Abstract
The nuclear factor kappa B (NF-κB) transcription factors-mediated transcription is the endpoint of a series of signal transduction events that are initiated by a vast array of stimuli. Both the proteolytic and non-proteolytic functions of ubiquitination are critically important for the regulation of NF-κB activation. Lys63-linked polyubiqutination of TAK1 is required for IL-1β-induced IKK/NF-κB activation. However, the lysine site that mediates Lys63-linked TAK1 polyubiquitination in IL-1β signaling is still controversial. Here we report that TAK1 lysine 158 but not lysine 209 is required for IL-1β-induced Lys63-linked TAK1 polyubiquitination and TAK1-mediated IKK, JNK, and p38 activation. Co-overexpression of TAK1 wild-type and K209R mutant with TAB1 induced Lys63-linked TAK1 polyubiquitination and NF-κB activation whereas TAK1 K158R mutant failed to do so. Furthermore, IL-1β induces polyubiqutination of TAK1 wild-type and K209R mutant but not K158R mutant. Reconstitution of TAK1-deficient mouse embryo fibroblast cells with wild-type, K158R mutant, or K209R mutant TAK1 reveals that TAK1 Lys-158 but not Lys-209 is required for IL-1β-induced IKK, p38 and JNK activation.
Keywords: IL-1β, TAK1, Lys63-linked polyubiquitination, Lysine 158
1. Introduction
The nuclear factor kappa B (NF-κB) transcription factors-mediated transcription is the endpoint of a complex series of reactions that are initiated by a vast array of stimuli, ranging from cellular stress to the engagement of receptors that mediate innate and adaptive immunity (1). Both the proteolytic and non-proteolytic functions of ubiquitination are critically important for the regulation of NF-κB (2). Ubiquitin is a 76-amino-acid protein containing seven lysine residues, Lys-6 , Lys-11, Lys-27, Lys-29,Lys-33, Lys-48, and Lys-63, and any one of these can participate in polyubiquitin chain formation (3). The topology of polyubiquitin chains can influence the fate of target proteins. For example, Lys48- and Lys11-linked polyubiquitination usually targets substrates for proteosomal degradation, whereas Lys63-linked polyubiquitin chains function as scaffolds to assemble signaling complexes and thereby participate in diverse cellular processes, including protein kinase activation, DNA repair and membrane trafficking (4).
Interleukin 1 beta (IL-1β) is a potent inflammatory cytokine that activates NF-κB and other signaling pathways which are critical for effective immune responses against microbial infection (5). IL-1β exerts its biological function through binding to its transmembrane receptor, Interleukin 1 receptor, type 1 (IL-1RI) that contains an intracellular Toll and IL1 receptor (TIR) domain. Upon binding of IL-1β, IL-1RI recruits the adaptor protein MyD88 that contains a TIR domain. MyD88 in turn recruits the IL-1 receptor associated kinases, IRAK4 and IRAK1. IRAK4 phosphorylates IRAK1 and release IRAK1 into the cytosol, where it forms a complex with TRAF6 (6, 7). IRAK1 and TRAF6 have been shown to be the targets of Lys63-linked polyubiquitination by TRAF6. Lys63-linked polyubiquitination of IRAK1 and TRAF6 may facilitate the recruitment of TAK1 and IKK complexes. The activated TAK1 then triggers the activation of the IKKβ, which leads to activation of transcription factors NF-κB and up-regulation of many genes encoding proinflammatory cytokines, chemokines, adhesion molecules, and proteolytic enzymes (8).
TAK1, a member of the MAPK kinase kinase family, is a crucial signaling intermediate in IL-1R1 signaling to NF-κB and AP-1, a function that derives from its ability to induce IKK and JNK activation (9). Although TAK1 is undoubtedly required for NF-κB activation by proinflammatory stimuli, how TAK1 mediates IKK activation remains to be clearly defined. Several reports suggest TAK1 Lys63-linked polyubiquitination is involved in the regulation of TAK1-mediated signaling pathways (10-12). However, direct experimental evidence supporting this assumption has been provided only recently. We and other two groups provided evidence that TAK1 Lys63-linked polyubiqutination is required for TNFα-, IL-1β- and TGFβ-induced NF-κB activation (13-15). We also found that TAK1 Lys63-linked polyubiquitination occurred at Lys-158 after TNFα and IL-1β stimulation, while Yamazaki reported IL-1β induced TAK1 Lys63-linked polyubiquitination at Lys-209 (13, 14). To our surprise, Sorrentino et al. reported that TGFβ induced TAK1 Lys63-linked polybuiquitination at Lys-34 (15). Recently, we compared biologic behavior of TAK1 Lys-158 and Lys-34 mutants after TGFβ stimulation in the same experimental setting and found TAK1 Lys-158 but not Lys-34 is required for TGFβ-induced NF-κB activation (16). Thus, it is urgent to compare biologic behavior of TAK1 Lys-158 and Lys-209 after IL-1β stimulation in the same experimental setting to clarify the discrepancy.
2. Materials and methods
2.1 Antibodies and reagents
The following antibodies and reagents were used: Flag (F3165), β-actin (A2228) and Nickel beads from Sigma; MYC (sc-40), HA (sc-7392), TAB1 (sc-6052) and A-agarose from Santa Cruz; phospho-TAK1 (4508S), TAK1 (4505), phospho-IKKα/β (2681S), IKKβ (2684), phospho-JNK (9251), JNK (9252L), phospho-p38 (9211S), p38 (9212), phosphor-ERK (9106S), ERK (9102), IκBα (9242L) and secondary antibodies conjugated to horseradish peroxidase from Cell Signaling; Recombinant IL-1β and TNFα from (R & D Systems).
2.2 Expression vectors
Full-length human TAK1 expression constructs were generated with a C-terminal V5-His tag or N-terminal Flag tag in pcDNA3.1 vector. TAK1 Lys-158 and Lys-209 mutations were created as described previously (13). Retroviral TAK1-K209R mutant construct was generated using pBabe-puro vector. Mammalian expression vectors for TAB1, TRAF6, IL-1bRI and HA-K63Ub were constructed as described previously (13). The NF-κB-dependent firefly luciferase reporter and pCMV promoter-dependent Renilla luciferase reporter plasmids were purchased from Clontech.
2.3 Cell culture and transfection
HeLa, HEK-293T and TAK1 deficient MEF cells were maintained in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% fetal calf serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 5% CO2. HeLa, HEK-293T and TAK1-deficient MEF cells were transfected with mammalian expression plasmids using Fugene HD, Fugene 6 (Roche Applied Science) and Lipofectamine 2000 reagents (Invitrogen), respectively.
2.4 Establishment of TAK1 stable cell lines
For generation of retrovirus particles, the pBabe-TAK1 wide type, or pBabe-TAK1-K158R, or pBabe-TAK1-K209R were co-transfected with retrovirus packing vector Pegpam 3e and PLC-ECO in HEK-293T cells to obtain retroviral supernatants. Viral supernatants were collected after 48 and 72 h. TAK1-deficient cells were incubated with retroviral supernatant in the presence of 4 μg/ml polybrene (hexadimethrine bromide; Sigma). Stable cell lines were established after 10 days of puromycin (3 μg/ml) selection.
2.5 Immunoprecipitation and Immunoblotting
Total cell lysates were prepared as following: cells were washed 3 times with ice-cold PBS on ice, then were lysed by lysis buffer. After centrifugation, cell lysates were incubated with anti-Flag antibody with rotation for 3 h at 4°C. The precipitates were washed four times in cold washing buffer (20 mM HEPES (pH 7.4), 50 mM NaCl, 2.5 mM MgCl2, and 0.05% Triton X-100), then the beads were resuspended in Loading buffer and boiled for 10 min. The immunoprecipitates or the whole cell lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). The membranes were probed with appropriate antibodies. The IgG horseradish peroxidase-conjugated antibodies were used as the secondary antibodies. The proteins were detected by the ECL-Plus Western blotting detection system.
2.6 Luciferase reporter assays
HEK-293T and different MEF cell lines were split at a concentration of 3 × 105 cells per well and cultured overnight in 6-well plates. NF-κB-dependent firefly luciferase reporter and effector plasmids were co-transfected along with the Renilla luciferase plasmid in HEK-293T cells and MEF cell lines. Forty-eight hours after transfection, cells were harvested in passive lysis buffer (Promega), and luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega). The relative luciferase activity was calculated by dividing the firefly luciferase activity by the Renilla luciferase activity. Data represent three independent experiments performed in triplicate.
3. Results
3.1 TAK1 Lys-158 but not Lys-209 is required for TAK1/TAB1 co-overexpression-induced TAK1 Lys63-linked polyubiquitination
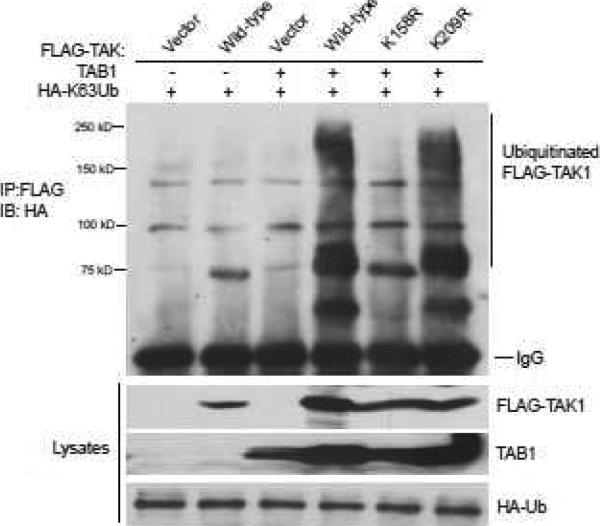
The biological function of TAK1 needs co-expression of several cofactors including TAB1, TAB2 and TAB3 since overexpression of TAK1 alone does not result in significant NF-κB activation (17, 18). TAB1 aids in TAK1 autophosphorylation, acting as an activating subunit in the TAK1 complex (19). In addition, overexpression of TAK1 with its regulatory subunit TAB1 leads to TAK1 K63-linked polyubiquitination and activation in the cells (13). To determine whether TAK1 Lys-158 or Lys-209 residue is required for TAB1 co-overexpression-induced TAK1 Lys63-linked polyubiquitination, Expression vectors encoding the N-terminal Flag-tagged TAK1 wild-type, K158R, K209R mutants were co-transfected with TAB1 into HEK-293T cells along with HA-K63Ub. The cell lysates were boiled in the presence of 1% SDS and immunoprecipitated with anti-Flag antibody, followed by SDS-PAGE and immunoblotting with anti-HA antibodies. In this assay, we found that only TAK1 K158R mutant inhibited TAB1 co-overexpression-induced TAK1 Lys63-linked polyubiquitination compared to TAK1 wild-type and K209R mutant (Figure 1).
Figure 1. TAK1 lysine 158 but not lysine 209 is required for TAK1/TAB1 co-overexpression-induced Lys63-linked TAK1 polyubiquitination.
Co-overexpression of TAK1/TAB1 induces Lys63-linked TAK1 polyubiquitination at Lysine 158. Expression vectors encoding mutant HA-ubiquitin in which all lysine residues except Lys 63 were mutated to arginine residue (HA-K63Ub) and TAB1 were co-transfected into HEK-293T cells with control vector or expression vectors encoding Flag-TAK1-wild-type, -K209R and -K158R mutants, respectively. The cell lysates were heated in the presence of 1% SDS and diluted with lysis buffer in order to disrupt non-covalent protein-protein interactions. Flag-TAK1 proteins in the transfected cells were precipitated with anti-Flag antibodies and immunoblotted with anti-HA antibodies to detect the presence of ubiquitinated Flag-TAK1.
3.2 TAK1 Lys-158 but not Lys-209 is required for TAK1/TAB1 co-overexpression-induced IKK/NF-κB activation
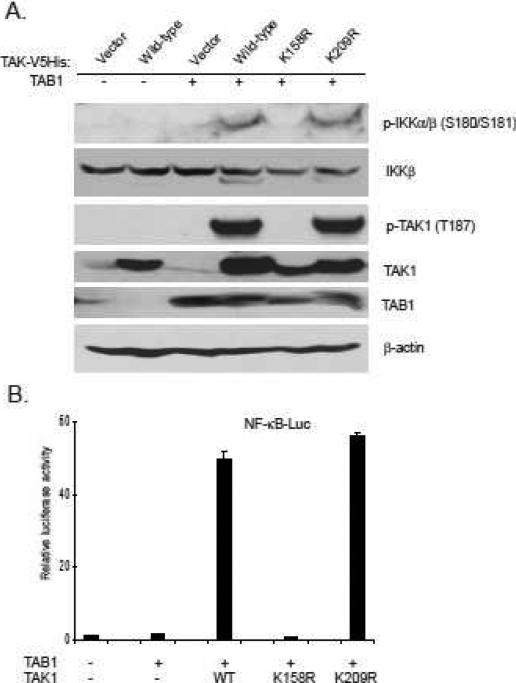
Growing evidence suggests that protein ubiquitination plays an essential role in the tight regulation of the NF-κB activation (22). K63-linked TAK1 polyubiquitination is critical for TAK1 activation (17, 23). To determine which lysine (Lysine 158 or Lysine 209) is required for TAK1 activation, we co-transfected TAB1 into 293T cells along with vector, TAK1 wild-type, K158R or K209R mutant respectively. Overexpression TAB1 with TAK1 wild-type and K209R mutant induced IKK phospholation, whereas TAK1 K158R mutant failed to do so (Figure 2A).
Figure 2. TAK1 lysine 158 but not lysine 209 is required for TAK1/TAB1 co-overexpression-induced IKK/NF-κB activation.
A. TAK1 lysine 158 is required for TAK1/TAB1 co-overexpression-induced TAK1 and IKKα/β phosphorylation. TAB1 expression vector was co-transfected with empty vector or expression vectors encoding TAK1-wild-type, -K209R and -K158R mutants into 293T cells, respectively. Cell lysates were immunoblotted with antibodies indicated.
B. TAK1 lysine 158 is required for TAK1/TAB1 co-overexpression-induced NF-κB activation. TAB1 expression vector, NF-κB luciferase reporter vector and control Renilla luciferase reporter vector were co-transfected into 293T cells with empty vector or expression vectors encoding TAK1-wild-type, -K209R and -K158R mutants, respectively. The relative luciferase activity was measured 48 h later and normalized with the Renilla activity. Error bars indicate ± standard deviation in triplicate experiments.
Consistent with these results, luciferase analysis with NF-κB-responsive reporters showed that TAK1 K158R mutant but not wild-type or K209R mutant blocked TAK1/TAB1 overexpression-induced reporter gene luciferase activity (Figure 2B). This result indicates that TAK1 Lys-158 but not Lys-209 residue is required for TAB1 co-overexpression-induced TAK1 K63-linked polyubiquitination and activation.
3.3 TAK1 Lys-158 but not Lys-209 is required for IL-1β-induced TAK1 Lys63-linked polyubiquitination
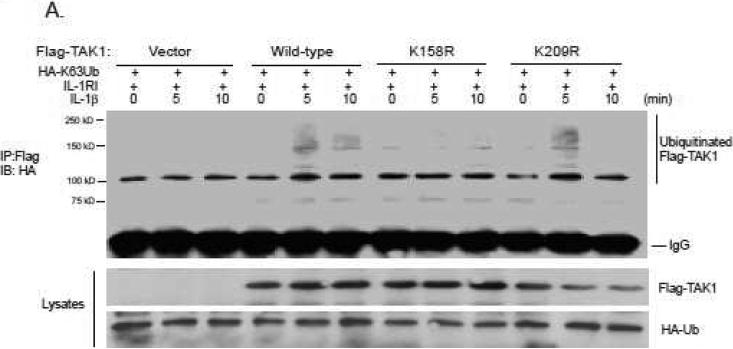
TAK1 Lys-158 but not Lys-209 residue is essential for TAB co-overexpression-induced TAK Lys63-linked polyubiqitination and activation. To determine which lysine (Lysine 158 or Lysine 209) in TAK1 is IL-1β-induced Lys63-linked polyubiquitination acceptor site, we co-transfected expression vectors encoding the N-terminal Flag-tagged TAK1 wild-type, K158R, K209R mutants with HA-K63Ub and IL-1RI into HEK-293T cells, respectively. Then the transfected cells were treated with IL-1β at the time points indicated. The cell lysates were boiled in the presence of 1% SDS and Flag-TAK1 proteins were immunoprecipitated with anti-Flag antibodies followed by SDS-PAGE and immunoblotting with anti-HA antibodies. We found that IL-1β induced Lys63-linked polyubiqutination of TAK1 wild-type and K209R mutant but not K158R mutant. This result suggests that TAK1 Lysine 158 is required for IL-1β-induced Lys63-linked TAK1 polyubiquitination in the cells.
3.4 TAK1 Lys-158 but not Lys-209 is required for IL-1β-and TNFα-induced IKK, p38 and JNK activation
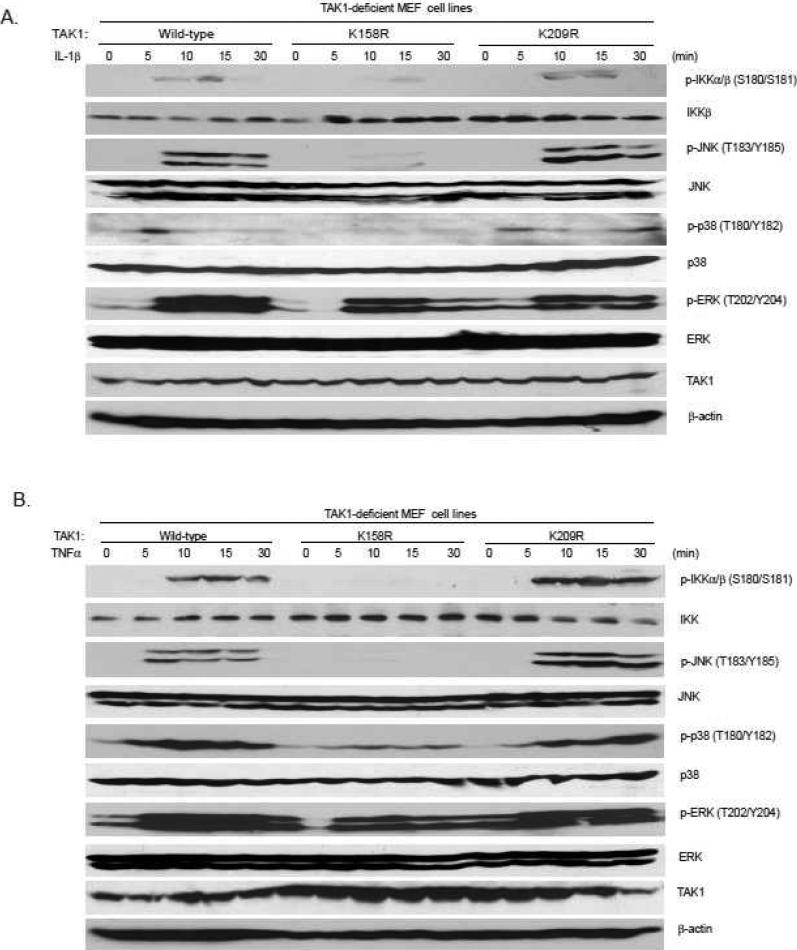
TAK1 mediates IL-1β-induced NF-κB activation via IKK phosphorylation (24). TAK1 also induces IL-1β-induced p38 and JNK activation. Therefore, we further analyzed the effect of TAK K158R and K209R mutations on the IL-1β-induced IKK phosphorylation, p38 and JNK activation. The TAK1 wild-type, or K158R, or K209R mutant expression vectors were stably introduced back into the TAK1-deficient MEF cells by a retroviral transduction system. Then TAK1 wild-type, K158R mutant and K209R mutant-reconstituted MEF cells were treated with IL-1β at different time points and then the cell lysates were immunoblotted with the indicated antibodies to examine IL-1β-induced JNK, p38, and IKK phosphorylation. In this assay, IL-1β-induced IKK, JNK and p38 phosphorylation were completely blocked in the TAK1 K158R mutant-reconstituted MEF cells, whereas IL-1β induced similar levels of IKK, JNK and p38 phosphorylation in the TAK1 wild-type or K209R mutant-reconstituted cells (Figure 4A). Similarly, TNFα-induced IKK, JNK and p38 phosphorylation were completely blocked in the TAK1 K158R mutant-reconstituted MEF cells, whereas TNFα induced similar levels of IKK, JNK and p38 phosphorylation in the TAK1 wild-type or K209R mutant-reconstituted cells (Figure 4B). These results suggest that TAK1 Lys-158 residue is required for IL-1β- and TNFα-induced IKK, p38 and JNK activation.
Figure 4. TAK1 lysine 158 but not lysine 209 is required for IL-1β- and TNFα-induced IKK, JNK and p38 activation.
Expression of the TAK1-K158R mutant, but not -K209R mutant, inhibits IL-1β- and TNFα-induced p38, JNK, IKK phosphorylation. TAK1-deficient MEF cells were transduced with the retrovirus encoding TAK1-wild-type, -K158R or -K209R mutant and subsequently selected with puromycin (3 μg/ml) to establish the TAK1-deficient MEF cell lines with the stable expression of either TAK1 wild-type, -K158R or –K209R mutant. The reconstituted TAK1-deficient MEF cells were untreated or treated with IL-1β (10 ng/ml) (A) or TNFα (10 ng/ml) (B) for the time points indicated and subsequently harvested. Whole cell extracts were subjected to SDS-PAGE and immunoblotted with antibodies indicated.
3.5 TAK1 Lys-158 but not Lys-209 residue is required for IL-1β-and TRAF6-induced NF-κB activation
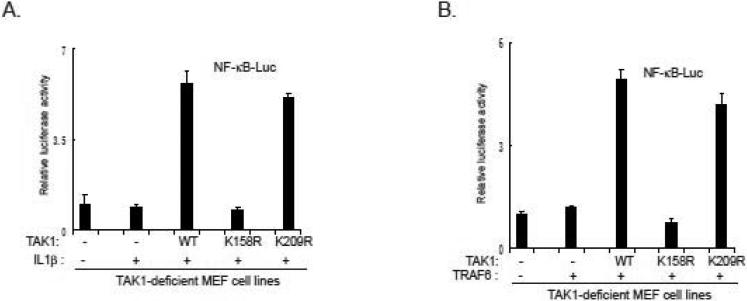
Consistent with above finding, we found IL-1β failed to induce NF-κB-responsive luciferase activity in TAK1 K158R mutant MEF cells, whereas IL-1β induced a similar level of luciferase reporter activity in both TAK1 wild-type and K209 mutant-reconstituted MEF cells (Figure 5A). In addition, TRAF6-induced NF-κB reporter activity was blocked in TAK1 Lys-158 but not TAK1 wild-type and Lys-209-reconstituted MEF cells (Figure 5B). This result suggests that TAK1 Lys-158 but not Lys-209 resudue is essential for TRAF6-mediated NF-κB activation.
Figure 5. TAK1 lysine 158 but not lysine 209 is required for IL-1β and TRAF6-induced NF-κB activation.
A. TAK1 Lys158 is required for IL-1β-induced NFκB activation. NF-κB luciferase reporter and control Renilla luciferase reporter vectors were co-transfected into TAK1-deficient MEF reconsititued with vector, TAK1 wild-type, K158R mutant or K209R mutant cells. Transfected cells will be treated with or without IL-1β. The relative luciferase activity was measured and normalized with the Renilla activity. Error bars indicate ± standard deviation in triplicate experiments.
B. TAK1 Lys158 is required for TRAF6-induced NFκB activation. NF-κB luciferase reporter and control Renilla luciferase reporter vectors along with TRAF6 expression vector were co-transfected into TAK1-deficient MEF reconsititued with vector, TAK1 wild-type, K158R mutant or K209R mutant cells. The relative luciferase activity was measured and normalized with the Renilla activity. Error bars indicate ± standard deviation in triplicate experiments.
4. Discussion
TAK1, a member of the MAPK kinase kinase family, is a crucial signaling intermediate in IL-1R1 signaling to IKK/NF-κB and JNK/AP-1 (9). Although TAK1 is undoubtedly required for NF-κB activation by proinflammatory stimuli, the mechanism of TAK1-mediated IKK remain to be fully characterized. TAK1 has been shown to function in vitro as an IKK kinase by phosphorylating IKKβ (17). Based on this observation, a model of TAK1-mediated IKK activation has been proposed in which TRAF6 K63-linked polyubiquitin chains recruit both TAK1 and IKK complexes (25). Through this interaction, IKK complex was bring into proximity with TAK1 at the TRAF6 complex, thereby facilitating IKK activation though direct phosphorylation by TAK1. Furthermore, TAK1 K63-linked polyubiquitination has been suggested to be involved in the regulation of TAK1-mediated signaling pathways (10-12). Recently, We and other two groups have provided direct experimental evidence supporting this assumption by reporting that TAK1 Lys63-linked polyubiquitination is required for TNFα, IL1β and TGFβ signaling (13-15). However, TAK1 Lys63-linked polyubiquitination acceptor lysine site remains to be controversial. We found that Lys63-linked polyubiquitination of TAK1 occurs at Lys-158 after stimulation of cells with TNFα and IL-1β (13). In contrast, Yamazaki reported that TAK1 Lys-209 is polyubiquitinated in IL-1β stimulated cells (14). Furthermore, Sorrentino reported that TGFβ induced TAK1 Lys63-linked polybuiquitination at Lys-34 (15). Different cell systems and experimental design may contribute this discrepancy. Therefore, it is critical to compare these lysine sites to clearly define the physiologic roles of these lysine residues in TAK1 k63-linked polyubiquitination. Recently, by comparing TAK1 Lys-158 and Lys-34 function in TGFβ-mediated signaling pathway, we concluded that TAK1 Lys-34 is not essential for TGFβ-induced TAK1 k63-linked polyubiquitination and activation (16). Herein, using the same experimental setting, we found that TAK1 Lys-209 is not required for TAK1/TAB1 co-overexpression- and IL-1β-induced TAK1 Lys63-linked polyubiquitination and TAK1-mediated IKK/ NF-κB activation.
IL-1RI is a prototypical TIR domain-containing receptor and recruits MyD88 through TIR domain after IL-1β stimulation. MyD88 has a modular structure, which consists of C-terminal TIR domain and an N-terminal death domain (DD) that performs homotypic interactions with downstream proteins of the IRAK family (IRAK4 and IRAK1) (26, 27). IRAK4 phosphorylates IRAK1, which promotes dissociation of IRAK1. Dissociated IRAK1 binds with TRAF6 and further recruits TAK1/TAB2/TAB3 complex and IKK complex via Lys63-linked polyubiquitination chains. Indeed, TRAF6, IRAK1, NEMO and the Pellino proteins are all reported targets of Lys63-linked polyubiquitination (28, 29). We and others reported that TAK1 Lys63-linked polyubiquitination is required for IL-1β signaling transduction. Nevertheless, the E3 enzymes responsible for activating NF-κB signaling downstream of IL-1RI are not completely characterized. TRAF6 and Pellino are both suggested to act as RING ligases to catalyze the formation of Lys63-linked polyubiquitination chains. Here, we found that TAK1 Lys-158 but not Lys-209 is required for TRAF6-mediated NF-κB activation. Together with our early report, our data support that TRAF6 acts as E3 ligase catalyze TAK1 K63-linked polyubiquitination at Lys-158.
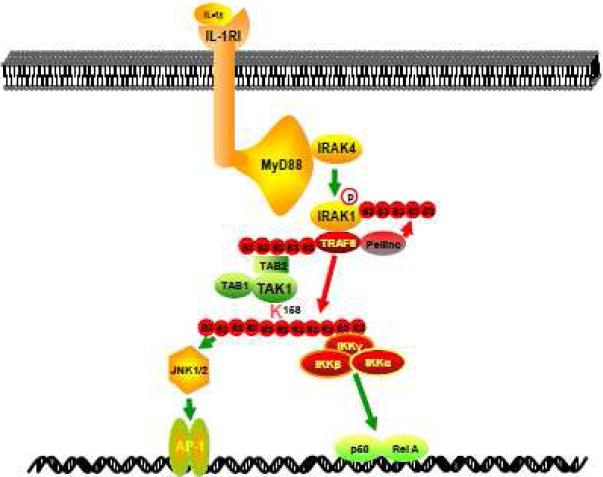
In view of the data presented here and in previous reports, we propose a working model (Figure 6) in which, upon IL-1β binding to their cognate receptors, the receptor-mediated signaling events induce TRAF6-mediated Lys63-linked TAK1 polyubiquitination at lysine 158 residue within the kinase domain which in turn triggers the activation of TAK1-mediated IKK/ NF-κB and JNK/AP-1 activation.
Figure 6.
Working Model: IL-1β binding to its cognate receptor triggers the receptor-mediated signaling events that induce TRAF6-mediated TAK1 polyubiquitination at the Lys-158 within the kinase domain which in turn causes TAK1-mediated IKK and JNK activation.
Figure 3. TAK1 lysine 158 but not lysine 209 is required for IL-1β-induced TAK1 Lys63-linked polyubiquitination.
293T cells transfected with expression vectors encoding IL-1RI and HA-K63Ub along with empty vector, Flag-TAK1wild-type, K158R mutant or K209R mutant respectively, were treated with IL-1β (10 ng/ml) for the time points indicated and subsequently lysated. The cell lysates were heated in the presence of 1% SDS and diluted with lysis buffer in order to disrupt non-covalent protein-protein interactions. Then, Flag-TAK1 proteins in the cell lysates were precipitated with anti-Flag antibodies and immunoblotted with anti-HA antibodies to detect the presence of ubiquitinated Flag-TAK1.
Acknowledgments
This work was supported in part by the grants from the NIH/NCI 1R21CA106513-01A2 (to J.Y.), the American Cancer Society grant RSG-06-070-01-TBE (to J.Y.), and the Virginia & L E Simmons Family Foundation Collaborative Research Fund (to J.Y.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Wertz IE, Dixit VM. Cold Spring Harb Perspect Biol. 2010;2:3350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiu YH, Zhao M, Chen ZJ. Chem Rev. 2009;109:1549. doi: 10.1021/cr800554j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Cell. 2009;137:133. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikeda F, Dikic I. EMBO Rep. 2008;9:536. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunne A, O'Neill LA. Sci STKE. 2003;3 doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 6.Kenny EF, O'Neill LA. Cytokine. 2008;43:342. doi: 10.1016/j.cyto.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter S, O'Neill LA. Biochem J. 2009;422:1. doi: 10.1042/BJ20090616. [DOI] [PubMed] [Google Scholar]
- 8.Hayden MS, Ghosh S. Genes Dev. 2004;18:2195. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 9.Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, Matsumoto K, Takeuchi O, Akira S. Nat Immunol. 2005;6:1087. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 10.Thiefes A, Wolf A, Doerrie A, Grassl GA, Matsumoto K, Autenrieth I, Bohn E, Sakurai H, Niedenthal R, Resch K, Kracht M. EMBO Rep. 2006;7:838. doi: 10.1038/sj.embor.7400754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koga T, Lim JH, Jono H, Ha UH, Xu H, Ishinaga H, Morino S, Xu X, Yan C, Kai H, Li JD. J Biol Chem. 2008;283:12546. doi: 10.1074/jbc.M710518200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiley WW, Jin W, Lee AJ, Wright A, Wu X, Tewalt EE, Leonard TO, Norbury CC, Fitzpatrick L, Zhang M, Sun SC. J Exp Med. 2007;204:1475. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan Y, Yu Y, Shi Y, Sun W, Xie M, Ge N, Mao R, Chang A, Xu G, Schneider MD, Zhang H, Fu SB, Qin J, Yang J. J Biol Chem. 285(2010):5347. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki K, Gohda J, Kanayama A, Miyamoto Y, Sakurai H, Yamamoto M, Akira S, Hayashi H, Su B, Inoue J. Sci Signal. 2009;2:66. doi: 10.1126/scisignal.2000387. [DOI] [PubMed] [Google Scholar]
- 15.Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, Zhang S, Heldin CH, Landstrom M. Nat Cell Biol. 2008;10:1199. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- 16.Mao R, Fan Y, Mou Y, Zhang H, Fu S, S, Yang J. Cell Signal. doi: 10.1016/j.cellsig.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. Nature. 2001;412:346. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 18.Ishitani T, Takaesu G, Ninomiya TJ, Shibuya H, Gaynor RB, Matsumoto K. Embo J. 2003;22:6277. doi: 10.1093/emboj/cdg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishimoto K, Matsumoto K, Ninomiya TJ. J Biol Chem. 2000;275:7359. doi: 10.1074/jbc.275.10.7359. [DOI] [PubMed] [Google Scholar]
- 20.Conze DB, Wu CJ, Thomas JA, Landstrom A, A., Ashwell JD. Mol Cell Biol. 2008;28:3538. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ordureau A, Smith H, Windheim M, Peggie M, Carrick E, Morrice N, Cohen P. Biochem J. 2008;409:43. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skaug B, Jiang X, Chen ZJ. Annu Rev Biochem. 2009;78:769. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 23.Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, Ninomiya TJ, Matsumoto K. Mol Cell. 2000;5:649. doi: 10.1016/s1097-2765(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 24.Delhase M, Hayakawa M, Chen Y, Karin M. Science. 1999;284:309. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 25.Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Veron M, Agou F, Israel A. Embo J. 2009;28:2885. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. Immunity. 1997;7:837. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 27.Muzio M, Ni J, Feng P, Dixit VM. Science. 1997;278:1612. [Google Scholar]
- 28.Jiang Z, Johnson HJ, Nie H, Qin J, Bird TA, Li X. J Biol Chem. 2003;278:10952. doi: 10.1074/jbc.M212112200. [DOI] [PubMed] [Google Scholar]
- 29.Moynagh PN. Trends Immunol. 2009;30:33. doi: 10.1016/j.it.2008.10.001. [DOI] [PubMed] [Google Scholar]