SUMMARY
Although most tissues in an organism are genetically identical, the biochemistry of each is optimized to fulfill its unique physiological roles, with important consequences for human health and disease. Each tissue’s unique physiology requires tightly regulated gene and protein expression coordinated by specialized, phosphorylation-dependent intracellular signaling. To better understand the role of phosphorylation in maintenance of physiological differences among tissues, we performed proteomic and phosphoproteomic characterizations of nine mouse tissues. We identified 12,039 proteins, including 6296 phosphoproteins harboring nearly 36,000 phosphorylation sites. Comparing protein abundances and phosphorylation levels revealed specialized, interconnected phosphorylation networks within each tissue while suggesting that many proteins are regulated by phosphorylation independently of their expression. Our data suggest that the ‘typical’ phosphoprotein is widely expressed, yet displays variable, often tissue-specific phosphorylation that tunes protein activity to the specific needs of each tissue. We offer this dataset as an online resource for the biological research community.
INTRODUCTION
Despite sharing identical genomes and overlapping transcription profiles, mammalian tissues exhibit diverse physiology. This specialization arises via variable protein expression and differential post-translational modifications that tune the activity of ubiquitous proteins to each tissue’s needs. The resulting biochemical idiosyncrasies can account for tissue-specific disease and drug resistance, with consequences for human health (Goh et al., 2007). Thus, although transcriptome and proteome profiling uncover physiological differences among tissues due to differential gene expression (Su et al., 2004) or protein abundance and subcellular localization (Kislinger et al., 2006), they do not address tissue-specific effects of post-translational regulation.
A fundamental mechanism for regulating protein activity is covalent post-translational modification of serine, threonine, and tyrosine residues with phosphate. Because phosphorylation is fast, reversible, and often highly specific, it is often employed for temporary modulation of protein function, serving to alternately induce or abolish enzyme activity, facilitate or disrupt protein interactions, alter protein conformations, or target proteins for destruction. Protein phosphorylation and dephosphorylation are catalyzed by over 500 kinases and 100 phosphatases, and are themselves regulated by phosphorylation, revealing the interconnections among cellular signaling pathways.
Many phosphorylation networks have been elucidated using model organisms and in vitro systems, providing generalized models of signal transduction. However, such models cannot account for specialized tissue physiology. Furthermore, these studies have typically used targeted methods, precluding exhaustive analysis of phosphorylation. Recently, phosphoproteomics has enabled large-scale identification of protein phosphorylation sites, benefiting from advances in phosphopeptide enrichment (Pinkse et al., 2004; Villen and Gygi, 2008) and improvements in mass spectrometry instrumentation and methods. However, many previous surveys of protein phosphorylation have used immortalized cell lines, which differ from their tissues of origin in gene and protein expression (Lukk et al., 2010; Pan et al., 2009). Furthermore, previous surveys have generally examined only a few tissues such as liver (Villen et al., 2007) and brain (Wisniewski et al., 2010), selected for their relevance to human disease. Further complicating analysis of these studies, observed phosphorylation changes may reflect differential protein expression rather than truly modified phosphorylation. To distinguish these scenarios, the relative abundance of each phosphorylation site must be compared with that of its parent protein to verify differential phosphorylation. Thus, a large-scale, multi-tissue survey of protein abundance combined with phosphorylation site identification would provide insight into phosphorylation-dependent signaling pathways and could be a critical first step in delineating the key proteins and pathways underlying specific tissue physiology.
Here we report the most thorough characterization of tissue-specific protein abundance and phosphorylation to date, including 12,000 proteins and 36,000 phosphorylation sites from 9 mouse tissues. These data revealed distinctive and complementary protein and phosphoprotein expression profiles that support each tissue’s unique physiology. Moreover, by combining protein abundance measurements with phosphorylation observations, we could distinguish tissue-specific phosphorylation of ubiquitous proteins from phosphorylation of tissue-specific proteins. Furthermore, most phosphoproteins integrate input from multiple kinases spanning diverse signaling pathways. Overall, the ‘typical’ phosphoprotein is broadly expressed, yet is variably phosphorylated to tune protein function to the needs of each tissue. We now present these protein abundance and phosphorylation data as a web-based resource (https://gygi.med.harvard.edu/phosphomouse/index.php) to aid analysis of existing biological data and inspire future biological investigations.
RESULTS
Identification of 12,000 proteins and 36,000 phosphorylation sites from 9 mouse tissues
To survey protein and phosphoprotein abundance, 9 organs were harvested from three-week-old male Swiss-Webster mice: brain, brown fat, heart, liver, lung, kidney, pancreas, spleen, and testis. After tissue homogenization, proteins were either digested in solution for subsequent strong cation exchange chromatography and phosphopeptide enrichment via IMAC (10 mg per tissue) or separated via SDS-PAGE (65 μg per tissue) followed by in-gel digestion (Supplemental Methods and Fig. S1). Protein and phosphoprotein extraction conditions were selected to minimize protease and phosphatase activity (Castellanos-Serra and Paz-Lago, 2002; Roche, 2004). This was most critical for pancreas, due to its high levels of endogenous proteases and phosphatases. Although assessing phosphatase activity is challenging, minimal protein degradation was observed via SDS-PAGE (Figure S2). Samples enriched with phosphopeptides (24 per tissue) were analyzed in duplicate using a hybrid linear ion trap Orbitrap mass spectrometer, while non-phosphorylated samples (12 per tissue) were analyzed once (Extended Experimental Procedures). The final dataset contained over 284,000 phosphopeptide identifications (Table S1), matching nearly 36,000 phosphorylation sites (Table S2) from 6296 proteins (Table S3) at peptide and protein-level false discovery rates (FDRs) of 0.15% and 1.7%, respectively.
Following peptide- and protein-level filtering, each site on every phosphopeptide was scored using the AScore algorithm to assess the confidence of phosphorylation site localization (Beausoleil et al., 2006). Sites scoring above 13 were considered localized (p < 0.05). 85% of sites were localized to a single amino acid and ranged from 89–93% for individual tissues (Figure 1B). A minimal list of phosphorylation sites was then assembled. Localized sites were counted once; non-localized sites were grouped when their regions of possible modification overlapped. Groups of non-localized sites were counted only when no localized sites could explain the observed phosphorylation.
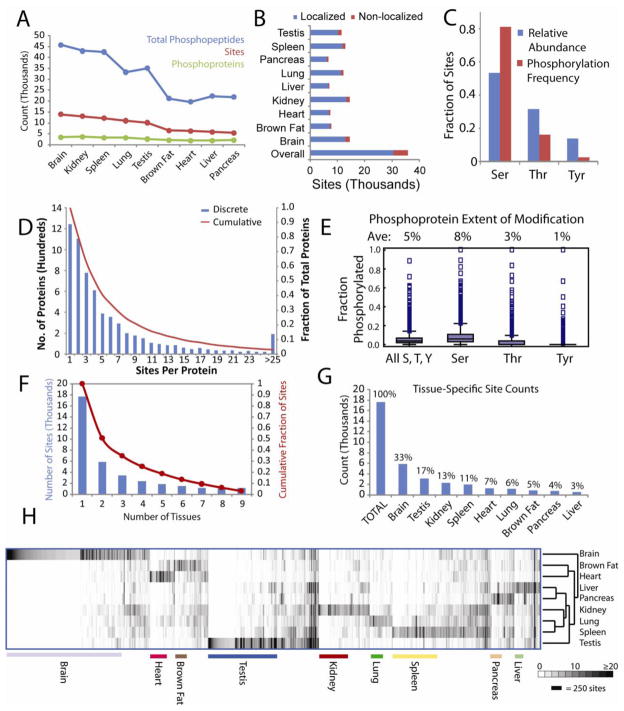
Figure 1. Overview of Tissue-specific Phosphorylation.
A. Numbers of phosphopeptides, sites, and phosphoproteins. B. Numbers of localized and non-localized phosphorylation sites. C. Relative frequencies of Ser, Thr, and Tyr within all phosphoproteins and their relative likelihood of phosphorylation. D. Histogram depicting numbers of sites observed per protein. E. Box plots indicating the relative extents of modification for residues prone to phosphorylation within all observed phosphoproteins. F. Histogram indicating numbers of sites detected in multiple tissues. G. Distribution of tissue-specific sites across organs. H. Hierarchical clustering of sites and tissues based on spectral counts. Only the 11,414 sites containing 5 or more spectral counts in at least one tissue are shown. See also Figures S1, S3, S4 and S7 and Tables S1–S3.
Importantly, over 50% of observed sites and phosphoproteins were previously unreported, based on comparison with the PhosphoSite database of known phosphorylation sites (Hornbeck et al., 2004) (Figure S3A). Similarly, most sites have not been reported in the Phospho.ELM database (Diella et al., 2008) (Figure S3B). Several factors contribute to the high proportion of unreported sites. First, we included tissues (brown fat, kidney) that have been less studied. Second, continual improvements in instrumentation and methodology have enhanced the sensitivity of phosphoproteomic analyses. Previously, our lab surveyed the mouse liver phosphoproteome, using similar techniques to characterize organs from mice of identical age, strain, and sex as used in this study. This present work encompasses far more phosphorylation sites, across all tissues and within the liver itself (Figure S3C). Interestingly, though the present study encompasses virtually all sites reported by Villen et al., some pTyr sites were not observed. These missing sites were detected via immunoprecipitation of pTyr-containing peptides from much larger amounts of digested peptides.
Without phosphopeptide enrichment, 894,041 peptide spectral matches corresponding to 10,102 proteins were made at a peptide and protein-level FDRs of 0.11% and 1.25% (Tables S3 and S4). Traditionally, control of peptide and protein FDRs from large datasets has posed significant challenges, risking accumulation of incorrect identifications to unacceptably high FDRs. To reliably estimate protein FDRs, we developed a method based on the target-decoy database search strategy (Elias and Gygi, 2007). Peptide identifications were filtered using a multivariate approach that used linear discriminant analysis to distinguish valid identifications from random matches, following training with target and decoy peptides as positive and negative training data. Peptides were subsequently assembled into proteins and filtered via several protein quality metrics (Extended Experimental Procedures). This extensive peptide- and protein-level filtering of both phosphorylated and non-phosphorylated data ensured the highest quality of all identifications.
Tissue Distribution of Phosphorylation Sites
We first examined the number of phosphorylated peptide spectral matches, the number of unique sites and the total phosphoproteins identified per tissue. These varied, reflecting differences in complexity and variable intracellular signaling within each tissue (Figure 1A). While their heterogeneity varies, each tissue contains many cell types that together create specialized physiology. The reported phosphorylation profiles are thus weighted averages that reflect signaling within all cell types in each tissue. Highest numbers of phosphopeptides, phosphorylation sites, and phosphoproteins were identified in brain, highlighting its unique cellular diversity and the specialized signaling networks these cells employ. In addition to brain, kidney, spleen, lung, and testis each contained over 30,000 phosphopeptides and 10,000 localized sites. Since spleen contains numerous immune cell populations, many phosphorylation-dependent signaling pathways are constitutively active, priming the young mice for rising immune challenges. Similarly, immune cells in lung contribute to its complex phosphorylation profile. Despite varying phosphopeptide numbers, phosphoprotein counts were similar across tissues, indicating that decreased phosphopeptide counts in tissues such as pancreas and heart reflect true differences in signaling and tissue heterogeneity, rather than varying instrument performance.
The diverse cell populations contributing to each phosphoproteome profile include cells specific to each tissue, as well as red blood cells and proteins within the vasculature of all tissues. To identify proteins and phosphorylation sites whose levels could be influenced by blood contamination, we compared our protein and phosphoprotein profiles with a proteomic survey of murine red blood cells (Pasini et al., 2008). A small fraction of proteins in each tissue were also seen in red blood cells (see Figure S3D). Overall, 445 of 12000 proteins detected in this study with or without phosphorylation were also seen in red blood cells. While some proteins such as hemoglobin are predominantly found in red blood cells, most, including actins and glycolytic enzymes, are found in virtually all cells, including cells within the tissues in question.
We next examined patterns among identified phosphorylation sites. Similar to previous studies (Olsen et al., 2006), we observed mostly Ser phosphorylation (83%), followed by Thr (15%) and Tyr (2%). This enrichment far exceeds the relative abundance of Ser among residues subject to phosphorylation within the phosphoproteins detected in this study, indicating a strong preference for Ser phosphorylation (Figure 1C). We then analyzed numbers of sites within each phosphoprotein. 80% of phosphoproteins contained multiple sites, while 50% were phosphorylated on four or more residues and 10% carried more than fourteen sites (Figure 1D). Though these multiple modifications do not necessarily occur simultaneously on individual protein molecules, such multiple phosphorylation could reflect regulation of a single protein function via multiple pathways, or could suggest that many of the protein’s cellular activities and interactions are independently regulated via phosphorylation at distinct sites. Indeed, examination of predicted structural elements within proteins using PsiPred (Jones, 1999) and VSL2 (Peng et al., 2006) revealed that phosphorylated Ser, Thr, and Tyr residues exhibited marked differences in structural classification compared to their unmodified counterparts (Figure S3E). Phosphorylated sites were predominantly predicted to reside in coiled and disordered regions rather than in ordered secondary structures. Although phosphorylation usually occurred in disordered regions, sites within kinase activation loops were a notable exception: most of the 120 observed activation loop sites were ordered, with elevated levels classified as strands. Virtually no phosphorylation sites were located in known or predicted α-helices.
While many proteins were multiply phosphorylated, generally only a small portion of Ser, Thr, and Tyr residues within each protein were modified (Figure 1E). Overall, 5% of these residues were modified, with some variability for each residue: Ser, 8%; Thr, 3%; Tyr, 1%. Nevertheless, some proteins bore extensive phosphorylation. Based on fractional modification (the number of potential sites versus the number of observed phosphorylated sites), the most heavily phosphorylated proteins included hemoglobin β1 (94% phosphorylated) as well as Marcksl1 (also known as MLP; 61% phosphorylated), which spans the protein kinase C and calmodulin signaling networks, and Hmgn1 (60% phosphorylated), which regulates DNA-histone interactions.
To assess overlap, we counted the number of tissues in which each site was observed (Figure 1F). 50% of sites were observed exclusively in single tissues, while 3% were found in all tissues and 18% were present in over half of examined tissues. Although tissue-specific sites were observed in all organs, they were not evenly distributed (Figure 1G). Most tissue-specific sites were found in brain (33%) and testis (17%), while lung contained only 6% and liver contributed 3%. These differences are not due to lower phosphopeptide counts in these tissues, since lung contained 95% of the total number of phosphopeptides as testis. To better assess tissue distributions, tissue enrichment was quantified for each site using Shannon’s entropy (Experimental Procedures) (Shannon, 1948). Selected tissue-specific phosphorylation sites are shown in Table 1. These sites come from variably abundant proteins, including Bassoon and Mtap1a which were highly expressed in brain, as well as Nexilin and the CXC chemokine receptor which were found in low abundance in heart and spleen, respectively. Many sites are previously unknown, with most of these sites identified in less frequently studied tissues. For comparison, proteins bearing global phosphorylation sites are listed in Table 2. Examples include Huntingtin, the protein implicated in Huntington’s disease, and kinases Mapk3 and Gsk3b. Few global sites are previously uncharacterized, presumably due to their ubiquity. Though some sites are globally modified, extensive tissue-specific phosphorylation underscores the importance of multi-tissue phosphoproteomics. First, even widely expressed proteins display dramatically different phosphorylation profiles across tissues. Even the heavily phosphorylated Srrm2 (310 sites) harbors an abundant testis-specific site (S1434). Second, many proteins are only expressed in a subset of tissues and could obviously only be phosphorylated in tissues where they are expressed. The proteins Speg (heart), calmegin (testis), and B-lymphocyte antigen CD-20 were only found in single tissues. Clearly, comprehensive phosphoproteomics requires analysis of many tissues.
Table 1. Abundant Tissue-Specific Sites, as Determined by Spectral Counts.
This table lists selected abundant phosphorylation sites that were only detected in one of the nine tissues characterized, grouped by protein and tissue of origin. Representative tissue-specific sites are listed for all nine tissues. See also Tables S1–S3.
| Major Tissue | Name | Annotation | IPI | Protein Level a | Tissue-enriched Expression?b | Novel?c | Site | Classd | Entropye | Brain | Brown Fat | Heart | Kidney | Liver | Lung | Pancreas | Spleen | Testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain | Camk2a | Ca/calmodulin dependent protein kinase type II α chain | 00621806 and 00420725 | Low | Yes | N | T334 | O | 0.05 | 146 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| N | S331 | B | 0.08 | 88 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| N | T337 | A | 0.08 | 86 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Bsn | Bassoon | 00134093 | Hi | Yes | N | S1114 | P | 0.06 | 117 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| N | S1108 | P | 0.07 | 96 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Mtap1a | Microtubule-associated protein 1A | 00408909 | Hi | Yes | N | S991 | P | 0.06 | 109 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| N | S1008 | P | 0.09 | 69 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Fat Brown | Flg2 | Filaggrin-2 | 00406870 | Med | No | N | S1204 | A | 0.22 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| N | S1205 | A | 0.29 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Nif3l1 | ngg1 interacting factor 3-like 1 | 00230615 | Hi | No | Y | T254 | B | 0.39 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Y | S258 | O | 0.42 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Heart | Pkp2 | Uncharacterized protein | 00132134 | Low | No | N | Y573 | T | 0.84 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Speg | Striated muscle-specific Ser/Thr-protein kinase | 00331223 | Hi | No | Y | S2171 | O | 0.35 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Y | T2488 | P | 0.42 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Lrrfip2 | Putative uncharacterized protein | 00659860 | Low | No | Y | S233 | B | 0.37 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Y | S261 | B | 0.42 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Kidney | Slc34a1 | Na-dependent phosphate transport protein 2A | 00121337 | Hi | Yes | Y | S623 | P | 0.08 | 0 | 0 | 0 | 90 | 0 | 0 | 0 | 0 | 0 |
| Y | T621 | P | 0.09 | 0 | 0 | 0 | 73 | 0 | 0 | 0 | 0 | 0 | ||||||
| Slco1a6 | Solute carrier organic anion transporter 1A6 | 00114950 | Med | No | Y | S634 | A | 0.14 | 0 | 0 | 0 | 43 | 0 | 0 | 0 | 0 | 0 | |
| Y | T632 | A | 0.17 | 0 | 0 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | ||||||
| Y | S635 | A | 0.20 | 0 | 0 | 0 | 28 | 0 | 0 | 0 | 0 | 0 | ||||||
| Pdzk1 | Na+/H+ Exchange Regulatory Cofactor | 00228883 | Hi | Yes | Y | T503 | B | 0.22 | 0 | 0 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | |
| N | S510 | O | 0.23 | 0 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | ||||||
| Liver | Hal | Histidine ammonia-lyase | 00118625 | Hi | Yes | N | S635 | P | 0.21 | 0 | 0 | 0 | 0 | 27 | 0 | 0 | 0 | 0 |
| Hao1 | Hydroxyacid oxidase 1 | 00123750 | Hi | Yes | N | S194 | P | 0.26 | 0 | 0 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | |
| Mtus1 | Mitochondrial tumor suppressor 1 homolog | 00480490 | Med | No | N | S385 | A | 0.33 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | |
| Lung | Ager | Advanced glycosylation end product-specific receptor | 00122231 | Hi | Yes | Y | S377 | A | 0.10 | 0 | 0 | 0 | 0 | 0 | 64 | 0 | 0 | 0 |
| Y | S390 | A | 0.73 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | ||||||
| Bcl11b | B-cell lymphoma/leukemia 11B | 00121608 | Low | Yes | N | T744 | P | 0.35 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | |
| Prx | Periaxin | 00469952 | Hi | Yes | Y | S1331 | P | 0.35 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | |
| Y | S1337 | P | 0.35 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | ||||||
| N | S1028 | P | 0.58 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | ||||||
| Pancreas | Sytl1 | Synaptotagmin-like protein 1 | 00118188 | Med | Yes | Y | S397 | P | 0.25 | 0 | 0 | 0 | 0 | 0 | 0 | 21 | 0 | 0 |
| Y | S120 | A | 0.31 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 0 | 0 | ||||||
| C77080 | Uncharacterized protein KIAA1522 | 00420527 | Low | No | Y | S17 | P | 0.29 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 0 | 0 | |
| Copa | Coatomer subunit alpha | 00229834 | Hi | No | Y | S402 | O | 0.35 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | |
| Spleen | Ms4a1 | B-lymphocyte antigen CD20 | 00115397 | Hi | Yes | Y | S29 | B | 0.24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23 | 0 |
| Hemgn | Hemogen | 00458120 | Hi | Yes | N | S158 | P | 0.33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | |
| Cxcr5 | C-X-C chemokine receptor type 5 | 00108764 | Low | No | Y | S361 | B | 0.31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 0 | |
| Testis | Clgn | Calmegin | 00123699 | Hi | Yes | Y | S582 | B | 0.09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 75 |
| Y | S592 | P | 0.09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 74 | ||||||
| Y | S595 | A | 0.19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31 | ||||||
| Srrm2 | Serine/arginine repetitive matrix protein 2 | 00225062 | Med | No | N | S1434 | B | 0.10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 62 |
Protein levels (Low, Medium, High) assigned using spectral counts observed in proteome assays without phosphopeptide enrichment (Table S4). Values correspond to the 0–33rd percentiles (0–5 counts), 34–66th percentiles (6–33 counts), and 67–100th percentiles (>33 counts).
Tissue-enriched expression (Yes/No) defined by those proteins whose spectral counts correspond to entropies less than 0.84 for both phosphorylation and non-phosphorylation datasets (see methods).
Novelty per phosphorylation site (Yes/No) was assigned according to whether the site is included in the PhosphoSite database of literature references to mouse protein post-translational modifications (www.phosphosite.org; 11/4/09).
Site classes assigned using a decision tree algorithm: A = Acidic; B = Basic; O = Other; P = Proline-directed; T = Tyrosine.
Entropy measured as described in methods.
Table 2. Abundant “Global” Phosphorylation Sites.
This table lists selected abundant phosphorylation sites that were detected in most or all of the mouse tissue characterized, grouped by protein. See also Tables S1–S3.
| Name | Annotation | IPI | Protein Abundancea | Novel?b | Site | Entropyc | Classd | Brain | Brown Fat | Heart | Kidney | Liver | Lung | Pancreas | Spleen | Testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trim28 | Transcription intermediary factor 1-β | 00312128 | Hi | N | S473 | 2.17 | B | 10 | 9 | 7 | 12 | 10 | 7 | 5 | 10 | 9 |
| Srrm1 | Serine/arginine repetitive matrix protein 1 | 00605037 | Low | N | S758 | 2.13 | P | 48 | 57 | 19 | 78 | 66 | 49 | 28 | 75 | 54 |
| N | S756 | 2.13 | P | 55 | 64 | 19 | 109 | 90 | 76 | 58 | 92 | 60 | ||||
| N | S461 | 2.11 | A | 4 | 2 | 3 | 7 | 5 | 8 | 3 | 6 | 8 | ||||
| N | S220 | 2.10 | P | 8 | 8 | 3 | 16 | 10 | 17 | 7 | 16 | 15 | ||||
| N | S915 | 2.08 | A | 13 | 18 | 6 | 21 | 27 | 13 | 16 | 13 | 40 | ||||
| N | S624 | 2.08 | P | 8 | 6 | 8 | 17 | 14 | 17 | 6 | 17 | 28 | ||||
| N | S626 | 2.06 | P | 9 | 6 | 8 | 19 | 14 | 17 | 4 | 15 | 27 | ||||
| N | S574 | 2.06 | P | 58 | 41 | 15 | 97 | 83 | 64 | 35 | 81 | 144 | ||||
| N | S572 | 2.05 | P | 58 | 41 | 15 | 104 | 83 | 67 | 35 | 82 | 145 | ||||
| N | S463 | 2.04 | A | 4 | 2 | 2 | 4 | 3 | 8 | 1 | 5 | 8 | ||||
| N | T913 | 2.04 | B | 9 | 12 | 8 | 25 | 40 | 16 | 24 | 18 | 47 | ||||
| Ahsg | Alpha-2-HS- glycoprotein | 00128249 | Hi | N | S309 | 2.15 | A | 22 | 52 | 43 | 47 | 59 | 38 | 44 | 27 | 27 |
| N | S312 | 2.14 | O | 30 | 54 | 53 | 48 | 27 | 74 | 27 | 43 | 29 | ||||
| N | S314 | 2.07 | A | 20 | 60 | 57 | 42 | 30 | 50 | 8 | 27 | 19 | ||||
| Nucks1 | Nuclear ubiquitous casein and cyclin-dependent kinase substrate | 00341869 | Med | N | S181 | 2.06 | P | 9 | 4 | 1 | 11 | 10 | 13 | 11 | 16 | 17 |
| N | S19 | 2.05 | A | 49 | 20 | 22 | 58 | 96 | 36 | 41 | 75 | 17 | ||||
| Sdpr | Serum deprivation- response protein | 00135660 | Hi | N | S359 | 2.06 | B | 7 | 67 | 26 | 27 | 18 | 46 | 29 | 46 | 24 |
| Nfia | Nuclear factor 1 A-type | 00131415 | Low | N | S310 | 2.16 | P | 13 | 25 | 16 | 19 | 22 | 16 | 13 | 24 | 10 |
| N | S323 | 2.12 | P | 9 | 5 | 6 | 10 | 5 | 8 | 2 | 11 | 8 | ||||
| N | S342 | 2.12 | P | 3 | 5 | 3 | 7 | 4 | 6 | 2 | 7 | 8 | ||||
| N | S303 | 2.10 | B | 9 | 10 | 18 | 12 | 16 | 9 | 3 | 15 | 5 | ||||
| Htt | Huntingtin | 00271166 | Hi | N | S397 | 2.15 | B | 11 | 6 | 8 | 9 | 17 | 15 | 8 | 12 | 15 |
| N | S412 | 2.14 | P | 7 | 5 | 8 | 7 | 12 | 8 | 3 | 11 | 7 | ||||
| Mapk3 | Mitogen-activated protein kinase 3 | 00230277 | Hi | N | T203 | 2.10 | A | 3 | 6 | 4 | 5 | 2 | 4 | 1 | 3 | 2 |
| N | Y205 | 2.10 | T | 3 | 6 | 4 | 5 | 2 | 4 | 1 | 3 | 2 | ||||
| Gsk3b | Glycogen synthase kinase-3 β | 00125319 | Hi | N | S219 | 2.15 | O | 7 | 6 | 4 | 5 | 5 | 7 | 2 | 4 | 4 |
| N | Y216 | 2.13 | T | 16 | 24 | 6 | 18 | 9 | 17 | 23 | 14 | 12 | ||||
| Vim | Vimentin | 00227299 | Hi | N | S51 | 2.12 | B | 2 | 4 | 2 | 1 | 2 | 2 | 1 | 2 | 3 |
| N | S39 | 2.09 | B | 2 | 6 | 4 | 3 | 1 | 4 | 2 | 5 | 2 | ||||
| Mtmr2 | Myotubularin-related protein 2 | 00128196 | Med | Y | S74 | 2.16 | B | 12 | 14 | 8 | 13 | 11 | 13 | 6 | 15 | 8 |
| N | S77 | 2.08 | O | 10 | 9 | 4 | 6 | 5 | 5 | 2 | 4 | 2 | ||||
| Mia3 | Melanoma inhibitory activity protein 3 | 00850156 | Hi | N | S1767 | 2.17 | O | 4 | 4 | 4 | 5 | 3 | 4 | 4 | 5 | 2 |
| N | S1755 | 2.08 | A | 3 | 1 | 2 | 3 | 1 | 2 | 1 | 1 | 4 | ||||
| Phf14 | PHD finger protein 14 | 00137250 | Low | N | S283 | 2.18 | O | 4 | 5 | 3 | 4 | 4 | 4 | 2 | 4 | 4 |
| N | T280 | 2.13 | A | 4 | 5 | 3 | 4 | 6 | 4 | 1 | 5 | 4 |
Protein levels (Low, Medium, High) assigned using spectral counts observed in proteome assays without phosphopeptide enrichment (Table S4). Values correspond to the 0–33rd percentiles (0–5 counts), 34–66th percentiles (6–33 counts), and 67–100th percentiles (>33 counts).
Novelty per phosphorylation site (Yes/No) was assigned according to whether the site is included in the PhosphoSite database of literature references to mouse protein post-translational modifications (www.phosphosite.org; 11/4/09).
Entropy measured as described in methods.
Site classes assigned using a decision tree algorithm: A = Acidic; B = Basic; O = Other; P = Proline-directed; T = Tyrosine.
To compare phosphorylation profiles for each tissue, we performed hierarchical clustering (Figure 1H). Total spectral counts (TSC) were used to approximate each site’s abundance within each tissue (Liu et al., 2004). Clustering of sites based on their tissue distributions highlights tissue-specific phosphorylation, especially in brain and testis. Furthermore, clustering tissues based on their phosphorylation profiles reveals that lung and spleen were most similar, likely reflecting immune cell signaling, whereas brain was most dissimilar.
Multiple kinases modify most phosphoproteins
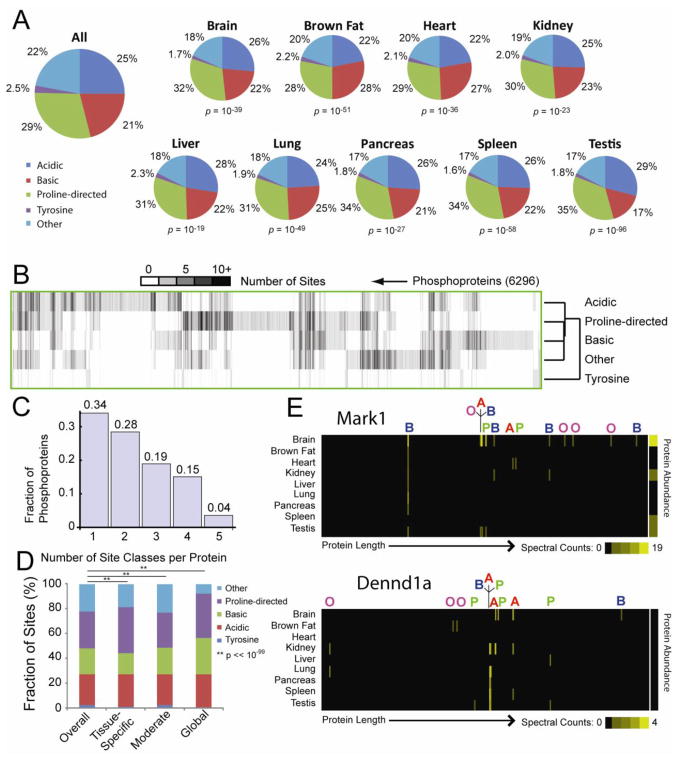
To investigate which kinase classes were likely responsible for observed phosphorylation events, we examined the amino acid motifs surrounding each site and broadly classified each as basic, acid, proline-directed, or tyrosine using a decision tree (Villen et al., 2007). Proline-directed sites were most common (29% of sites) (Figure 2A), while only 2.5% of sites corresponded to tyrosines. Statistically significant variations in frequencies of these classes were found across tissues, suggesting that specific tissues rely on distinct kinases to maintain specialized signaling. Proline-directed sites were elevated in spleen, testis and pancreas, while brown fat exhibited increased basic sites. Furthermore, when sites were divided into tissue-specific, moderate, and globally abundant groups based on entropy filtering, each showed distinct proportions of the 5 site classes (Figure 2D). Proline-directed sites were more frequently classified either tissue-specific or global, while basic sites were enriched among global events. Both tyrosine sites and those classified as “other” were decreased among tissue-specific and global phosphorylation events.
Figure 2. Distribution of Phosphorylation Site Classes across Tissues and Phosphoproteins.
Phosphorylation sites were classified as acidic, basic, proline-directed, tyrosine, or other as described (Villen et al., 2007). A. The relative frequencies with which each class is observed overall and for each tissue are plotted as pie charts. P-values reflect the likelihood that the tissue-specific sites were drawn randomly from a population with frequencies matching the entire dataset (χ2 Test). B. The heat map presents the numbers of sites of each class observed for 6,296 phosphoproteins. Proteins and site classes have been clustered to highlight similarities. C. Histogram indicating proportions of phosphoproteins containing phosphorylation sites from variable numbers of classes. D. Bar graph indicating relative proportions of tissue-specific, moderate, and global phosphorylation sites in each class. E. Heat maps indicating the tissue distributions and relative abundances of phosphorylation sites along the lengths of proteins Mark1 and Dennd1a. Sites are labeled according to their classes: A = acidic; B = basic; O = other; P = proline-directed; T = tyrosine. See also Figures S3, S4 and S7 and Tables S1–S3.
We next examined the distribution of phosphorylation classes within each phosphoprotein (Figure 2B). Hierarchical clustering revealed that Ser/Thr classes were similar, while Tyr sites diverged. 66% of phosphoproteins contained sites from multiple kinase classes and 4% harbored sites from all classes (Figure 2C). Two variably phosphorylated proteins are Mark1, a kinase involved with cytoskeletal dynamics (Timm et al., 2008b) and Dennd1a, a protein that acts in synaptic endocytosis (Allaire et al., 2006) (Figure 2E). Each was phosphorylated across its length and contained sites targeted to 4 site classes (neither contained pTyr). Individual sites showed distinct tissue profiles. In some cases, pairs of sites within the same class showed similar phosphorylation patterns; however, even within the same protein, different sites within the same class often showed variable patterns of modification. Overall, the presence of multiple site classes and the distinctive tissue-specific profiles seen across sites within most phosphoproteins suggest that the ‘typical’ phosphoprotein sits at the crossroads of multiple signaling pathways, where its activity depends upon many intracellular and extracellular influences.
A representative protein spanning multiple signaling networks is the kinase GSK3β, which regulates glycogen synthesis, microtubule dynamics, apoptosis, and cell proliferation (Forde and Dale, 2007). We found 4 sites on GSK3β, from 3 classes: S9 (basic), S25 (other), Y216 (Tyrosine), and S219 (other). Multiple kinases catalyze these phosphorylations, allowing multiple networks to modulate GSK3β activity. Specifically, Y216 phosphorylation activates GSK3β, and results from autocatalytic activity or Pyk2 action. In contrast, S9 phosphorylation inhibits GSK3β and results from activity of PKB, PKA, and S6K, as well as through auto-inhibition (Forde and Dale, 2007). Though sites S25 and S219 have been seen in multiple previous studies (Hornbeck et al., 2004), the kinase(s) responsible for their phosphorylation are unknown.
Combining protein abundance and phosphorylation measurement identifies true differential protein phosphorylation
Differential phosphorylation can reflect changes in protein abundance, as well as changes in a particular site’s phosphorylation. To distinguish these factors, we also performed a proteomic analysis of the 9 tissues examined in our phosphoproteomic experiments. Altogether we identified 12,039 proteins, 36% of which were identified both with and without phosphopeptide enrichment (Figure 3A), an overlap that was consistent across tissues (Figure 3B). 5,745 proteins were only identified without phosphopeptide enrichment, while 1,937 proteins were detected in the phosphorylation data alone, indicating that normally, these proteins are of low abundance, resisting detection via our shotgun proteomic approach. Phosphopeptide enrichment provides an excellent means to access proteins that are invisible to other fractionation methods.
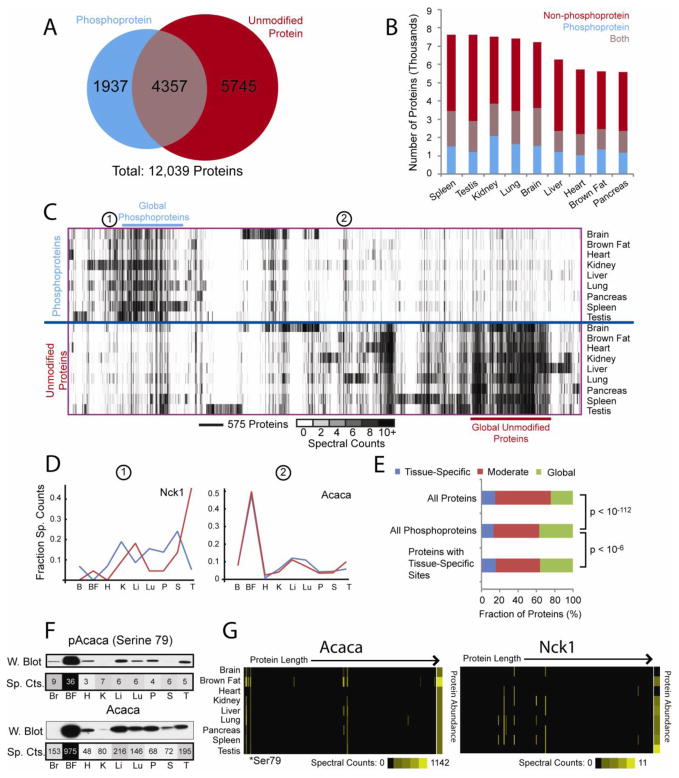
Figure 3. Cross-Tissue Comparison of Protein and Phosphoprotein Expression.
Overlap among phosphoproteins and proteins, overall (A) and in each tissue (B). C. Clustering of proteins based on spectral counts with and without phosphopeptide enrichment. Columns represent 12,000 observed proteins, while rows represent tissues with or without phosphopeptide enrichment. D. Phosphorylated and non-phosphorylated abundance profiles for selected proteins. E. Bar charts reflecting proportions of global, moderate, and tissue-specific proteins identified in non-phosphorylated form. F. Western blotting confirms spectral counting quantification of proteins and phosphorylation sites (Figure S4). G. Heat maps depicting spectral counts observed across tissues for sites along the length of each protein, revealing variable phosphorylation within proteins (Figure S3F). Abundances for each non-phosphorylated protein are also displayed. See https://gygi.med.harvard.edu/phosphomouse for plots of all proteins. See also Figures S2, S4–S6, S7 and Tables S1–S4).
To explore their expression and phosphorylation, proteins were clustered based on spectral counts within each tissue, with and without phosphopeptide enrichment and plotted as a heat map (Figure 3C). As with individual sites (Figure 1H), phosphorylated and non-phosphorylated protein profiles ranged from tissue-specific to global expression. Again, most tissue specificity was in brain and testis; however, unmodified proteins were more consistently expressed across tissues, indicating that protein expression is less variable than phosphorylation.
Perhaps most striking are differences among phosphorylated and non-phosphorylated profiles. Though many abundant, ubiquitous proteins were identified in the non-phosphorylated dataset, these proteins showed little phosphorylation. Similarly, the most abundant and globally phosphorylated proteins were sparsely observed without phosphorylation. Generally, there is little correlation between protein abundance and phosphorylation levels, either for the entire dataset, or for individual proteins. After spectral counts were normalized, comparison of each protein’s expression and phosphorylation profiles frequently revealed large differences. For example, the abundances of Nck1 with and without phosphorylation were very distinct (Figure 3D-1). In contrast, high concordance was observed for phosphorylated and non-phosphorylated Acaca (Figure 3D-2). Nevertheless, considerable heterogeneity was observed for both proteins’ individual sites across tissues (Figure 3G) indicating that these fluctuations are not due to changes in substrate protein abundance and thus reflect true differential tissue-specific phosphorylation. Since this analysis relies upon accurate quantitation of proteins and phosphoproteins via spectral counting, we investigated its reproducibility by comparing duplicate analyses of non-phosphorylated brown fat (Figure S4A) and found strong agreement. We also confirmed agreement between TSC and protein abundance using Western blots of selected proteins and their phosphorylation sites (Figure 3F; Figure S4B). Finally, we compared our protein expression profiles with those reported in a previous proteomic survey of several mouse tissues (Kislinger et al., 2006). Though only a subset of tissues was included in this prior study, excellent agreement was observed for the 3202 proteins shared among these datasets (Figure S4C–E).
Tissue-Specific Phosphorylation does not Imply Tissue-Specific Expression
To assess the relationship between tissue-specific phosphorylation and protein expression, proteins identified without phosphopeptide enrichment were classified as “tissue-specific”, “moderate”, or “global” based on entropy filtering (Figure 3E, “All Proteins”). Next, those proteins also identified with one or more phosphorylation sites were selected (“All Phosphoproteins”). Phosphoproteins were more likely to be “globally” expressed in their non-phosphorylated forms (24% to 37%; p < 10−112, χ2 test). When this list was further filtered to include only proteins for which one or more tissue-specific sites were observed (“Proteins with Tissue-Specific Sites”), a subtle increase in the fraction of tissue-specific proteins was observed (13% to 15%; p < 10−6, χ2 test). Thus proteins containing tissue-specific sites are only slightly more likely to display tissue-specific expression in non-phosphorylated form. In contrast, the vast majority (85%) of proteins that contained tissue-specific sites were expressed across multiple tissues in non-phosphorylated form, and 36% were globally expressed. Most tissue-specific phosphorylation is not due to tissue-specific protein expression, and instead reflects the independent influence of tissue-specific signaling.
Biological Classification of Global and Tissue-Specific Proteins and Phosphoproteins
To explore their biological roles, proteins and phosphoproteins were classified as “global” or “tissue-enriched” via entropy filtering. Each of these classes was then compared with all identified proteins and phosphoproteins to detect enriched Gene Ontology (GO) categories and Protein Information Resource (PIR) classifications using DAVID (Dennis et al., 2003). Enriched GO categories (Ashburner et al., 2000) and PIR classifications (Wu et al., 2003) were then clustered based on p-values reflecting enrichment in each class, following log transformation and z-transformation (Figure S5). Global proteins were enriched for protein synthesis and degradation as well as mitochondrial function, nucleotide binding and ligase activity, while ubiquitin ligase activity and phosphoproteins were enriched among global phosphoproteins. GO and PIR enrichments for each tissue generally agreed with expectations. Brain-specific proteins and phosphoproteins were enriched with neuron differentiation and vesicle transport classes, while heart-specific proteins and phosphoproteins were enriched with classes specific to muscle and cardiac tissue. Some tissue-specific proteins and phosphoproteins displayed complementary enrichment patterns. Testis-specific phosphoproteins were enriched in meiosis and cell cycle as well as DNA damage and repair while testis-specific non-phosphorylated proteins were enriched in spermatogenesis and microtubule-based movement. This suggests that distinct regulatory strategies govern these testis-specific functions.
Tissue-Specific Expression of Phospho-Transfer Proteins
To better understand variable phosphorylation across tissues, we examined proteins involved with phospho-transfer: kinases, kinase inhibitory proteins, phosphatases, and phosphatase inhibitory proteins (Figure S6A, B). Proteins were classified based on GO classifications and clustered. We identified 416 of 556 kinases (Figure S6A), with 57% detected in both phosphorylated and non-phosphorylated forms, as well as 11 of 21 kinase inhibitory proteins. Though mostly globally expressed, tissue-specific kinases were found in brain, lung, spleen, and testis. In contrast, notwithstanding a few brain-specific inhibitors, most kinase inhibitory proteins were widely expressed. Of 151 phosphatases, we identified 112 (Figure S6B), with tissue-specific phosphatases observed in brain and testis. A significant fraction (43%) of phosphatases were not detected in phosphorylated form, despite nearly ubiquitous expression. We also identified 17 of 18 phosphatase inhibitory proteins, with most being widely expressed across tissues.
Tissue-Specific Expression and Phosphorylation within Protein Interaction Networks
One effect of phosphorylation is to regulate physical interactions among proteins. Therefore, mapping phosphoproteomic data onto networks of known interacting proteins can reveal tandem phosphorylation that regulates the proteins’ shared biological activities. We created a high-confidence interaction map of the mouse proteome using protein-protein interactions in the STRING database (Jensen et al., 2009) and superimposed onto this network protein phosphorylation and abundance data from each tissue. Figure 4 shows 3 networks composed of the nearest neighbor interactors for the proteins Syk, Vamp1, and Bad.
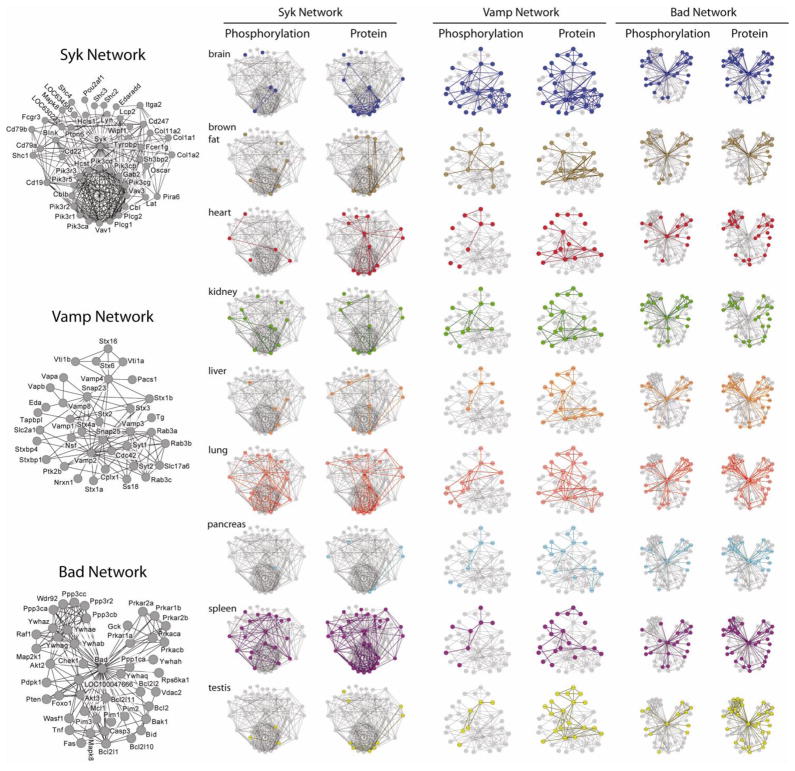
Figure 4. Mapping Protein Expression and Phosphorylation Data onto Protein Interaction Networks.
Protein observations with and without phosphopeptide enrichment were mapped onto the mouse STRING database of known protein interactions (Jensen et al., 2009). Only high confidence interactions (score > 0.7) were considered. Proteins that interact with Syk, Vamp, and Bad are depicted in networks above. Labeled diagrams are provided on the left that indicate the identities of all proteins. Each network is displayed twice for each tissue: the network on the left reflects phosphorylation, while the network on the right depicts protein expression. Proteins detected in phosphorylated or nonphosphorylated form in each tissue are represented as colored nodes, with colored edges connecting detectable proteins. Interacting proteins within the STRING database that were not detected in each tissue are shown in gray. See also Tables S1–S4.
Each interaction network in Figure 4 displays distinct protein expression and phosphorylation patterns. Syk (spleen tyrosine kinase) and its interactors display tissue-specific phosphorylation that mostly correlates with protein expression. Syk is a tyrosine kinase that is active in B and T cells during immune responses and is also expressed in kidney, heart, brain, and lung (Duta et al., 2006; Ulanova et al., 2005). Accordingly, the most phosphorylation was found in spleen and lung, which also contain the most expressed proteins from this network; in contrast, liver, pancreas, brown fat, and testis show low network expression and phosphorylation. The high phosphorylation observed for Syk and its interactors in spleen and lung reflect immune activities of splenic lymphocytes and airway epithelia. Furthermore, many network proteins, including Syk, were expressed and phosphorylated in heart, while kidney showed both expression and phosphorylation of network proteins, but not Syk itself.
In contrast to Syk, Vamp1 and its interactors are expressed in all tissues, though brain shows dramatically increased network phosphorylation. Various Vamp isoforms are expressed in nearly every tissue, where they participate in vesicular trafficking; however, Vamp1 and Vamp2 are specific to brain and participate in neurotransmitter release (Chen and Scheller, 2001). The extensive phosphorylation of Vamps and interacting proteins in brain suggests that phospho-regulation has enabled adaptation of widely distributed cellular machinery to support neural functions.
While the previous networks display variable and tissue-specific protein expression and phosphorylation, Bad (Bcl2-associated agonist of cell death) and its interactors exhibit remarkably consistent expression and phophorylation. Bad is a pro-apoptotic protein that regulates mitochondrial metabolism and when un-phosphorylated can trigger cell death (Danial, 2009). Because apoptotic machinery is found in essentially every cell type, ubiquitous detection of this network is not surprising. Furthermore, the uniformly high phosphorylation is consistent with healthy, mature tissues whose cells are unlikely to undergo apoptosis.
Combining phosphorylation data with signaling network maps reveals ztissue-specific differences
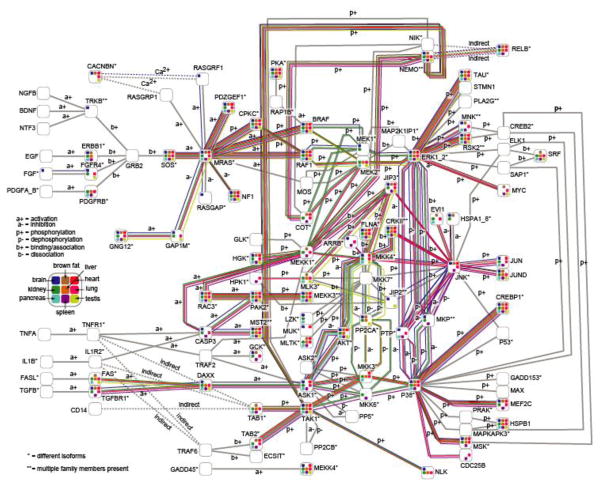
Most cellular signaling networks rely on sequential and coordinated phosphorylation of constituent pathway proteins to relay and amplify the initial signal; these pathways are found in virtually all cells and are required for sensing and responding to environmental cues. We investigated one of the most ubiquitous kinase cascades, the MAP kinase pathway, as it mediates cellular responses to growth factors and other survival and proliferation cues. To survey differences in MAPK signaling among tissues, we overlayed each tissue’s phosphoproteomic profile onto the KEGG database (Kanehisa et al., 2010) MAPK pathway (Figure 5). As expected for a central signaling pathway, much of the network was globally utilized; however, tissue-specific patterns were also apparent. Although signaling from Mras to Erk1 was found in almost all tissues, Mek1 was phosphorylated in brain and kidney, while Mek2 was modified in liver, lung, pancreas, and testis. These differences are post-translationally controlled, as unmodified Mek1 and Mek2 were detected in most tissues. These observations suggest avenues for future study that will elucidate how tissue-specific phenotypes are achieved through ubiquitous pathways.
Figure 5. Integrating Phosphorylation Data with Known Signaling Pathways.
Shown above are all members of the MAPK pathway as recorded in KEGG (Kanehisa et al. 2010). Each protein is represented as a node containing up to nine squares, each indicating presence of phosphorylation in one tissue. Edges represent interactions among proteins in the MAP Kinase pathway and are colored when their proteins were phosporylated in each tissue. See also Tables S1–S4.
DISCUSSION
After comparing expression and phosphorylation for thousands of proteins and phosphorylation sites across several mammalian tissues, several trends emerged.
Most Phosphoproteins Span Multiple Phosphorylation Networks
Most phosphoproteins contain several independent sites that frequently belong to different structural classes and display dramatically different phosphorylation across tissues. Multiple events allow modulation of each protein’s activity by kinases integrating multiple signaling pathways.
One example is Mark1, a kinase that phosphorylates Tau and other microtubule-associated proteins and regulates cytoskeletal dynamics (Figure 2E). Mark1 is alternately induced by phosphorylation at T215 by Markk/Tao-1 (Timm et al., 2003) or LKB1 (Lizcano et al., 2004) and inhibited by GSK3b-catalyzed phosphorylation at S219 (Timm et al., 2008a). We observed 13 sites within Mark1, spanning 4 site classes with variable phosphorylation across tissues. Its activation site, T215, was nearly ubiquitously phosphorylated, suggesting wide activity. Because the remaining sites occupy distinct motifs and display dramatically different phosphorylation profiles, Mark1 activity is likely regulated by multiple kinases representing discrete signaling networks.
Tissues Possess Specialized Phosphorylation Networks
When phosphorylation profiles are compared across tissues, the differences are striking. i) Half of all sites are unique to a single tissue, and few sites are globally phosphorylated. ii) Classes of sites are differentially enriched in different tissues. iii) Distinct kinases and phosphatases are present in each tissue. iv) Both individual proteins and entire protein complexes show tissue-specific phosphorylation. Clearly, phosphorylation networks have been optimized to support each tissue’s unique physiological functions.
Some of the most obvious evidence of tissue-specific phosphorylation appears when phosphorylation data are mapped onto protein interaction networks (Figure 4). Yet even individual proteins show combinations of tissue-specific and global phosphorylation. The proteins Mtap1a, Mtap2, and Tau have been primarily studied in neurons, where they bind to microtubules and regulate their stability and interactions with numerous cytoskeletal, membrane-bound, and enzymatic cellular components (Dehmelt and Halpain, 2005; Halpain and Dehmelt, 2006). However, we detected phosphorylation and expression of these proteins in nearly all tissues. The majority of Mtap1a’s 97 sites are brain-specific, though some were also seen in other tissues. Mtap2 also displays extensive brain-specific phosphorylation, though several C-terminal sites are widely phosphorylated. In contrast to Mtap1a and Mtap2, Tau displays few brain-specific phosphorylation events. The more widespread phosphorylation of Mtap2 and especially Tau suggests that these proteins play general roles regulating cytoskeletal dynamics. Although all cells rely upon microtubules, neurons have adapted them for their unique structural, transport, and signal transduction needs; thus it is appropriate that these proteins demonstrate a mixture of multi-tissue and brain-specific phosphorylation.
Tissue-Specific Protein Expression and Phosphorylation are Independent
Phosphorylated and non-phosphorylated proteins display markedly different expression patterns. Phosphoproteins are more often expressed globally, suggesting that tissue-specific phosphorylation allows tuning of ubiquitous proteins to optimize cell performance. Together, complementary protein expression and phosphorylation maintain the unique properties of distinct tissues.
While phosphorylation is generally specific and tightly-regulated, it has been proposed that some phosphorylation events may be non-functional byproducts of aberrant kinase activity (Lienhard, 2008). While we cannot directly address questions of biological function for individual sites from our data, the phosphorylation patterns we observe within individual proteins and across the proteome strongly suggest that non-specific modifications account for little of observed phosphorylation. First, only the minority of residues prone to modification were observed to be phosphorylated. Among proteins that were phosphorylated at least once and thus were demonstrably accessible for kinase activity, only 5% of serines, threonines, and tyrosines were modified (Figure 1E). Even allowing for some potential sites to be inaccessible due to protein topology, if non-specific phosphorylation were rampant, one would expect more even distribution of modifications across the surfaces of accessible proteins.
The strongest evidence against non-specific phosphorylation is the independence among protein abundance and phosphorylation. Non-specific phosphorylation would occur most frequently on highly abundant and accessible proteins. The minimal phosphorylation we observe among the most abundant proteins suggests that aberrant kinase activity accounts for little mammalian phosphorylation.
Complementary Protein Expression and Phosphorylation Maintain Tissue-Specific Signaling
Together, complementary protein expression and phosphorylation maintain the unique biochemistry of each tissue, as demonstrated by the kinase PKA and several of its substrates. We detected multiple sites on PKA, as well as its unmodified form. Because PKA phosphorylation at Thr198 is required for activity (Steinberg et al., 1993), our data suggest that PKA is active in all tissues, with highest activity in brain and brown fat. Within brown fat, PKA mediates hormone-stimulated lipolysis under fasting conditions. By phosphorylating Perilipin (Miyoshi et al., 2007), PKA enables HSL and ATGL to initiate lipolysis (Watt and Steinberg, 2008). Furthermore, PKA phosphorylates HSL, potentiating its activity. Using these proteins’ phosphorylation profiles, together with predictive kinase motif software (Obenauer et al., 2003), we identified several known PKA sites on each protein, along with numerous predicted PKA sites that have not been reported (Figure S7). Each of these sites is most abundant in brown fat, matching each protein’s expression as well as PKA expression and activity. Intriguingly, though no direct link has been identified between PKA and ATGL, the rate-limiting enzyme for initiation of triglyceride lipolysis (Haemmerle et al., 2006), we identified a putative PKA phosphorylation site on ATGL (Ser406) that could participate in lipolytic regulation.
PKA is also active in developing murine brain, where it phosphorylates cAMP response element binding protein (CREB), which mediates transcription of genes essential for nervous system development. PKA also phosphorylates proteins involved in neurotransmitter release and cytoskeletal organization, including microtubule-associated protein 2 (Mtap2) and synapsin 1 (Syn1). Phosphorylation of Mtap2 by PKA alters dendritic tree morphology, possibly modifying its physiological activity (Itoh et al., 1997), while synapsins are the primary presynaptic targets of PKA and represent one of several substrates that enable PKA to modulate synaptic transmission (Kao et al., 2002). We identified several known sites of PKA phosphorylation, along with numerous predicted PKA phosphorylation sites within each protein (Figure S7). PKA and its substrates demonstrate how ubiquitous kinases can participate in tissue-specific biological processes through carefully regulated activity coupled with tissue-specific protein expression. Since most kinases and phosphatases are widely expressed (Figure S6), such mechanisms likely play an important role maintaining tissue-specific phosphorylation.
Future Applications for Protein Expression and Phosphorylation Data
Our proteomic survey ranks among the largest reported in mice, while our phosphorylation survey is among the largest accrued. Furthermore, all data have been collected and analyzed using the same state-of-the-art techniques, ensuring results of consistently high quality. These data represent a first-of-its-kind murine phosphorylation atlas, recording patterns of expression and phosphorylation for thousands of proteins in healthy tissues. By providing detailed views of protein expression and phosphorylation across several mammalian tissues via an intuitive online interface, these data will provide a firm basis for future targeted research to better understand the biological roles of each protein.
In addition to providing insight into biology of individual proteins, these phosphorylation data will also be a valuable resource to the bioinformatics community. Recently algorithms have been developed to predict phosphorylation sites and motifs in uncharacterized proteins based on amino acid sequence and other properties (Miller et al., 2008; Schwartz et al., 2009). Many of these algorithms must be trained using known sites, and are only as reliable and comprehensive as their training data allow; our phosphorylation survey, including many previously unreported sites from several tissues, will enable better training of models, ultimately providing better predictions.
Challenges Remain for Mammalian Phosphoproteomics
While this investigation has expanded knowledge of mammalian phosphorylation, some aspects cannot yet be measured comprehensively from intact tissues via bottom-up proteomics techniques. One of these is connectivity: which phosphorylation sites occur simultaneously on the same protein molecules. This information is largely lost during tryptic digestion, leaving only connectivities among sites within the same peptides to be observed. The second aspect is site occupancy: what fraction of each site is phosphorylated. Previously phosphorylation site occupancy has been measured using targeted techniques. However, a proteomic strategy for measurement of phosphorylation site occupancies from cultured cells has been reported (Olsen et al., 2010). Measuring these properties in intact tissues would further advance our understanding of tissue-specific phosphorylation.
Although this survey has provided an expansive view of murine phosphorylation, it has not addressed many biological variables that influence phosphorylation. Our intention was to provide an overview of phosphorylation, initially focusing on a homogeneous population of relatively young and healthy male mice. However, it remains to be determined how physiological variables such as age, sex, strain, and diurnal cycles influence the phosphoproteome. Furthermore, many diseases alter phosphorylation. While it does not directly address these issues, our present work provides a foundation for subsequent studies by demonstrating effective methods for large-scale multi-tissue surveys of phosphorylation. This phosphoproteomic profile can also serve as a basis of comparison to explore changes in phosphorylation that occur in many physiological and pathological states.
We have presented a large-scale survey of protein expression and phosphorylation spanning multiple murine tissues, and have mined these data to better understand the biochemical basis of tissue specificity. These data suggest that the ‘typica’ phosphoprotein is widely expressed across tissues, yet displays variable, often tissue-specific phosphorylation sites from multiple kinases that tune protein activity to the specific needs of each tissue. We now offer these data as a resource, in the hope that they will inspire further targeted research.
EXPERIMENTAL PROCEDURES
Brief descriptions of key experimental procedures are provided below. For complete details, see Expanded Experimental Procedures. Data may be downloaded from the ProteomeCommons.org Tranche network. Specific Tranche keys for each dataset are listed in the Expanded Experimental Procedures.
Phosphopeptide Enrichment and Mass Spectrometry
9 organs were harvested from three-week-old male Swiss-Webster mice: brain, brown fat, heart, liver, lung, kidney, pancreas, spleen, and testis. Mice were sacrificed after overnight feeding, 6 hours after lights were turned on and eating ceased. Following tissue homogenization and protein extraction, samples containing 10 mg of protein per tissue were digested with trypsin and the resulting peptides fractionated via strong cation exchange chromatography, prior to phosphopeptide enrichment via immobilized metal affinity chromatography (Villen and Gygi, 2008). Phosphopeptides were analyzed in duplicate via LC-MS/MS on an LTQ-Orbitrap mass spectrometer. Peptides were identified using Sequest (Eng et al., 1994) and filtered to a 1% peptide FDR via the target-decoy approach, using a linear discriminant function to score each peptide based on parameters such as Xcorr, ΔCn, and precursor mass error (Huttlin et al. manuscript in preparation). Individual phosphorylation sites were scored using AScore (Beausoleil et al., 2006) and the resulting dataset was further filtered to achieve an estimated 1.7% final protein FDR (final peptide FDR: 0.15%). MS/MS spectra have been annotated for all 36,000 phosphorylation sites and are available online (http://gygi.med.harvard.edu/phosphomouse) with matching SEQUEST. out files.
Protein Isolation and Mass Spectrometry
Protein extracts from 9 tissues were separated via SDS-PAGE (65 μg per tissue) and digested in-gel with trypsin. The resulting peptides were then analyzed via LC-MS/MS on an LTQ-Velos-Orbitrap mass spectrometer. As before, peptides were identified by Sequest and filtered to a 1% peptide FDR. Proteins were further filtered to achieve a 1.25% final protein FDR (final peptide FDR: 0.11%).
Identification of Tissue-Enriched and Global Proteins and Phosphorylation Sites
The extent to which proteins and phosphorylation sites exhibited tissue-enriched or global tissue distributions was quantified using Shannon’s entropy (Shannon, 1948). A single pseudocount was divided across tissues for all sites to avoid problems with counts of zero. Sites predominantly found in a single tissue give small entropies, while sites that are evenly expressed across tissues give large entropies. We define sites with entropy values below 0.838 as tissue-enriched; this corresponds to a site with 4 spectral counts observed in one tissue and zero in all others. Those with entropies above 2.038, corresponding to observation in at least 7 of 9 tissues, are globally expressed.
Hierarchical Clustering
Unless noted otherwise, clustering was performed using centroid linkage with Pearson correlation as a distance metric.
Supplementary Material
Acknowledgments
The authors thank Xue Li and Abir Mukherjee for help with phosphorylation studies, and acknowledge Julian Mintseris, Deepak Kolippikam, Noah Dephoure, Younghao Yu, and other Gygi lab members for insightful discussions. This work was funded in part by a grant from the NIH to SPG (HG3456) and by an industry-sponsored research project with ThermoFisher Scientific.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allaire PD, Ritter B, Thomas S, Burman JL, Denisov AY, Legendre-Guillemin V, Harper SQ, Davidson BL, Gehring K, McPherson PS. Connecdenn, a novel DENN domain-containing protein of neuronal clathrin-coated vesicles functioning in synaptic vesicle endocytosis. J Neurosci. 2006;26:13202–13212. doi: 10.1523/JNEUROSCI.4608-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Castellanos-Serra L, Paz-Lago D. Inhibition of unwanted proteolysis during sample preparation: evaluation of its efficiency in challenge experiments. Electrophoresis. 2002;23:1745–1753. doi: 10.1002/1522-2683(200206)23:11<1745::AID-ELPS1745>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- Danial NN. BAD: undertaker by night, candyman by day. Oncogene. 2009;27(Suppl 1):S53–70. doi: 10.1038/onc.2009.44. [DOI] [PubMed] [Google Scholar]
- Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2005;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites--update 2008. Nucleic Acids Res. 2008;36:D240–244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duta F, Ulanova M, Seidel D, Puttagunta L, Musat-Marcu S, Harrod KS, Schreiber AD, Steinhoff U, Befus AD. Differential expression of spleen tyrosine kinase Syk isoforms in tissues: Effects of the microbial flora. Histochem Cell Biol. 2006;126:495–505. doi: 10.1007/s00418-006-0188-z. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Forde JE, Dale TC. Glycogen synthase kinase 3: a key regulator of cellular fate. Cell Mol Life Sci. 2007;64:1930–1944. doi: 10.1007/s00018-007-7045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- Halpain S, Dehmelt L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006;7:224. doi: 10.1186/gb-2006-7-6-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- Itoh TJ, Hisanaga S, Hosoi T, Kishimoto T, Hotani H. Phosphorylation states of microtubule-associated protein 2 (MAP2) determine the regulatory role of MAP2 in microtubule dynamics. Biochemistry. 1997;36:12574–12582. doi: 10.1021/bi962606z. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HT, Song HJ, Porton B, Ming GL, Hoh J, Abraham M, Czernik AJ, Pieribone VA, Poo MM, Greengard P. A protein kinase A-dependent molecular switch in synapsins regulates neurite outgrowth. Nat Neurosci. 2002;5:431–437. doi: 10.1038/nn840. [DOI] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Lienhard GE. Non-functional phosphorylations? Trends Biochem Sci. 2008;33:351–352. doi: 10.1016/j.tibs.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkila J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. A global map of human gene expression. Nat Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Perfield JW, 2nd, Souza SC, Shen WJ, Zhang HH, Stancheva ZS, Kraemer FB, Obin MS, Greenberg AS. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J Biol Chem. 2007;282:996–1002. doi: 10.1074/jbc.M605770200. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics. 2009;8:443–450. doi: 10.1074/mcp.M800258-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini EM, Kirkegaard M, Salerno D, Mortensen P, Mann M, Thomas AW. Deep coverage mouse red blood cell proteome: a first comparison with the human red blood cell. Mol Cell Proteomics. 2008;7:1317–1330. doi: 10.1074/mcp.M700458-MCP200. [DOI] [PubMed] [Google Scholar]
- Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- Roche. The Complete Guide For Protease Inhibition. Mannheim, Germany: Roche Applied Science; 2004. [Google Scholar]
- Schwartz D, Chou MF, Church GM. Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Mol Cell Proteomics. 2009;8:365–379. doi: 10.1074/mcp.M800332-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423. 623–656. [Google Scholar]
- Steinberg RA, Cauthron RD, Symcox MM, Shuntoh H. Autoactivation of catalytic (C alpha) subunit of cyclic AMP-dependent protein kinase by phosphorylation of threonine 197. Mol Cell Biol. 1993;13:2332–2341. doi: 10.1128/mcb.13.4.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm T, Balusamy K, Li X, Biernat J, Mandelkow E, Mandelkow EM. Glycogen synthase kinase (GSK) 3beta directly phosphorylates Serine 212 in the regulatory loop and inhibits microtubule affinity-regulating kinase (MARK) 2. J Biol Chem. 2008a;283:18873–18882. doi: 10.1074/jbc.M706596200. [DOI] [PubMed] [Google Scholar]
- Timm T, Li XY, Biernat J, Jiao J, Mandelkow E, Vandekerckhove J, Mandelkow EM. MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 2003;22:5090–5101. doi: 10.1093/emboj/cdg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm T, Marx A, Panneerselvam S, Mandelkow E, Mandelkow EM. Structure and regulation of MARK, a kinase involved in abnormal phosphorylation of Tau protein. BMC Neurosci. 2008b;9(Suppl 2):S9. doi: 10.1186/1471-2202-9-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanova M, Puttagunta L, Marcet-Palacios M, Duszyk M, Steinhoff U, Duta F, Kim MK, Indik ZK, Schreiber AD, Befus AD. Syk tyrosine kinase participates in beta1-integrin signaling and inflammatory responses in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2005;288:L497–507. doi: 10.1152/ajplung.00246.2004. [DOI] [PubMed] [Google Scholar]
- Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt MJ, Steinberg GR. Regulation and function of triacylglycerol lipases in cellular metabolism. Biochem J. 2008;414:313–325. doi: 10.1042/BJ20080305. [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. Brain Phosphoproteome Obtained by a FASP-Based Method Reveals Plasma Membrane Protein Topology. J Proteome Res. 2010 doi: 10.1021/pr1002214. [DOI] [PubMed] [Google Scholar]
- Wu CH, Yeh LS, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, et al. The Protein Information Resource. Nucleic Acids Res. 2003;31:345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.