of growth by low adenylate energy charge.
In both of these studies, mutants defective in transitions following nutrient addition but not steady state growth were critical to discovering these dynamic regulatory mechanisms. In each case, the regulatory mechanisms deal in part with “indicator” metabolites glutamine as a sign of nitrogen sufficiency and AMP of energy insufficiency. The mechanisms are specifically designed to function in a crowded cytosolic environment filled with many look-alike metabolites. In former case, this crowding helps glutamine properly drive metabolic flux. In the latter case, energy is spent to avoid the possibility of AMP or ADP detrimentally dwelling in active sites meant for ATP, their more energy rich cousin. The carefully regulated responses of metabolite concentrations during nutrient shifts suggest tight evolutionary optimization of these transitions.
Integrated flux regulation
While active site occupancy, allostery, and protein covalent modification enable fast metabolic responses, metabolism is also regulated on longer time scales by changes in enzyme concentrations. As bioengineering efforts typically involve sustained growth of genetically modified microbes, understanding the interplay of transcriptional and faster acting regulatory mechanisms in control of metabolism is especially industrially relevant. Two recent studies in yeast shed light on this topic, by probing how metabolism adapts when enzyme levels are dysregulated due to transcription factor knockout.
Fendt et al. measured transcript, enzyme, and metabolite concentrations in wild-type yeast and a GCR2 transcription factor mutant [35•]. This mutant exhibits decreased glycolytic and enhanced TCA cycle enzyme expression. The resulting changes in enzyme levels are buffered by changes in substrate concentrations so as to maintain metabolic flux a property essential for cell growth. Interestingly, this observed anti-correlation (enzyme down, substrate up, and vice versa) was stronger for glycolytic and TCA cycle compounds than for reaction cofactors (e.g., NAD+). This is in keeping with recent data from E. coli, which suggests that cofactor sites are usually substrate-saturated, whereas sites for glycolytic and TCA metabolites often are not [3]. Changing glycolytic and TCA metabolite levels therefore is the more straightforward road to modulating flux than varying cofactors. In follow-up experiments, expression of specific glycolytic enzymes was reduced using a Tet-repressible promoter system in place of the more globally perturbed GCR2 mutant. Consistent with flux compensation through greater substrate active-site occupancy, this brought about local accumulation of the enzyme substrates. This accommodation of altered enzyme levels by substrate concentration changes is in accordance with flux control being distributed over multiple pathway enzymes, as argued by metabolic control analysis [36]. It is also possible, however, that flux control resides outside of glycolysis or the TCA cycle per se, e.g., in demand for ATP or in regulation of glucose uptake. Resolving such issues is of more than academic interest. For example, if glycolytic flux is limited by demand for ATP, then even over-expression of the full glycolytic pathway would not enhance production of the biofuel ethanol; however, expressing a general ATP hydrolyase might do so.
Amino acid biosynthesis in yeast provides such a case where flux control typically resides outside of a metabolic pathway. Except in cases (such as glycine) where a single amino acid can be made multiple different ways, steady-state pathway flux must match consumption by protein synthesis. Moxley et al. probed the means by which yeast lacking the GCN4 transcription factor maintain amino acid production when enzyme levels are inappropriately low [37•]. An inhibitor of histidine biosynthesis was titrated into cultures of wild type and mutant yeast to induce matched growth rates. This inhibitor caused amino acid starvation and thus, in wild-type cells, GCN4-induced expression of amino acid biosynthetic enzymes. In the absence of such expression, amino acid levels were lower, and this appeared to restore flux by relief of feedback inhibition by metabolite end products.
Glycine biosynthesis presented an interesting exception to the norm of metabolic flux conservation. In the absence of GCN4 activation, yeast synthesize glycine primarily from serine; upon its activation, glycine is instead made from threonine. While Moxley et al. found no overall correlation between transcript levels and biosynthetic fluxes, the GCN4-driven shift in the route of glycine biosynthesis was accompanied by matching changes in enzyme transcript levels.
These two studies examined different aspects of yeast metabolism under experimental conditions that demanded flux conservation. In the face of deranged transcription, yeast maintained flux through altered levels of metabolic substrates [35•] and of allosteric effectors [37•]. When an overt shift in pathway usage occurred (i.e., of routes of glycine biosynthesis), this was driven by transcriptional regulation. In a biotechnology setting, the flux maintenance would have been a depressing outcome a failure of metabolic engineering. In a more natural setting, rather than maintaining similar flux, the transcription-defective mutants would have been out-competed for scarce nutrients and ended up flux-deficient. Thus, while these studies highlight the potential for tradeoffs between metabolite and enzyme levels, further work is needed to understand how these tradeoffs play out in natural and industrial environments. To enable effective engineering, identifying points of flux control is particularly important. Figuring out the best way to do this via metabolomics and other modern methods is an open area of research.
Homeostasis maintenance
There are two overarching goals of integrated flux regulation: optimizing fluxes and maintaining metabolite levels within acceptable homeostatic bounds. Simultaneously meeting these goals is facilitated by the acceptable homeostatic bound often being broad, e.g., greater than 10-fold. For example, yeast abruptly transitioned to nitrogen starvation quickly manifest lower amino acid levels but do not slow growth for at least one generation [38]. Similarly, E. coli lacking the pentose phosphate pathway enzyme transaldolase (talAB) grow at only a slightly reduced rate on xylose, despite 40-fold accumulation of sedoheptulose-7-phosphate [39•].
For genetic deletions, the ability to maintain homeostasis reflects intrinsic robustness of the metabolic network. For most of central metabolism, when one pathway is knocked out, another automatically assumes its function. As microbes have not evolved to deal with genetic changes, they do not manifest a substantial adaptive transcriptional response. For example in E. coli, knockout of glucose-6-phosphate dehydrogenase eliminates the normal route for making the required nucleic acid component ribose-5-phosphate. Its level accordingly falls very low [40]. Independent of changes in enzyme levels, this metabolite concentration change results in reversal of flux through the non-oxidative pentose phosphate pathway. The “backwards” flux reroutes carbon from fructose-6-phosphate to ribose-5-phosphate, providing the essential metabolite. Such flux reversal requires changes in metabolite concentrations sufficient to shift the ΔG for the entire pathway from positive to negative.
Recently, Nakahigashi et al. have found evidence for promiscuity of certain central carbon metabolic enzymes. Such promiscuity provides another potential source of metabolic network robustness [39•]. For example, in the transaldolase knockout mentioned above, sedoheptulose-7-phosphate accumulates to sufficient levels to compete with fructose-6-phosphate for the active site of phosphofructokinase. Ensuing phosphorylation produces sedoheptulose-1,7-bisphosphate, which is then hydrolyzed to dihydroxyacetone phosphate and erythrose-4-phosphate by fructose-bisphosphate aldolase. This provides a route to catabolize xylose in the absence of transaldolase activity. This alternate activity can support around 90% of the wild type growth rate, an impressive amount given that phosphofructokinase likely evolved to minimize, rather than maximize, use of sedoheptulose-7-phosphate as its substrate. An intriguing possibility is that such enzyme promiscuity could be put to use for bioengineering purposes.
In contrast to genetic ablations, changing environmental conditions have been strong drivers of microbial evolution. Accordingly, both transcriptional and post-translational regulatory mechanisms for producing appropriate metabolic responses are extensive. These mechanisms are critically needed, both to optimize long-term fluxes and to avoid immediate toxicity due to excessive metabolome alteration. For example, E. coli lacking the ability to inactivate glutamine synthetase by covalent modification suffer toxic metabolic derangements when shifted into a nitrogen-rich environment [31•].
Despite these homeostatic mechanisms, the response of the metabolome to changing environmental conditions is rapid and specific [41]. The speed reflects the short lifetime of most metabolites. The specificity reflects the direct ties between intracellular metabolites and the nutrient environment. For example, in yeast, glutamine is a direct product of ammonia and a sentinel of nitrogen status; ATP is a direct product of inorganic phosphate and a sentinel of phosphorous status [42]. Together, this quickness and specificity enables metabolites to serve as reliable signals of the nutrient environment to ever-vigilant cells. Metabolite concentrations modulate metabolic flux, trigger signaling cascades, regulate transcription factor activity, and alter biomass production rates.
Towards integrative models
Given dynamic interplay between metabolites, fluxes, proteins, and transcripts, where does this leave us in terms of making use of metabolomic data? Currently, metabolomic data is used mainly for two purposes: to get a general impression of metabolome changes and to pick out a few interesting compounds for further study. New approaches are much needed to capitalize fully on both the data’s comprehensive and quantitative nature. To this end, FBA could provide a useful starting point for global quantitative analysis. Recently, Yizhak et al. devised a framework to augment FBA with enzyme and metabolite concentration data [43•]. Their method refines the underdetermined flux space of traditional FBA with estimates of fluxes based on Michaelis-Menten type kinetics. The requisite parameters are derived using metabolomic data to estimate enzyme active site saturation and proteomic data to estimate changes in enzyme Vmax values. As highlighted above, these metrics are directly relevant to metabolic control. Their method predicted the flux effects of enzyme knockouts in E. coli substantially better than FBA alone.
Dynamic models are an important complement to constraints-based models like FBA, as they capture mechanisms of flux control. A difficulty of such models is that they typically involve a large number of unmeasured parameters [44]. As shown in Yuan et al., metabolomic data can be used for parameter identification as well as for model testing and refinement [31•]. Because dynamic models can quickly become intractable, it is most promising, initially, to develop and validate sub-network models, e.g., of specific pathways. Validated sub-network models can then be joined to simulate larger and larger systems. Metabolites at the sub-networks’ interfaces are candidates for regulating integration between them. A key question is whether such joining together of subnetwork models will yield larger predictive models.
Future horizons
A great deal remains to be learned to understand entirely the metabolism of any microbe, even one as simple and genetically tractable as E. coli. A pivotal milestone will be reaching a degree of understanding sufficient to enable quantitative prediction, based on underlying biochemistry, of metabolome-wide response to perturbations. For prokaryotes, the most important barrier in this regard is probably in quantitative data integration. While better data more precise measurements of more metabolites in more conditions would surely help, existing methods are largely adequate.
For eukaryotes, to deal with their extensive compartmentation, improved experimental methods are a more pressing need. All current methods for quenching metabolism also result in mixing of metabolites between subcellular compartments; accordingly, standard fractionation approaches, common in proteomics, are not viable for metabolomics. There is no direct solution to this critical problem on the horizon. Instead, the most immediate hope seems to lie in fluorescence-based measurements of metabolite concentrations in live cells and subcellular compartments [45]. The expansive chemical diversity of metabolites necessitates a correspondingly vast collection of specific protein sensors, whose scarcity currently limits these techniques. But advances in artificial evolution and design of binding proteins should accelerate these in situ metabolite measurements over the coming decade such that double-digit numbers of metabolites concentrations may be resolved spatially in real time.
While yeast and E. coli happily grow unaccompanied in a test tube, most ecologically and medically interesting microbiology involves multiple organisms. There have already been informative metabolomic studies of multi-organism systems, which range in complexity from a defined virus-host cell pair [46–48] to diverse organisms colonizing the mammalian gut [49–51]. Such systems highlight the need for spatially localized metabolite measurements as well as for computational methods of integrating multifarious data sets. As the relevant techniques evolve, the next decade should see substantial breakthroughs in understanding the metabolic interchanges in such microcosms.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edwards JS, Ibarra RU, Palsson BO. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nature Biotechnology. 2001;19:125–130. doi: 10.1038/84379. http://www.ncbi.nlm.nih.gov/pubmed/11175725. [DOI] [PubMed]
- 2.Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: A comprehensive view of Escherichia coli biology. Nucleic Acids Research. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. http://www.nar.oxfordjournals.org/cgi/doi/410.1093/nar/gkn1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nature Chemical Biology. 2009;5:593–599. doi: 10.1038/nchembio.186. http://www.nature.com/doifinder/510.1038/nchembio.1186. [DOI] [PMC free article] [PubMed]
- 4.Büscher JM, Czernik D, Ewald JC, Sauer U, Zamboni N. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Analytical Chemistry. 2009;81:2135–2143. doi: 10.1021/ac8022857. http://www.ncbi.nlm.nih.gov/pubmed/19236023. [DOI] [PubMed]
- 5.Coen M, Holmes E, Lindon JC, Nicholson JK. NMR-Based Metabolic Profiling and Metabonomic Approaches to Problems in Molecular Toxicology. Chemical Research in Toxicology. 2008;21:9–27. doi: 10.1021/tx700335d. [DOI] [PubMed] [Google Scholar]
- 6.Fiehn O. Extending the breadth of metabolite profiling by gas chromatography coupled to mass spectrometry. TrAC Trends in Analytical Chemistry. 2008;27:261–269. doi: 10.1016/j.trac.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohashi Y, Hirayama A, Ishikawa T, Nakamura S, Shimizu K, Ueno Y, Tomita M, Soga T. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Molecular BioSystems. 2008;4:135. doi: 10.1039/b714176a. http://xlink.rsc.org/?DOI=b714176a. [DOI] [PubMed]
- 8.Ikeda TP, Shauger AE, Kustu S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. Journal of Molecular Biology. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. http://www.ncbi.nlm.nih.gov/pubmed/8683567. [DOI] [PubMed]
- 9.Mashego MR, Wu L, Van Dam JC, Ras C, Vinke JL, Van Winden WA, Van Gulik WM, Heijnen JJ. MIRACLE: mass isotopomer ratio analysis of U-13C-labeled extracts. A new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnology and Bioengineering. 2004;85:620–628. doi: 10.1002/bit.10907. http://doi.wiley.com/610.1002/bit.10907. [DOI] [PubMed]
- 10.Rabinowitz JD. Cellular metabolomics of Escherchia coli. Expert Review of Proteomics. 2007;4:187–198. doi: 10.1586/14789450.4.2.187. [DOI] [PubMed] [Google Scholar]
- 11.Rabinowitz JD, Kimball E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Analytical Chemistry. 2007;79:6167–6173. doi: 10.1021/ac070470c. http://www.ncbi.nlm.nih.gov/pubmed/17630720. [DOI] [PubMed]
- 12.van der Werf MJ, Overkamp KM, Muilwijk B, Coulier L, Hankemeier T. Microbial metabolomics: toward a platform with full metabolome coverage. Analytical Biochemistry. 2007;370:17–25. doi: 10.1016/j.ab.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Buescher JM, Moco S, Sauer U, Zamboni N. Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Analytical Chemistry. 2010;82:4403–4412. doi: 10.1021/ac100101d. http://www.ncbi.nlm.nih.gov/pubmed/20433152. [DOI] [PubMed]
- 14.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic Analysis via Reversed-Phase Ion-Pairing Liquid Chromatography Coupled to a Stand Alone Orbitrap Mass Spectrometer. Analytical Chemistry. 2010;82:3212–3221. doi: 10.1021/ac902837x. http://pubs.acs.org/doi/abs/3210.1021/ac902837x. [DOI] [PMC free article] [PubMed]
- 15.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography mass spectrometry. Journal of Chromatography A. 2007;1147:153–164. doi: 10.1016/j.chroma.2007.02.034. http://linkinghub.elsevier.com/retrieve/pii/S0021967307002907. [DOI] [PubMed]
- 16.Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nature Biotechnology. 2003;21:692–696. doi: 10.1038/nbt823. http://www.nature.com/doifinder/610.1038/nbt1823. [DOI] [PubMed]
- 17.Saito N, Robert M, Kitamura S, Baran R, Soga T, Mori H, Nishioka T, Tomita M. Metabolomics Approach for Enzyme Discovery. Journal of Proteome Research. 2006;5:1979–1987. doi: 10.1021/pr0600576. http://pubs.acs.org/doi/abs/1910.1021/pr0600576. [DOI] [PubMed]
- 18.Heinemann M, Sauer U. Systems biology of microbial metabolism. Current Opinion in Microbiology. 2010;13:337–343. doi: 10.1016/j.mib.2010.02.005. http://linkinghub.elsevier.com/retrieve/pii/S1369527410000263. [DOI] [PubMed]
- 19.Saghatelian A, Cravatt B. Discovery metabolite profiling forging functional connections between the proteome and metabolome. Life Sciences. 2005;77:1759–1766. doi: 10.1016/j.lfs.2005.05.019. http://linkinghub.elsevier.com/retrieve/pii/S0024320505005072. [DOI] [PubMed]
- 20.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Analytical Chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. http://pubs.acs.org/doi/abs/710.1021/ac051437y. [DOI] [PubMed]
- 21.Ewald JC, Heux Sp, Zamboni N. High-Throughput Quantitative Metabolomics. Workflow for Cultivation, Quenching, and Analysis of Yeast in a Multiwell Format. Analytical Chemistry. 2009;81:3623–3629. doi: 10.1021/ac900002u. http://pubs.acs.org/doi/abs/3610.1021/ac900002u. [DOI] [PubMed]
- 22.Amador-Noguez D, Feng X-J, Fan J, Roquet N, Rabitz H, Rabinowitz JD. Systems-level metabolic flux profiling elucidates a complete, bifurcated TCA cycle in Clostridium acetobutylicum. Journal of Bacteriology. 2010 doi: 10.1128/JB.00490-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olszewski KL, Mather MW, Morrisey JM, Garcia BA, Vaidya AB, Rabinowitz JD, Llinás M. Branched tricarboxylic acid metabolism in Plasmodium falciparum. Nature. 2010;466:774–778. doi: 10.1038/nature09301. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Sheldon MT, Mistrik R, Croley TR. Determination of Ion Structures in Structurally Related Compounds Using Precursor Ion Fingerprinting. Journal of the American Society for Mass Spectrometry. 2009;20:370–376. doi: 10.1016/j.jasms.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Loh KD, Gyaneshwar P, Markenscoff Papadimitriou E, Fong R, Kim K-S, Parales R, Zhou Z, Inwood W, Kustu S. A previously undescribed pathway for pyrimidine catabolism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5114–5119. doi: 10.1073/pnas.0600521103. http://www.ncbi.nlm.nih.gov/pubmed/16540542. [DOI] [PMC free article] [PubMed]
- 26.Kim K-S, Pelton JG, Inwood WB, Andersen U, Kustu S, Wemmer DE. The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. Journal of Bacteriology. 2010 doi: 10.1128/JB.00201-10. http://www.ncbi.nlm.nih.gov/pubmed/20400551. [DOI] [PMC free article] [PubMed]
- 27••.Peyraud R, Kiefer P, Christen P, Massou S, Portais JC, Vorholt JA. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proceedings of the National Academy of Sciences. 2009;106:4846–4851. doi: 10.1073/pnas.0810932106. http://www.pnas.org/cgi/doi/4810.1073/pnas.0810932106Metabolomics was used to validate the ethylmalonyl-CoA pathway in Methylobacterium extorquens AM1. This novel pathway provides a more efficient stoichiometry for methanol assimilation.l. [DOI] [PMC free article] [PubMed]
- 28.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proceedings of the National Academy of Sciences. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ninfa A, Jiang P. PII signal transduction proteins: sensors of ? -ketoglutarate that regulate nitrogen metabolism. Current Opinion in Microbiology. 2005;8:168–173. doi: 10.1016/j.mib.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annual Review of Microbiology. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 31•.Yuan J, Doucette CD, Fowler WU, Feng X-J, Piazza M, Rabitz HA, Wingreen NS, Rabinowitz JD. Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli. Molecular Systems Biology. 2009;5 doi: 10.1038/msb.2009.60. http://www.nature.com/doifinder/10.1038/msb.2009.60In response to changing nitrogen availability, metabolome changes in E. coli revolved around two central nitrogen metabolites. Kinetic modeling of the data identified the relevance of active site competition for controlling nitrogen assimilation flux. [DOI] [PMC free article] [PubMed]
- 32.Kresnowati MTAP, van Winden WA, Almering MJH, ten Pierick A, Ras C, Knijnenburg TA, Daran-Lapujade P, Pronk JT, Heijnen JJ, Daran JM. When transcriptome meets metabolome: fast cellular responses of yeast to sudden relief of glucose limitation. Molecular Systems Biology. 2006;2 doi: 10.1038/msb4100083. http://www.nature.com/doifinder/10.1038/msb4100083. [DOI] [PMC free article] [PubMed]
- 33.Wu L, van Dam J, Schipper D, Kresnowati MTAP, Proell AM, Ras C, van Winden WA, van Gulik WM, Heijnen JJ. Short-Term Metabolome Dynamics and Carbon, Electron, and ATP Balances in Chemostat-Grown Saccharomyces cerevisiae CEN.PK 113-7D following a Glucose Pulse. Applied and Environmental Microbiology. 2006;72:3566–3577. doi: 10.1128/AEM.72.5.3566-3577.2006. http://aem.asm.org/cgi/doi/3510.1128/AEM.3572.3565.3566-3577.2006. [DOI] [PMC free article] [PubMed]
- 34••.Walther T, Novo M, Rössger K, Létisse F, Loret M-O, Portais J-C, François J-M. Control of ATP homeostasis during the respiro-fermentative transition in yeast. Molecular Systems Biology. 2010;6 doi: 10.1038/msb.2009.100. http://www.nature.com/doifinder/10.1038/msb.2009.100Quantitative measurement of metabolome dynamics uncovered a role for purine recycling in avoiding build-up of AMP after glucose upshift in yeast. This new mechanism of regulating energy charge solved a long-standing mystery in yeast physiology. [DOI] [PMC free article] [PubMed]
- 35•.Fendt S-M, Buescher JM, Rudroff F, Picotti P, Zamboni N, Sauer U. Tradeoff between enzyme and metabolite efficiency maintains metabolic homeostasis upon perturbations in enzyme capacity. Molecular Systems Biology. 2010;6 doi: 10.1038/msb.2010.11. http://www.nature.com/doifinder/10.1038/msb.2010.11In a yeast GCR2 transcription factor mutant with attenuated enzyme levels across glycolysis, metabolomics revealed compensatory increases in substrate concentrations to maintain pathway flux. [DOI] [PMC free article] [PubMed]
- 36.Fell D. Understanding the control of metabolism. 1. London ;;Miami ;Brookfield VT: Portland Press ;;Distributed by Ashgate Pub. Co. in North America; 1997. [Google Scholar]
- 37•.Moxley JF, Jewett MC, Antoniewicz MR, Villas-Boas SG, Alper H, Wheeler RT, Tong L, Hinnebusch AG, Ideker T, Nielsen J, et al. Linking high-resolution metabolic flux phenotypes and transcriptional regulation in yeast modulated by the global regulator Gcn4p. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6477–6482. doi: 10.1073/pnas.0811091106. http://www.ncbi.nlm.nih.gov/pubmed/19346491 Metabolomics was paired with fluxomics and transcriptomics to interrogate the effects of knockout of the GCN4 transcription factor, which regulates biosynthetic enzyme levels in yeast. In the GCN4 knockout strain, reduction of feedback inhibition by decreased end product concentrations counteracted diminished enzyme concentrations. [DOI] [PMC free article] [PubMed]
- 38.Brauer MJ, Yuan J, Bennett BD, Lu W, Kimball E, Botstein D, Rabinowitz JD. Conservation of the metabolomic response to starvation across two divergent microbes. Proceedings of the National Academy of Sciences. 2006;103:19302–19307. doi: 10.1073/pnas.0609508103. http://www.pnas.org/cgi/doi/19310.11073/pnas.0609508103. [DOI] [PMC free article] [PubMed]
- 39•.Nakahigashi K, Toya Y, Ishii N, Soga T, Hasegawa M, Watanabe H, Takai Y, Honma M, Mori H, Tomita M. Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Molecular Systems Biology. 2009;5 doi: 10.1038/msb.2009.65. http://www.nature.com/doifinder/10.1038/msb.2009.65The phenotypes of multi-enzyme knockouts in E. coli were computationally predicted by flux balance analysis and then experimentally measured. In the case of one discrepancy, it was found that an enzymatic side-activity made relevant by drastic metabolite accumulation in the knockout strain provided a new pathway that enabled cell growth. [DOI] [PMC free article] [PubMed]
- 40.Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, et al. Multiple High-Throughput Analyses Monitor the Response of E. coli to Perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. http://www.sciencemag.org/cgi/doi/510.1126/science.1132067. [DOI] [PubMed]
- 41.Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, Selbig J, Willmitzer L. Metabolomic and transcriptomic stress response of Escherichia coli. Molecular Systems Biology. 2010:6. doi: 10.1038/msb.2010.18. http://www.nature.com/doifinder/10.1038/msb.2010.18. [DOI] [PMC free article] [PubMed]
- 42.Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. Growth-limiting Intracellular Metabolites in Yeast Growing under Diverse Nutrient Limitations. Molecular Biology of the Cell. 2009;21:198–211. doi: 10.1091/mbc.E09-07-0597. http://www.molbiolcell.org/cgi/doi/110.1091/mbc.E1009-1007-0597. [DOI] [PMC free article] [PubMed]
- 43•.Yizhak K, Benyamini T, Liebermeister W, Ruppin E, Shlomi T. Integrating quantitative proteomics and metabolomics with a genome-scale metabolic network model. Bioinformatics. 2010;26:i255–i260. doi: 10.1093/bioinformatics/btq183. http://www.bioinformatics.oxfordjournals.org/cgi/doi/210.1093/bioinformatics/btq1183 A flux balance model was constrained using Michalis-Menten kinetics with parameters drawn from metabolomics and proteomics datasets. This improved prediction of flux phenotypes in enzyme knockouts. [DOI] [PMC free article] [PubMed]
- 44.Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP. Universally Sloppy Parameter Sensitivities in Systems Biology Models. PLoS Computational Biology. 2007;3:e189. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fehr M, Frommer WB, Lalonde S. Visualization of maltose uptake in living yeast cells by fluorescent nanosensors. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9846–9851. doi: 10.1073/pnas.142089199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the Cellular Metabolome during Human Cytomegalovirus Infection. PLoS Pathogens. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodgers MA, Saghatelian A, Yang PL. Identification of an Overabundant Cholesterol Precursor in Hepatitis B Virus Replicating Cells by Untargeted Lipid Metabolite Profiling. Journal of the American Chemical Society. 2009;131:5030–5031. doi: 10.1021/ja809949r. http://pubs.acs.org/doi/abs/5010.1021/ja809949r. [DOI] [PMC free article] [PubMed]
- 48.Wikoff WR, Pendyala G, Siuzdak G, Fox HS. Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. Journal of Clinical Investigation. 2008:118. doi: 10.1172/JCI34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin F-P, Rezzi S, Ross A, Kochhar S, Holmes E, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Molecular Systems Biology. 2008:4. doi: 10.1038/msb.2008.56. http://www.nature.com/doifinder/10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed]
- 50.Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA. Human gut microbiome adopts an alternative state following small bowel transplantation. Proceedings of the National Academy of Sciences. 2009;106:17187–17192. doi: 10.1073/pnas.0904847106. http://www.pnas.org/cgi/doi/17110.11073/pnas.0904847106. [DOI] [PMC free article] [PubMed]
- 51.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]