Abstract
Although dermatophytes are the most common cause of fungal infections in the world, their basic biology is not well understood. The recent sequencing and annotation of the genomes of five representative dermatophyte species allows for the creation of hypotheses as to how they cause disease and have adapted to their distinct environments. An understanding of the microbiology of these strains will be essential for testing these hypotheses. This study is the first to generally characterize these five sequenced strains of dermatophytes for their microbiological aspects. We measured the growth rate on solid medium and found differences between species, with Microsporum gypseum CBS118893 having the fastest growth and Trichophyton rubrum CBS118892 the slowest. We also compared different media for conidia production and found that the highest numbers of conidia were produced when dermatophytes were grown on MAT agar. We determined the Minimum Inhibitory Concentration (MIC) of nine antifungal agents and confirmed susceptibility to antifungals commonly used as selectable markers. Finally, we tested virulence in the Galleria mellonella (wax moth) larvae model but found the results variable. These results increase our understanding of the microbiology and molecular biology of these dermatophyte strains and will be of use in advancing hypothesis-driven research about dermatophytes.
Keywords: dermatophytes, Galleria, antifungal, drug susceptibilities
1. Introduction
Dermatophytes are the most common cause of fungal infections worldwide, affecting approximately 20% of the population (Marques et al. 2000). Despite their prevalence, the basic biology of these organisms is not well understood (White et al. 2008). Although current treatments are able to control infections, there is a major problem of recurrence, and it is unclear whether this is due to acquisition of a new infection or reactivation of the previous infection Gupta and Cooper 2008).
Recently, the genome sequences of five dermatophyte species with distinct ecological niches White et al. 2008) were released with annotation (The Broad Institute 2010). Trichophyton rubrum is human-adapted and is the most common cause of human dermatophyte infections worldwide. T. tonsurans is also human adapted and is the major cause of tinea capitis (scalp ringworm) in the Americas. Scalp ringworm is most prevalent in urban centers, and T. tonsurans was found in a recent survey to be present in more than 30% of students in some grade levels at certain U.S. schools (Abdel-Rahman et al. 2010). T. equinum is a close relative of T. tonsurans which can also cause human infections, but unlike T. tonsurans it is primarily associated with horses. Microsporum canis is another zoophile (associated with animals) and is the most common cause of tinea capitis in Europe. M. gypseum is a geophile (soil-dweller) and is the cause of gardener’s ringworm, an acute infection.
The strains selected for sequencing of these species are all clinically relevant (associated with human disease), and have had limited passage in the lab (White et al. 2008). They are available from the Centraalbureau voor Schimmelcultures (CBS) and have been classified morphologically as well as by sequencing of the rRNA ITS (White et al. 2008). The availability of sequence information now allows researchers to develop hypotheses relating to how dermatophytes cause disease as well as the differences between species that have adapted to distinct environmental niches. Characterization of the microbiology of these specific strains will be useful for researchers wishing to pursue experiments with the sequenced organisms.
Microbiology and molecular biological techniques are essential for experiments that will test hypotheses about dermatophyte virulence and evolution. It is therefore important to characterize the sequenced strains for growth rate and the ability to form conidia. Dermatophyte conidia usually contain a single nucleus, whereas hyphae may contain heterokaryons (hyphae with genetically distinct nuclei), making the ability to produce conidia an important tool for genetic transformation. Furthermore, it is of use to know the minimum inhibitory concentration (MIC) of drugs commonly used for selection. This will aid in the ability to use selectable markers to perform genetic manipulations of the genome, such as creating gene deletions or overexpression constructs.
Another beneficial tool in molecular biology is the use of a non-mammalian model system. Larvae of the greater wax moth Galleria mellonella have recently been developed as a model for fungal pathogens, including Candida, Cryptococcus, and the mould Aspergillus, which is related to the dermatophytes (Cotter et al. 2000; Cowen et al. 2009; Jackson et al. 2009; Mowlds and Kavanagh 2008; Mylonakis et al. 2005; Reeves et al. 2004; St Leger et al. 2000). The Galleria model has several advantages (Kavanagh and Reeves 2004; Mylonakis 2008). It is inexpensive, and there are no animal use regulations. The assay is quick and easy to perform, and the results are easy to score. Infection is done by injecting a fungal suspension directly into the hemocoel using a Hamilton syringe. The larvae are pale yellow when healthy and melanize if their immune response is stimulated. Both percent survival and degree of melanization are used to define virulence. Importantly, Galleria has an immune system with some characteristics similar to the human innate immune system, including antimicrobial peptides and phagocytic cells called hemocytes (Kavanagh and Reeves 2004; Kavanagh 2007; Mylonakis 2008). Galleria have been reported to die after incubation on a lawn of M. gypseum (Li et al. 2009), but dermatophyte virulence has not been assessed in Galleria using the standard injection method. If the Galleria model is applicable for studying dermatophyte pathogenesis, it would allow researchers to directly probe the interactions between dermatophytes and the innate immune system.
The aim of this study was to further characterize the five newly-sequenced dermatophyte strains for microbiological aspects including (i) comparing growth rates, (ii) determining the best growth medium to induce conidiation, (iii) comparing susceptibility to antifungal agents and confirming susceptibility to commonly used antifungal markers, and (iv) determining if these species are virulent in the Galleria mellonella model system.
2. Methods
2.1 Strains and growth conditions
The strains used in this analysis and their sources are listed in Table 1. The dermatophyte strains whose genomes have recently been sequenced (The Broad Institute 2010) include: T. rubrum CBS118892); T. tonsurans (CBS112818); T. equinum (CBS127.97); M. canis (CBS113480); M. gypseum (CBS118893).
Table 1.
Strains used in this analysis
| Species | Strain | Source1 |
|---|---|---|
| M. canis | CBS 1134802 | (White et al. 2008) |
| TIMM 4092 | (Yamada et al. 2006) | |
| TIMM 4092 dnr-1 deletion | (Yamada et al. 2006) | |
| M. gypseum | ATCC24163 | (Li et al. 2009) |
| ATCC48982 | (Li et al. 2009) | |
| CBS118893 | (White et al. 2008) | |
| T. equinum | CBS 127.97 | (White et al. 2008) |
| T. rubrum | CBS 118892 | (White et al. 2008) |
| H6 | (Fachin et al. 2006) | |
| Tru mdr2 | (Fachin et al. 2006) | |
| MR 816 | Barton, Li and Heitman, unpublished3 | |
| MR 850 | Barton, Li and Heitman, unpublished3 | |
| MR1458 | Barton, Li and Heitman, unpublished3 | |
| MR 1459 | Barton, Li and Heitman, unpublished3 | |
| MR 1463 | Barton, Li and Heitman, unpublished3 | |
| T. tonsurans | CBS 112818 | (White et al. 2008) |
| CMHT 001 | Abdel-Rahman, unpublished4 | |
| CMHT 002 | Abdel-Rahman, unpublished4 | |
| CMHT 003 | Abdel-Rahman, unpublished4 | |
| CMHT 004 | Abdel-Rahman, unpublished4 |
We thank all these investigators for making these strains available to us.
CBS = Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands
Strains were isolated and identified by R. Barton, University of Leeds and made available by W. Li and J. Heitman, Duke University.
trains were isolated, identified, and made available by S. Abdel-Rahman, Children’s Mercy Hospital, Kansas City, MO.
Malt extract agar (MEA), Sabauroud, and Oatmeal Agar media were obtained in powdered form from Difco and reconstituted according to the manufacturer’s instructions. MAT agar was prepared by autoclaving a 10% solution of Sabauroud medium and adding 0.1% (final volume) MgSO4-7H2O and KH2PO4 (Takashio 1972). YEPD (10% yeast extract, 20% peptone, 2% dextrose) was made using Difco and Fisher reagents. Agar (BactoAgar, Difco) was added at 20 g per liter where required.
2.2 Growth Rates
Radial growth was measured by placing mycelial plugs (approx 1.8 mm diameter) onto the center of a 60 × 15 mm Petri dish containing YEPD growth medium. Plates were incubated at 30°C. Calipers were used to measure the outer diameter each day for 10 days, always selecting the longest possible diameter. The experiment was performed in triplicate, with the average result. Error bars represent the standard deviation.
2.3 Conidia production on different media
Strains were streaked onto 150 × 15 mm Petri dishes for each test medium and were incubated at 30°C for the indicated number of days (10–28). To harvest conidia, a mixture of conidia and mycelia were collected by scraping the surface of the plate with 20% tween 80. The liquid was filtered through a layer of miracloth (Calbiochem) to remove mycelia and then centrifuged (3,000 × g, 5 min), washed twice in PBS, and resuspended in 1 ml PBS (Gibco). Numbers of conidia plate−1 were determined by: (1) counting the number of conidia using a hemacytometer and (2) determining CFU ml−1 with plate dilution assays. Results shown are the average of six plates per media type, representing at least three independent experiments in which strains were streaked, incubated, harvested, and counted. The error bars represent the standard deviation.
2.4 Susceptibility to antifungal agents
Susceptibility of dermatophyte species to different antifungal agents was tested by determining the MICs of these drugs according to the M38-A microdilution technique for filamentous fungi (CLSI/NCCLS. 2002; Fernandez-Torres et al. 2002). Briefly, conidia were added to a final concentration of 104 CFU ml−1 in RPMI 1640 (BioWhittaker) in a 96-well plate format. RPMI was pH 6.9 for all drugs tested except FOA, for which it was lowered to pH 4.4 (Cusker 2010). Drug concentration ranges were: 0.5–256 µg ml−1 (Fluconazole, FLC); 0.0156–8 µg ml−1 (Miconazole, MCN); 0.03–16 µg ml−1 (Ketoconazole, KTC); 0.0156–8 µg ml−1 (Itraconazole, ITC); 1–512 µg ml−1 (G418); 16–8192 µg ml−1 (Nourseothricin, NAT); 16–8192 µg ml−1 (Hygromycin, HYG); 4–2048 µg ml−1 (5-Fluoroorotic Acid, FOA); 0.125–64 µg ml−1 (Caspofungin, CAS). The final volume in each well was 200 µl. Each experiment was performed in triplicate and included controls for sterility (no cells, no drugs) and growth (no drugs). Plates were incubated without shaking at 30°C for 7 days, following which the MIC-0, defined as the lowest drug concentration in which no growth occurs, was determined visually. Results of replicates did not differ by more than one well.
To determine the ability of dermatophytes to grow on solid medium containing antifungals, 105 conidia of each of the five species were plated on YEPD containing 0, 100, 200, or 500 µg ml−1 NAT or HYG. Plates were incubated at 30°C and checked for growth after four days.
2.5 Galleria mellonella virulence assay
Galleria mellonella larvae in the final larval stage were purchased from Vanderhorst Wholesale, Inc. (St. Marys, OH), stored at room-temperature (RT) in the dark, and used within 48-hr of arrival for either the injection or plate crawl method of infection.
For injections, 10 larvae (0.3–0.4 g body weight) per condition were immobilized at 4°C for 30 min and then injected with conidia in 5 µl PBS (Gibco). A 10 µl Hamilton syringe was used to inject 5 µl of conidial suspension, containing the indicated number of conidia, into the lower left proleg of the larvae. Injection of 5 µl PBS was used as a control for mortality related to trauma. The syringe was sequentially washed with 10% bleach, ethanol, distilled water (dH2O), and phosphate buffered saline (PBS) in between fungal samples and after every 5–6 injections. Conidia were harvested from MAT plates as described above. Heat-killed conidia were prepared by incubating the conidia at 90°C for 10 min. Germinated conidia were prepared by incubating 106 conidia in MAT medium at 30°C with shaking for 3, 6, or 24 hours prior to injections. Germination was confirmed microscopically. Activation and some germlings were observed by 6 hr, and mycelia had begun forming by 24 hr. Infected larvae were incubated in Petri dishes at 30°C in the dark. Survival and melanization were monitored daily. Larvae were considered dead if they did not move when touched. The experiment was performed three times, with representative results shown. GraphPad Prism (v5.2) was used for graphing and statistical analysis (Mantel-Cox test).
For the “plate crawl” method, 10 larvae (0.25–0.3 g body weight) were placed on a Petri dish containing a lawn of dermatophytes approximately 1-week old on YEPD, MAT, or Sabouraud medium. The plates with larvae were incubated at RT in the dark for 10 days. Survival and melanization were monitored daily. The experiment was performed three times, with results shown for plate crawls on dermatophyte lawns grown on Sabouraud medium. GraphPad Prism (v5.2) was used for graphing and statistical analysis (Mantel-Cox test).
Other external inoculation methods tested included rolling the larvae on a lawn of dermatophytes or in a solution containing 106 conidia ml−1 and then moving them to a clean Petri dish. In both cases, 10 larvae per condition were used, and survival and melanization were monitored daily.
3. Results
All strains were obtained from the CBS. They have previously been classified using standard culture and morphological methods (White et al. 2008). They have also been classified by sequencing of the rRNA ITS (White et al. 2008). These species and strains are part of the Dermatophyte Genome Project, and their annotated genomic sequences have recently been released (The Broad Institute 2010).
3.1 Growth Rates
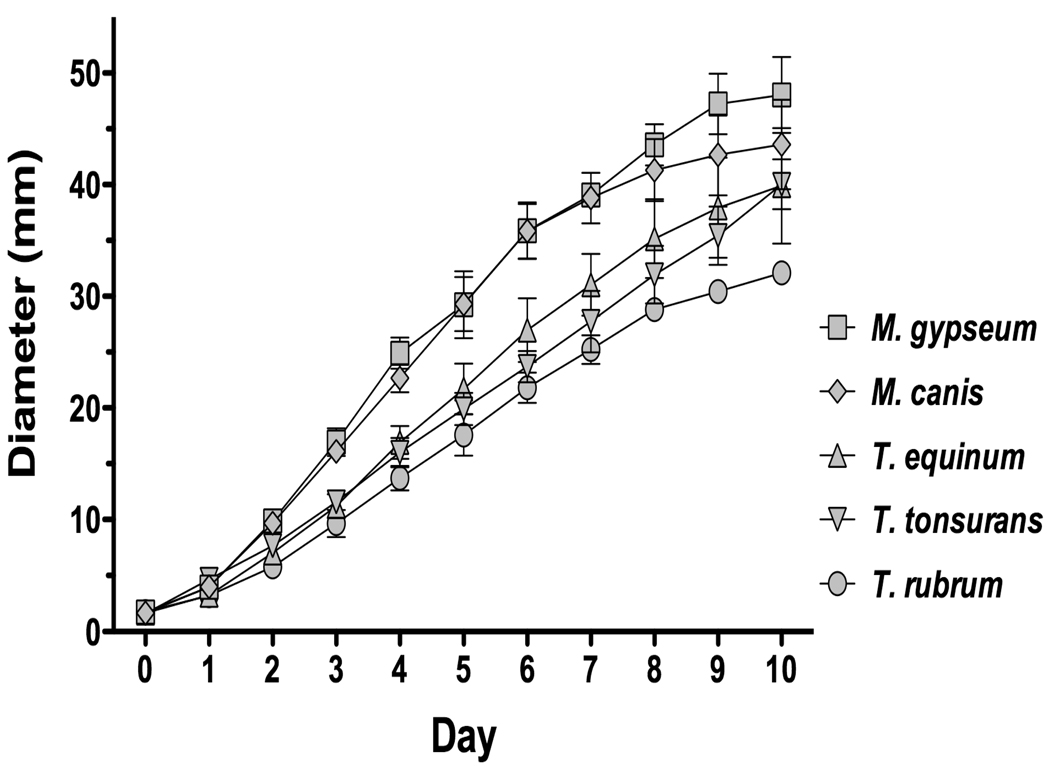
Radial growth remains a common way to compare growth of molds (for example (Arendrup et 2010; Rangel et al. 2010; Rodriguez-Jasso et al. 2010)). We therefore used the diameter of growth of a mycelial plug on an agar surface over time as a measurement of the growth rate (Fig. 1). We found that M. gypseum CBS118893 and M. canis CBS113480 had the most rapid growth, while T. rubrum CBS118892 had the slowest growth. This is consistent with previous work that found that M. canis had a faster growth rate than T. rubrum (Alio et al. 2005).
Figure 1. Growth of sequenced strains over time.
The outer diameter of mycelial plugs was measured each day using calipers. The experiment was performed in triplicate, with the average result shown. For M. gypseum, one of the three plugs reached the outer diameter of the plate by day nine.
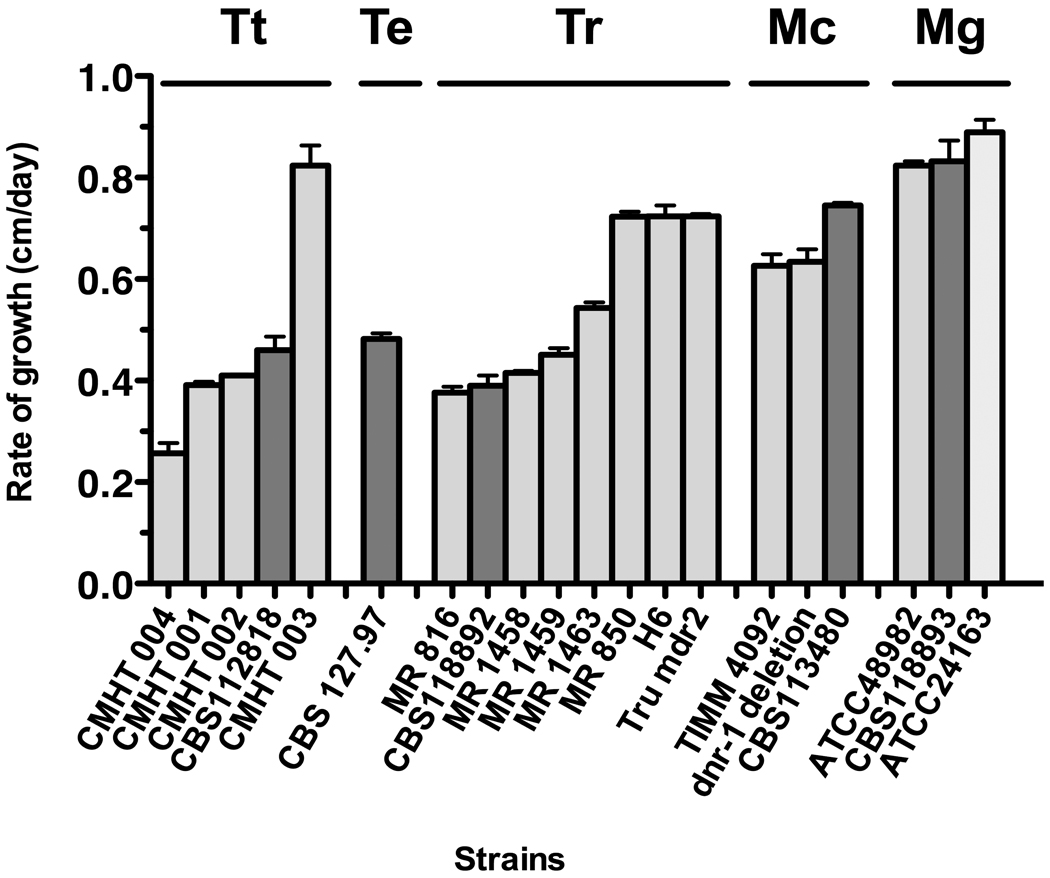
We compared the growth rates of these 5 strains to the growth rates of 15 other strains from four the five species. Additional strains of T. equinum were not immediately available. We found that the growth rates of the five sequenced strains were consistent to the growth rates of other strains in each of the four species (Fig 2).
Figure 2. Growth rates of 20 strains.
The growth rates of 20 strains (Table 1), including the 5 sequenced strains, were calculated for days 2 to 5, during which time the rates were linear over time, without reaching the outside of the petri plate. The rates are expressed as cm/day. The strains are grouped into species: T. tonsurans (Tt); T. equinum (Te), T. rubrum (Tr), M canis (Mc) and M. gypseum (Mg). Results are the average of three independent experiments with standard deviations. The darker bars in each species are the sequenced strain.
3.2 Conidiation in different media
The use of conidia is essential for experiments in which a known quantity of fungi is required. Microconidia are also useful for transformation techniques, since they are known to contain just one nucleus. Therefore, we examined several media types to identify the best growth medium for conidia formation. Dermatophytes can produce different types of conidia including spindle-shaped, knobbed macroconidia, club- or pear-shaped microconidia, segmented arthroconidia, and enlarged chlamydoconidia. Macroconidia are multinucleate, while other conidia are usually uninucleate. In this study, conidia were isolated by passage thru Miracloth, which traps hyphae and may also trap macroconidia. Since one of the purposes of the conidiation studies was to maximize production of uninucleate conidia for molecular analyses such as transformation, the loss of multinucleate macroconidia during isolation is an advantage, as transformation of multinucleate conidia complicate further analyses. Still, all types of conidia were enumerated after isolation through Miracloth.
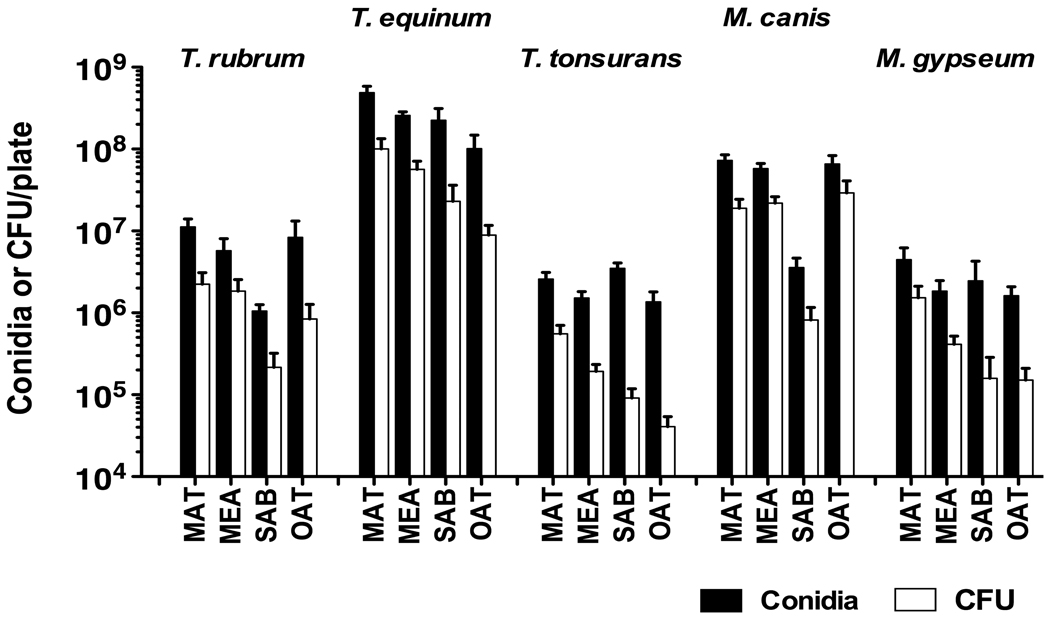
We tested different growth times to optimize conidia formation for each species. T. equinum CBS127.97 formed high levels of conidia after incubation for just 10 days at 30°C, whereas T. tonsurans CBS112818, M. gypseum CBS118893, and M. canis CBS113480 required 2–3 weeks of growth. T. rubrum CBS118892 had the most conidia after four weeks of growth.
For all species, conidia production (as determined by visual counts) and numbers of viable conidia (as determined by CFU) were highest when grown on MAT agar (Fig. 3). Growth on Sabouraud or Oatmeal agar generally had the lowest numbers of conidia. We also noticed a difference in conidia production between species, with T. equinum CBS127.97 producing the most conidia by far, and M. gypseum CBS118893 and T. tonsurans CBS112818 producing the fewest conidia.
Figure 3. Conidiation in different media.
Conidia were harvested from 150 × 15 mm Petri dishes containing MAT, MEA, Sabouraud (SAB), or Oatmeal (OAT) agar. Number of conidia per plate was determined using a hemocytometer (solid bars), or CFU per plate was determined by plate dilutions (striped bars). Data represent the average of six plates from at least three independent experiments. Error bars show the standard deviation.
3.3 Susceptibility to antifungal agents
Knowing the susceptibility of an organism to a particular drug is useful for clinical practice as well as molecular biology techniques that require selectable markers. However, widespread characterization of drug susceptibilities is not available for many dermatophyte strains. We therefore measured the MICs of multiple drugs for the five species and strains of dermatophytes recently sequenced (Table 2). We found that the MICs varies with species of dermatophyte, as has been reported previously for other strains (Coelho et al. 2008). The MICs of MCN, KTC, and ITC were fairly similar for all species (0.5 – 1 µg ml−1, 2 – 4 µg ml−1, and 0.5 – 1 µg ml−1, respectively) while that of FLC, G418, NAT, and HYG had more variability between species see Table 2). For example, the MICs of NAT and HYG for T. rubrum CBS118892, and of HYG for T. equinum CBS127.97, is more than 10-fold higher than the MICs of those agents for T. tonsurans CBS112818.
Table 2.
Antifungal activity against five species of dermatophytes as determined by MIC 0 (µg/ml).1
| Drug3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Species2 | FLC | MCN | KTC | ITC | G418 | NAT | HYG | FOA | CAS |
| T. rubrum | 64 | 0.5 | 2 | 1 | 256 | 1024 | 1024 | 256 | >64 |
| T. equinum | 128 | 1 | 2 | 0.5 | 64 | 256 | 1024 | 512 | 64 |
| T. tonsurans | 256 | 1 | 4 | 0.5 | 32 | 64 | 64 | 512 | 64 |
| M. canis | 32 | 0.5 | 2 | 1 | 256 | 256 | 512 | 512 | >64 |
| M. gypseum | 64 | 1 | 4 | 0.5 | 256 | 256 | 512 | 512 | >64 |
MICs were performed in triplicate. MIC are representative, and variation did not differ by more than one 2-fold dilution.
Strains used in this analysis were the sequenced strains.
FLC, fluconazole; MCN, miconazole; KTC, ketoconazole; ITC, itraconazole; NAT, nourseothricin; HYG, hygromycin; FOA, 5-fluoroorotic acid; CAS, caspofungin
NAT and HYG are commonly used as selectable markers. At least one species had an MIC to NAT or HYG of 1024 µg ml−1,. Therefore, we thought it was relevant to determine the minimum amount of drug required to prevent growth on an agar plate. 105 conidia of each species were plated onto YEPD agar containing 100, 200, or 500 µg ml−1 of NAT or HYG. YEPD with no drug added was included as a positive control for growth. All strains grew on the no-drug control plate. No strains were able to grow on the 500 µg ml−1 plate for either drug.
3.4 Galleria mellonella virulence assay
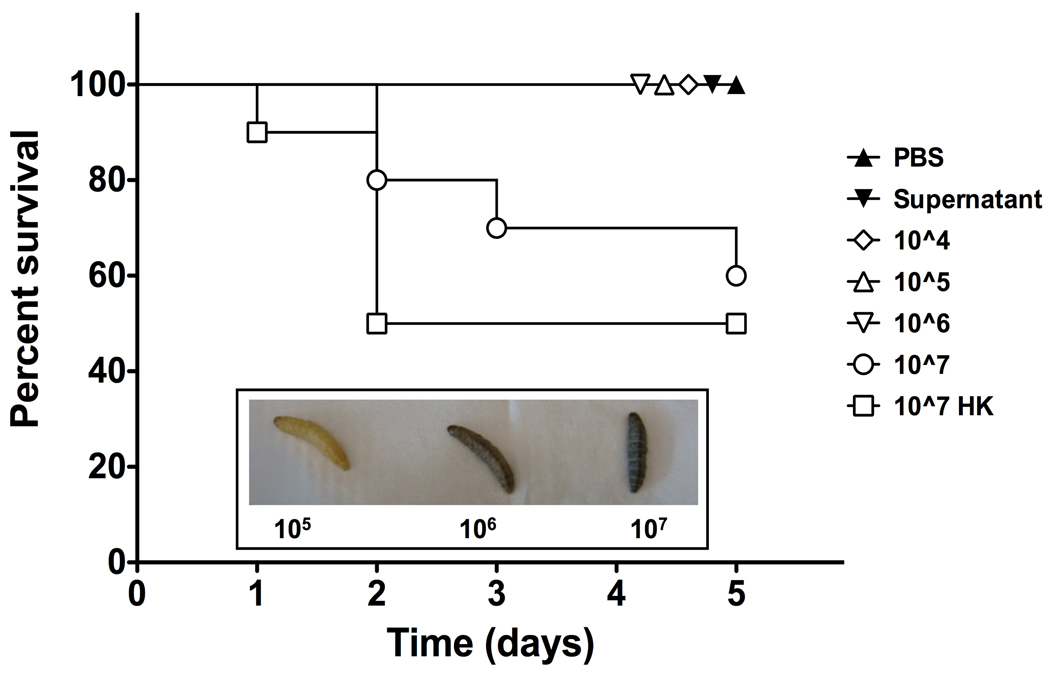
Galleria mellonella larvae are a well-established virulence assay for several fungi. Although Galleria have been reported to die after incubation on a lawn of M. gypseum (Li et al. 2009), no one has tested whether dermatophytes kill Galleria using the standard injection method. We performed a pilot experiment using T. equinum CBS127.97 conidia to determine the optimal numbers of conidia to inject per larvae. Melanization occurred rapidly following injection of conidia (Fig. 4 insert). Larvae had high survival when injected with buffer, supernatant from the conidial suspension, or conidial concentrations up to and including 106 conidia (Fig. 4). Larval death occurred after injection with 107 conidia, but similar death was observed with 107 heat-killed conidia, suggesting this was due to something resembling “septic shock” rather than dermatophyte virulence (Fig. 4). We therefore elected to use 106 conidia per larvae in further experiments.
Figure 4. Galleria survival after injection with T. equinum.
Larvae were injected with different concentrations of T. equinum conidia and monitored for melanization and survival. The experiment was performed three times with 10 larvae per condition. A representative graph of larval survival is shown. Day 5 data points are separated graphically for better visualization. Insert: Close-up view of melanization of larvae injected with 105, 106, or 107 conidia.
Survival of larvae injected with 106 conidia of T. tonsurans CBS112818, T. rubrum CBS118892, and M. canis CBS113480 was high, with over 80 % survival for larvae injected with any of the species (data not shown). This confirmed the observation made with T. equinum CBS127.97 that dermatophytes are not virulent when injected into Galleria (Fig 4). It has been demonstrated that germinated conidia of Aspergillus fumigatus are more virulent in the Galleria model than ungerminated conidia (Renwick et al. 2006). We tested germinated versus ungerminated conidia and did not find a difference in virulence for the dermatophytes (data not shown).
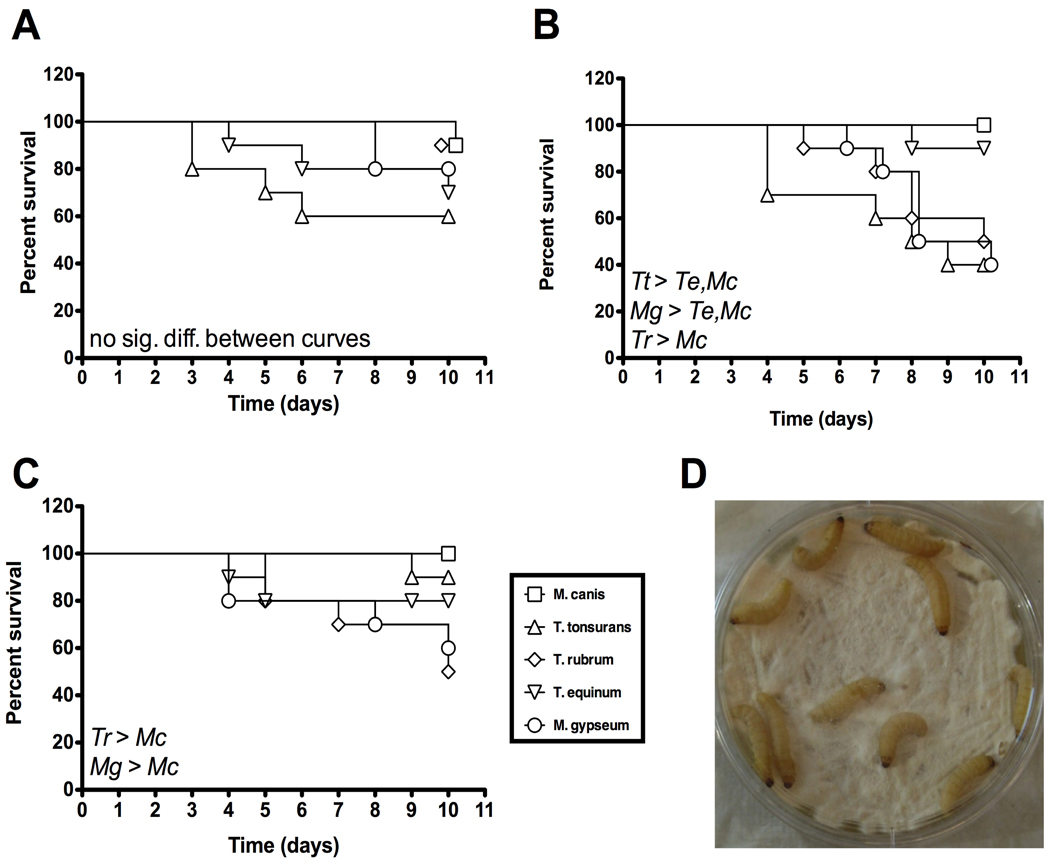
Dermatophytes cause superficial mycoses, suggesting the possibility that dermatophytes might be able to infect Galleria via the cuticle or mouth. We found high survival among larvae that were incubated on an empty Petri dish after being rolled either on a lawn of dermatophytes or in a solution containing a known quantity of conidia (data not shown). When Galleria larvae were incubated directly on the dermatophyte lawn, referred to as the “plate crawl” method (Fig. 5D), differences in virulence between the dermatophyte species were observed (Fig. 5 A–C). Different media types were tested. In the presence of larvae, dermatophyte lawns grown on YEPD had other microbial growth (bacterial or yeast). Larvae on these plates often had a high mortality rate, but we do not believe it was due to dermatophyte infection of the larvae. Rather, it seemed to be a result of presence of non-dermatophyte fungi and bacteria growing on the plate. In contrast, when larvae were incubated on dermatophyte lawns grown on MAT plates, the dermatophyte lawn was eliminated (presumably eaten or attached to larval cuticle) by the second day of incubation, but no additional fungal or bacterial growth was observed on the plates. Larval survival on these plates was high. Incubating the larvae on dermatophyte lawns grown on Sabouraud medium allowed for the most reproducible results, although the results from three different experiments were still highly variable (Fig. 5). This is in agreement with previously published work examining the survival of larvae on lawns of M. gypseum (Li et al. 2009). Larvae on Sabouraud plates (with no dermatophyte lawn) often had a high mortality rate (data not shown). We believe this was due to growth of non-dermatophyte fungi and bacteria that were brought in by the larvae. Growth of non-dermatophyte fungi and bacteria was much less prevalent when there was a dermatophyte lawn. For dermatophyte lawns grown on either Sabouraud or YEPD, T. tonsurans CBS112818 was regularly the most virulent strain. However, we still noted a high degree of variability in these experiments.
Figure 5. Galleria survival during plate crawl experiment.
Larvae were incubated on dermatophyte lawns for 10 days at RT in the dark. Survival was monitored daily. Tr, T. rubrum; Tt, T. tonsurans; Te, T. equinum; Mg, M. gypseum; Mc, M. canis. (A), (B), and (C) represent three experiments each using 10 larvae per plate. The Mantel-Cox test was used to determine statistical significance. (A) Day 10 data points are separated graphically for better visualization. No statistically significant difference between survival curves. (B) M. gypseum data points on have been shifted to the right for better visualization. M. gypseum and T. tonsurans are each significantly more virulent than T. equinum and M. canis. T. rubrum is significantly more virulent than M. canis. (C) M. gypseum and T. rubrum are each significantly more virulent than M. canis. (D) Experimental set-up for the “plate crawl” method of external inoculation.
4. Discussion and Conclusions
Of the 5 sequenced strains, M. gypseum CBS118893 had the fastest growth on solid medium, while T. rubrum CBS118892 had the slowest (Fig. 1). Compared to the other 15 strains, the 5 sequenced strains showed growth rates consistent with other strains of their species, although there is considerable variation. Two strains of M. canis were compared to the sequenced strains. One, a dnr1-deletion, is a derivative of the wild type strain TIMM 4092 (Yamada et al. 2006). It is clear from these results that the two strains do show the same growth rates by this assay. Similarly in T. rubrum, the mdr2 mutant TruMDR2 and its wild type parental strain H6 have similar growth rates, consistent with previous reports (Fachin et al. 2006).
We found that MAT agar is the best medium for inducing conidiation in all sequenced strains, that the timing of conidiation varies between strains and that different strains produce differing amounts of conidia. The high conidiation frequency of T. equinum CBS127.97 and M. canis CBS113480 make them particularly amenable to future studies. T. rubrum CBS118892 also had fairly high conidiation (Fig. 3), but a slower growth rate (Fig. 1). M. gypseum CBS118893, although it had the fastest growth on solid medium, produced few conidia.
This is the first comprehensive examination of antifungal susceptibilities for these sequenced dermatophyte strains. We observed a great deal of variance of susceptibilities between species, as has been described before for dermatophytes (Coelho et al. 2008). Our results for ITC are consistent for the few cases where published MICs for the same dermatophyte species are available. An MIC of 0.8 – 1 µg ml−1 for ITC against dermatophytes has been reported (Coelho et al. 2008; Fernandez-Torres et al. 2002). Similarly, we found the ITC MIC-0 to be 1 µg ml−1 (T. rubrum CBS118892 and M. canis CBS113480) or 0.5 µg ml−1 (T. tonsurans CBS112818, T. equinum CBS127.97, and M. gypseum CBS118893). For T. tonsurans CBS112818, our results for FLC are higher than previously published results (Coelho et al. 2008). This cannot be explained by strain differences, since both studies used T. tonsurans strain CBS112818. However, the previously published study incubated for 5 days before determining the MIC-0 to FLC, whereas we incubated for 7 days as recommended by Fernàndez-Torres et al. (Fernandez-Torres et al. 2002). A shorter incubation time would be expected to have less fungal growth and therefore a lower MIC. A different study reported the MIC-0 of FLC against M. canis and T. rubrum as 64–128 and 16–64 µg ml−1, respectively (Barchiesi et al. 2009). These ranges are similar to our findings (32 µg ml−1 for M. canis; 64 µg ml−1 for T. rubrum) even though different strains were used.
The antifungal agents HYG, NAT, and G418 have been proposed as markers for genetic manipulation of dermatophytes (Gonzalez et al. 1989; Yamada et al. 2008; Yamada et al. 2005). Our results confirm susceptibility of all five strains to G418 compared to the concentrations used for genetic manipulation. The strains differ in their susceptibilities to HYG and NAT, where T. rubrum CBS118892 and T. equinum CBS127.97 have elevated MIC-0 for HYG, and T. rubrum CBS118892 has an elevated MIC-0 for NAT, suggesting a high level of intrinsic resistance for these species. As with FLC discussed above, Coelho et al. reported a lower MIC for HYG against T. rubrum and T. equinum than reported here (Coelho et al. 2008). Again the difference could be due to the length of incubation, but may also be due to instability of HYG at 37° C, or alterations in HYG activity at different pH. These results emphasize the need for standardized testing methods.
Despite these elevated MIC-0 for HYG, successful transformation of T. rubrum using HYG as a selectable marker has been reported (Dobrowolska and Staczek 2009; Fachin et al. 2006). We found that growth of 105 conidia plated onto YEPD was inhibited by lower concentrations of NAT or HYG than described by the broth microdilution method. Thus the broth microdilution method does not necessarily mimic growth on solid medium that is traditionally used for genetic manipulation. Therefore, these markers remain useful for selection on solid medium, provided appropriate negative controls are included.
Non-mammalian model systems are of use in studying the molecular basis of pathogenesis. Galleria mellonella (wax moth) larvae have been established as a virulence assay for several pathogenic fungi, including Aspergillus, which is related to the dermatophytes (Cotter et al. 2000; Reeves et al. 2004; Renwick et al. 2006; St Leger et al. 2000). It has been demonstrated that Galleria larvae die when incubated on a lawn of M. gypseum (Li et al. 2009). However, no one has determined whether this virulence model is appropriate for other species of dermatophytes or whether dermatophytes cause larval death when injected directly into the hemocoel. Our results demonstrate that dermatophytes cause melanization but not death when low levels of conidia are injected directly into the hemocoel. At high concentrations, the larvae are killed even if the conidia have been inactivated by heat-killing. For the plate crawl method, we observed larval death during incubation on dermatophyte lawns, but the method proved inconsistent and most species did not exhibit a significant number of deaths on the plate. We therefore do not recommend the use of the Galleria model system for future dermatophyte virulence studies.
This study is the first to characterize generally the five newly sequenced dermatophyte species and strains for microbiological aspects including growth, optimal conditions for conidiation, susceptibility to antifungal agents, and pathogenesis in a commonly used non- mammalian model system. This information will prove useful for performing molecular biology and advancing hypothesis-driven research into the biology and pathogenic mechanisms of dermatophytes.
Acknowledgements
The authors acknowledge Kelsey Fowlkes, Mesa DeMeritt, and Kien-Thiet Phung Nguyen for technical assistance. We thank Drs. R. Barton (U. Leeds), W Li and J. Heitman (Duke U.), and S. Abdel-Rahman (Children’s Mercy Hospital, Kansas City, MO) for strains used in this work. This work was supported by R21-AI081235 awarded to T. C. White.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of Conflict of Interests
No conflict of interests.
References
- Abdel-Rahman SM, et al. The prevalence of infections with Trichophyton tonsurans in schoolchildren: the CAPITIS study. Pediatrics. 2010;125:966–973. doi: 10.1542/peds.2009-2522. [DOI] [PubMed] [Google Scholar]
- Alio AB, et al. Dermatophytes growth curve and in vitro susceptibility test: a broth micro-titration method. Med Mycol. 2005;43:319–325. doi: 10.1080/13693780500092947. [DOI] [PubMed] [Google Scholar]
- Arendrup MC, et al. Development of azole resistance in Aspergillus fumigatus during azole therapy associated with change in virulence. PLoS One. 2010;5:e10080. doi: 10.1371/journal.pone.0010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchiesi F, et al. In vitro susceptibility of dermatophytes to conventional and alternative antifungal agents. Med Mycol. 2009;47:321–326. doi: 10.1080/13693780802641920. [DOI] [PubMed] [Google Scholar]
- CLSI/NCCLS. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard., NCCLS document M38-A NCCLS, 940 West Valley Road, Suite 1400, Wayne Pennsylvania 19087-1898 USA. 2002 [Google Scholar]
- Coelho LM, et al. In vitro antifungal drug susceptibilities of dermatophytes microconidia and arthroconidia. J Antimicrob Chemother. 2008;62:758–761. doi: 10.1093/jac/dkn245. [DOI] [PubMed] [Google Scholar]
- Cotter G, et al. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunology and Medical Microbiology. 2000;27:163–169. doi: 10.1111/j.1574-695X.2000.tb01427.x. [DOI] [PubMed] [Google Scholar]
- Cowen LE, et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci U S A. 2009;106:2818–2823. doi: 10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusker MC. Yeast and Baceterial Media Recipes. 2010 http://www.duke.edu/web/microlabs/mccusker/Resources/MediaRecipes.pdf.
- Dobrowolska A, Staczek P. Development of transformation system for Trichophyton rubrum by electroporation of germinated conidia. Curr Genet. 2009;55:537–542. doi: 10.1007/s00294-009-0264-8. [DOI] [PubMed] [Google Scholar]
- Fachin AL, et al. Role of the ABC transporter TruMDR2 in terbinafine, 4-nitroquinoline N-oxide and ethidium bromide susceptibility in Trichophyton rubrum. J Med Microbiol. 2006;55:1093–1099. doi: 10.1099/jmm.0.46522-0. [DOI] [PubMed] [Google Scholar]
- Fernandez-Torres B, et al. Collaborative evaluation of optimal antifungal susceptibility testing conditions for dermatophytes. J Clin Microbiol. 2002;40:3999–4003. doi: 10.1128/JCM.40.11.3999-4003.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R, et al. Transformation of the dermatophyte Trichophyton mentagrophytes to hygromycin B resistance. Infect Immun. 1989;57:2923–2925. doi: 10.1128/iai.57.9.2923-2925.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AK, Cooper EA. Update in antifungal therapy of dermatophytosis. Mycopathologia. 2008;166:353–367. doi: 10.1007/s11046-008-9109-0. [DOI] [PubMed] [Google Scholar]
- Jackson JC, et al. Conidiation color mutants of Aspergillus fumigatus are highly pathogenic to the heterologous insect host Galleria mellonella. PLoS One. 2009;4:e4224. doi: 10.1371/journal.pone.0004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K, Reeves EP. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol Rev. 2004;28:101–112. doi: 10.1016/j.femsre.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Kavanagh KaR, E P. Insect and Mammalian Innate Immune Responses are much alice. Microbe. 2007;2:596–599. [Google Scholar]
- Li W, et al. Organization and evolutionary trajectory of the mating type (MAT) locus in the dermatophyte and dimorphic fungal pathogens. Eukaryot Cell. 2009 doi: 10.1128/EC.00259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques SA, et al. Mycoses associated with AIDS in the Third World. Medical Mycology. 2000;1:269–279. [PubMed] [Google Scholar]
- Mowlds P, Kavanagh K. Effect of pre-incubation temperature on susceptibility of Galleria mellonella larvae to infection by Candida albicans. Mycopathologia. 2008;165:5–12. doi: 10.1007/s11046-007-9069-9. [DOI] [PubMed] [Google Scholar]
- Mylonakis E. Galleria mellonella and the Study of Fungal Pathogenesis: Making the Case for Another Genetically Tractable Model Host. Mycopathologia. 2008;165:1–3. doi: 10.1007/s11046-007-9082-z. [DOI] [PubMed] [Google Scholar]
- Mylonakis E, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infection And Immunity. 2005;73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel DE, et al. Thermotolerance of germlings and mycelium of the insect-pathogenic fungus Metarhizium spp. and mycelial recovery after heat stress. J Basic Microbiol. 2010;50:344–350. doi: 10.1002/jobm.200900430. [DOI] [PubMed] [Google Scholar]
- Reeves EP, et al. Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia. 2004;158:73–79. doi: 10.1023/b:myco.0000038434.55764.16. [DOI] [PubMed] [Google Scholar]
- Renwick J, et al. Susceptibility of larvae of Galleria mellonella to infection by Aspergillus fumigatus is dependent upon stage of conidial germination. Mycopathologia. 2006;161:377–384. doi: 10.1007/s11046-006-0021-1. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Jasso RM, et al. Fucoidan-Degrading Fungal Strains: Screening, Morphometric Evaluation, and Influence of Medium Composition. Appl Biochem Biotechnol. 2010 doi: 10.1007/s12010-010-8992-2. [DOI] [PubMed] [Google Scholar]
- St Leger RJ, et al. Lack of host specialization in Aspergillus flavus. Appl Environ Microbiol. 2000;66:320–324. doi: 10.1128/aem.66.1.320-324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashio M. Sexual reproduction of some Arthroderma and Nannizzia on diluted Sabouraud agar with or without salts. Mykosen. 1972;15:11–17. doi: 10.1111/j.1439-0507.1972.tb02419.x. [DOI] [PubMed] [Google Scholar]
- The Broad Institute. Dermatophyte Comparative Genome Database. 2010 http://www.broadinstitute.org/annotation/genome/dermatophyte_comparative/MultiHome.html.
- White TC, et al. Generating and testing molecular hypotheses in the Dermatophytes. Eukaryot Cell. 2008;7:1238–1245. doi: 10.1128/EC.00100-08. PMCID: PMC2519771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, et al. Isolation, characterization, and disruption of dnr1, the areA/nit-2-like nitrogen regulatory gene of the zoophilic dermatophyte, Microsporum canis. Med Mycol. 2006;44:243–252. doi: 10.1080/13693780500410909. [DOI] [PubMed] [Google Scholar]
- Yamada T, et al. Genetic transformation of the dermatophyte, Trichophyton mentagrophytes, based on the use of G418 resistance as a dominant selectable marker. J Dermatol Sci. 2008;49:53–61. doi: 10.1016/j.jdermsci.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Yamada T, et al. Reproducible genetic transformation system for two dermatophytes, Microsporum canis and Trichophyton mentagrophytes. Med Mycol. 2005;43:533–544. doi: 10.1080/13693780500057619. [DOI] [PubMed] [Google Scholar]